Abstract
Background
The aryl hydrocarbon receptor (AhR) is commonly known as an environmental sensor. Polymorphisms in AhR gene have been implicated in susceptibility to cancer. However, the results were controversial. This study was conducted to quantitatively summarize the association between AhR polymorphisms and cancer risk by meta-analysis.
Methods
Relevant reports were searched in four databases (Embase, PubMed, Wanfang, and China National Knowledge Infrastructure). We used pooled odds ratio (OR) and 95% confidence interval (95% CI) to evaluate the strength of the association in both standard and cumulative meta-analysis. Subgroup and sensitivity analysis was also performed, and between-study heterogeneity and publication bias were checked.
Results
A total of seventeen studies referring to three AhR polymorphisms (rs2066853, rs7796976, and rs2074113) were identified, and 9557 cases and 10038 controls were included. There was no statistically significant association of AhR rs2066853 polymorphism with cancer risk in the overall population, and the negative results were repeated in subgroup analysis by the ethnicity and cancer type. Concerning AhR rs7796976 or rs2074113 polymorphism, no significant correlation was detected. Moreover, these non-significant findings were stable in sensitivity analysis, and the cumulative meta-analysis indicated a trend of no significant link between this three AhR polymorphisms and cancer risk as more data accumulated over time.
Conclusion
This meta-analysis provides evidence that the rs2066853, rs7796976, or rs2074113 polymorphism in AhR gene is not a susceptible predictor of cancer. Further clinical and functional investigation between AhR polymorphisms and cancer susceptibility are needed.
Similar content being viewed by others
Introduction
It is well recognized that cancer is a major public health issue, and an estimated 18.1 million new cancer cases and 9.6 million cancer deaths occurred in 2018 globally based on the GLOBOCAN data [1]. What is worse, the rising trend of cancer incidence and mortality will not slow down [1]. The exact mechanism of carcinogenesis is complex and not fully elucidated, but it has become clear that cancer links to the interaction between genetic factors and environmental exposure, such as lifestyle and chemical contaminants. Polycyclic aromatic hydrocarbons (PAHs) belong to the common environmental contaminants, and numerous PAHs are known as carcinogens [2]. The carcinogenic risk of environmental substances depends not only on the exposure dose, but also on personal susceptibility to the carcinogens [3]. Moreover, many investigations have confirmed that malignancies have some underlying genetic commonalities, and then, the hypothesis about some common susceptibility genes for the onset of cancer has been proposed [4].
In recent years, more and more studies have focused on identifying genetic causes of cancer. The human aryl hydrocarbon receptor (AhR) gene is located on chromosome 7p1 5[5], whose corresponding protein is a cytoplasmic ligand-activated transcription factor [6]. AhR is commonly known as an environmental sensor [7] and could be triggered by many immunological mediators to an activator of xenobiotics metabolism [6]. Namely, AhR gene is involved in the carcinogenic response against environmental compounds, such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and PAHs [7, 8]. Also, as a number of physiological AhR ligands are often formed during the adaptive and innate immune response, more and more data have described the role of AhR gene in tumor immune surveillance and enhanced tumorigenesis [9]. Actually, AhR protein expresses in most tissues, and increased level and activity of AhR and its nuclear localization have been detected in tumor microenvironment compared with surrounding non-malignant tissues [9]. The findings indicate that the chronic activation of AhR may facilitate cancer development [9, 10]. Recently, accumulating evidence has showed that AhR could influence the major stages of tumorigenesis, such as initiation, progression, and metastasis [11]. Taken together, AhR could be considered as a candidate susceptibility gene for cancer based on its biological functional role.
The AhR gene is highly polymorphic [12, 13], and the relationship between multiple AhR polymorphisms and cancer risk has been investigated [14]. Nevertheless, inconsistent results were reported in different cancers and ethnicities. Meta-analysis is a valuable quantitative means to pool data from different studies and enhance statistical power, thereby overcoming the limitations of individual studies and achieving more reliable conclusions. Although two meta-analyses of AhR polymorphisms with cancer risk have been carried out [15, 16], the following reasons impelled us to update them: (1) one previous meta-analysis merely focused on the correlation between AhR rs2066853 polymorphism and the risk of breast cancer using three original studies [15]; (2) several case-control, cohort, or cross-sectional studies published in recent years have not been included in either of the above meta-analyses; (3) there was no subgroup analysis of ethnicity in the previous meta-analyses [15, 16], probably because of inadequate original studies; and (4) cumulative meta-analysis, which could estimate the trend and robustness of the pooled results, has not been performed in either of the previous meta-analyses. Accordingly, in the present study, meta-analysis with the most updated data was performed to explore whether AhR polymorphisms confer susceptibility to cancer.
Materials and methods
Identification of eligible studies
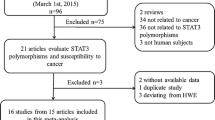
We conducted a systematic search of the literature in Embase, PubMed, and two Chinese databases (Wanfang and China National Knowledge Infrastructure) to identify relevant reports until July 2020. The following search terms were used: (“AhR” or “aryl hydrocarbon receptor”) and (variants or polymorphisms or variant or polymorphism or mutation) and (cancer or tumor or neoplasm or carcinoma or oma or leukemia). Moreover, the reference lists of relevant reviews were perused to acquire the previous unfound publications. Eligible studies had to fulfill all the following inclusion criteria: (1) an unrelated case-control, cross-sectional, or cohort design was used, (2) they examined the relationship between AhR polymorphisms and cancer risk, and (3) the genotype and allele frequencies could be available. Accordingly, publications were removed if either of the following conditions applied: (1) reports with overlapping or duplicate data or (2) literature with no useful data such as letters, abstracts, comments, or reviews. For the studies with repeated or overlapping data, the one with most subjects was selected.
Data extraction
Based on the above selection criteria, two authors (Li and Luo) assessed all abstracts of relevant publications and reviewed the full manuscripts of included studies, independently. The following information was collected from each included study: the first author, year of publication, original country, ethnicity of the study population, age, type of cancer, total number of cases and controls, specific AhR polymorphisms investigated in the original studies, and genotype frequencies. The above authors extracted and compared the required information independently and then input it into the predesigned form. Any disagreement was resolved through checking the original publications and adjudicated by a third author (Wang). Ultimately, the data came to a consensus.
Statistical analysis
Data management and statistical analysis was performed using the Comprehensive Meta-Analysis software, version 2.2 (Biostat, Englewood, New Jersey). This meta-analysis only included the polymorphisms of AhR gene which were investigated in at least three studies. A chi-squared test was applied to evaluate whether genotype frequencies in controls conformed to Hardy–Weinberg equilibrium (HWE). Meta-analysis was carried out in four genetic models: (1) the allelic, (2) dominant, (3) recessive, and (4) codominant model. The point estimates of odds ratio (OR) and 95% confidence interval (95% CI) were calculated to assess the strength of the link between AhR polymorphisms and cancer risk. Between-study heterogeneity was checked by the Q test, and P value < 0.10 was considered statistically significant. According to the absence (PQ ≥ 0.10) or presence (PQ < 0.10) of heterogeneity, the summary OR was calculated with the fixed- or random-effect model, respectively. Also, I2 statistics (ranging from 0 to 100%) were adopted to quantify heterogeneity, and their values of 0%, 25%, 50%, and 75% suggest no, low, moderate, and high heterogeneity, respectively [17]. The significance of pooled OR was determined with the Z test. Stratification was performed based on the ethnicity and cancer type, and if the similar data from at least three studies could be available in one group, other subgroup analysis was conducted.
Sensitivity analysis was adopted to validate the credibility of the findings with two methods: (i) removing a single study each time and (ii) excluding the studies that deviate from HWE in controls. Furthermore, cumulative meta-analysis was used to test the trend and robustness of the pooled results by sorting of the included studies in the descending order of publication year. Both Egger’s regression and Begg’s rank correlation methods were employed to evaluate the publication bias. Except for Q test, P < 0.05 was defined as the criterion of statistical significance.
Results
Main characteristics of included studies
The procedure for study identification is presented in Fig. 1. Finally, a total of seventeen relevant studies were obtained through a systematic search (Table 1), including 9557 cases and 10038 controls. The included studies encompassed breast cancer (n = 4) [18,19,20,21], lung cancer (n = 6) [13, 14, 22,23,24,25], bladder cancer (n = 2) [5, 26], colorectal cancer (n = 1) [27], lymphoma (n = 2) [28, 29], glioma (n = 1) [12], and leukemia (n = 1) [30]. Nine studies were performed in Asians, three studies concerned Caucasians, two studies focused on South Americans, and three studies paid attention to mixed ethnicities (Table 1). Three AhR polymorphisms (rs2066853, rs7796976, and rs2074113) investigated in at least three original studies were identified. Disease-specific meta-analysis was carried out for lung cancer and breast cancer, and ethnicity-specific meta-analysis was performed in Chinese, Asians, and Caucasians, as there were at least three studies in the above subgroups. No other subgroup analysis was carried out due to less than three studies in the corresponding groups. The selected characteristics of the original studies and genotype distributions of AhR polymorphisms were illustrated in Tables 1 and 2, respectively.
Meta-analysis of AhR rs2066853 polymorphism and cancer risk
Overall analysis
For AhR rs2066853 polymorphism, 9496 cases and 9977 controls from seventeen studies were included in meta-analysis. In the dominant model (AA+GA vs. GG), the between-study heterogeneity for all 17 studies was checked by Q test and the corresponding P value was less than 0.0001. Therefore, the random-effect model was chosen for synthesizing the data. The pooled OR for the 17 studies was 1.008 (95% CI = 0.898–1.131, P = 0.899; Fig. 2, Table 3), suggesting no significant link between AhR rs2066853 polymorphism and cancer risk in the dominant model. Similarly, combined data demonstrated no statistically significant relationship between this polymorphism and cancer risk in the recessive (Additional file 1: Fig. S1), codominant (Additional file 2: Fig. S2), or allelic model (Additional file 3: Fig. S3). All results were summarized in Table 3.
Subgroup analysis
Subgroup analysis by ethnicity detected the borderline association between rs2066853 polymorphism and cancer risk among Caucasians in the recessive (Additional file 4: Fig. S4a) and codominant (Additional file 5: Fig. S5a) models. Nevertheless, there was no significant association of rs2066853 polymorphism with cancer risk among Asians (AA+GA vs. GG: OR = 1.093, 95%CI = 0.885–1.351, P = 0.408, Fig. 3a; other models, Additional file 4, 5, and 6: Fig. S4a-S6a), Chinese (AA+GA vs. GG: OR = 1.168, 95%CI = 0.846–1.614, P = 0.345, Fig. 3a; other models, Additional file 4, 5, and 6: Fig. S4a-S6a)
Stratification analysis by cancer type indicated that AhR rs2066853 polymorphism was not significantly associated with the risk of lung cancer (AA+GA vs. GG: OR = 0.904, 95%CI = 0.721–1.134, P = 0.384, Fig. 3b; other models, Additional file 4, 5, and 6: Fig. S4b-S6b) or breast cancer (AA+GA vs. GG: OR = 1.014, 95%CI = 0.791–1.300, P = 0.912, Fig. 3b; other models, Additional file 4, 5, and 6: Fig. S4b-S6b) in any of the models. The main results of subgroup analysis were shown in Table 3.
Meta-analysis of AhR rs7796976 and rs2074113 polymorphisms and cancer risk
There were three studies of rs7796976 polymorphism (2489 cases and 2415 controls) and three studies of rs2074113 polymorphism (2194 cases and 2095 control), respectively. The overall analysis displayed no significant association of rs7796976 (Fig. 4) or rs2074113 (Fig. 5) polymorphism with cancer risk in any of the genetic models (Table 3). For rs7796976 or rs2074113 polymorphism, no stratification analysis was carried out owing to less than three studies in any of the potential subgroups.
Sensitivity analysis and cumulative meta-analysis
We further executed sensitivity analysis to examine the influence of individual studies on the pooled results. Regarding AhR rs2066853, rs7796976, or rs2074113 polymorphism, the non-significant findings in the overall or subgroup analysis were not materially altered when sequentially removing individual studies or excluding studies that deviated from HWE. Concerning the borderline association of AhR rs2066853 polymorphism among Caucasians in the recessive and codominant models, sensitivity analysis demonstrated that this trend was unstable and turned to no significance after excluding the study by Cotterchio et al. [27] or Figueroa et al. [26].
Cumulative analysis in the chronologic order was performed. For the relationship between AhR rs2066853 polymorphism and cancer risk, a stable trend of no significant results was identified among the overall sample in any of genetic models (Fig. 6). As shown in Fig. 6, the 95% CI became narrower as more data were accumulated. Furthermore, we carried out cumulative analysis in the subgroups, a stable tendency toward no significant association was repeated in the subgroups of lung cancer, breast cancer, Caucasians, Asians, or Chinese (figures not shown). With regard to the association of AhR rs7796976 or rs2074113 polymorphism with cancer risk, a tendency toward no significant link was also observed.
Heterogeneity and publication bias
Between-study heterogeneity in meta-analysis was demonstrated in Table 3. For rs2066853 polymorphism, I2 statistics revealed no to high between-study heterogeneity in the overall and subgroup analysis; however, no obvious between-study heterogeneity was observed concerning rs7796976 or rs2074113 polymorphism. Neither Begg’s nor Egger’s test detected significant publication bias on AhR rs2066853, rs7796976, or rs2074113 polymorphism (Table 4).
Discussion
Several lines of evidence have suggested that multiple factors and multi-procedures play a crucial role in the pathogenesis of cancer, such as genetic susceptibility, environmental factors, and immunity. Intriguingly, AhR has been proven to be an environmental sensor and a key regulator of T cell differentiation on which cancer immune surveillance is dependent, so it is reasonable to infer that AhR gene is implicated in carcinogenesis [9, 31]. Kolluri et al. considered that AhR may be a drug target for cancer [11], and AhR inhibitors can synergize with a immunotherapeutic treatment (anti-PD-L1 antibody) to enhance antitumor activity in the mouse model of lung cancer [32]. However, AhR deficient mice had a more rapid progress of colon carcinoma compared to wild-type mice [8]. The above facts indicate that there is a degree of complexity and discrepancies concerning the role of AhR gene in malignancy, as not only protumorigenic but also antitumorigenic activities of AhR signaling have been reported [9]. Intriguingly, AhR is involved in inducing the transcription of several cytochrome P450 enzymes such as CYP1A1, which is critical in metabolizing and bioactivating environmental compounds to highly carcinogenic metabolites [9]. One previous study found that induced CYP1A1 activity was higher in the variant allele A carriers of AhR rs2066853(G>A) polymorphism than those with GG genotype in a Caucasian population [33]. Therefore, some polymorphisms of AhR gene have been suggested as potential causal variants conferring susceptibility to cancer.
On the basis of 17 eligible studies, we have conducted this comprehensive meta-analysis to evaluate the relationship between three AhR polymorphisms and cancer susceptibility. To the best of our knowledge, this is the first meta-analysis that investigated the association of AhR rs7796976 and rs2074113 sites with cancer risk. The overall analysis failed to find significant link between AhR rs2066853 polymorphism and cancer susceptibility (Table 3 and Fig. 2), and stratified analysis upon type of cancer and ethnicity was consistent with this negative results (Table 3 and Fig. 3). The sensitivity analysis and cumulative meta-analysis further strengthened this evidence (Fig. 6). The usage of accumulative meta-analysis and the relatively larger sample size made the current meta-analysis more precise and comprehensive, but the results of rs2066853 polymorphism were in agreement with those in the previous meta-analysis [15]. The rs2066853 site, located in the transactivation domain of AhR gene, could result in a nonsynonymous amino acid substitution and influence the function of AhR protein [12]. One previous study reported that the variant allele (A) of rs2066853 polymorphism had an enhanced ability in inducing the transcription of CYP1A1 [34], but another research found that rs2066853 polymorphism did not affect the role of AhR gene in regulating CYP1A1-driven transcription [35]. Regarding rs2066853 polymorphism, conflicting evidence also existed in the studies of gene–disease association. While the mutant genotypes (AA or GA) of AhR rs2066853 polymorphism conferred statistically significant higher risk of glioma [12], lung cancer [13], acute leukemia [30], and breast cancer [18], some previous studies found that subjects with the same mutant genotypes (AA or GA) of rs2066853 site were more prone to have reduced risk for lung cancer [22, 25] and breast cancer [20, 25]. Thus, we speculate that no significant link between rs2066853 polymorphism and cancer risk may be ascribed to the contradictory effect of this polymorphism on the development of cancer. Actually, an estimated two thirds of cancer patients are linked to environmental factors, such as carcinogenic compounds [12], and different procarcinogens can be activated or detoxicated through phase I enzymes (mainly cytochromes P450 CYP1A1), which could be mediated by activated AhR [29]. The facts that AhR rs2066853 polymorphism might affect the two-way biological process (activation or detoxification) of procarcinogens regulated by CYP1A1 enzyme suggest that the mutant allele (A) and genotype (AA and GA) of this polymorphism is possibly associated with the indeterminate (increased or decreased) susceptibility to cancer, which could be used to explain the negative results in meta-analysis. Another plausible explanation of the negative findings on rs2066853 polymorphism is that the solitary polymorphism in AhR merely influences the transcription of CYP1A1 gene but does not play a vital role in the subsequent pathway to cancer development.
Regarding the rs7796976 (promoter region) or rs2074113 (intronic region) polymorphism, the current meta-analysis demonstrated no significant correlation with cancer susceptibility (Figs. 4 and 5) and no obvious between-study heterogeneity indicated by I2 values (Table 3). These findings probably reflect that these polymorphisms play no role in cancer development. Noticeably, the negative results of rs7796976 or rs2074113 polymorphism should be interpreted with caution due to a small number of studies (n = 3). The rs7796976 (promoter region) and rs2074113 (intronic region) polymorphisms may potentially alter expression regulation of AhR gene and produce alternative splicing owing to the regulatory function of the intron and promoter [24, 29], but some other researchers considered that these polymorphisms could not interfere in splicing due to their distance from intron/exon junctions where splice sites are detected [29]. Actually, no effect of these two polymorphisms on AhR gene regulation has been revealed. It is possible that the contribution of these polymorphisms to cancer susceptibility is not great enough so that no significant correlation can be detected. More genetic association studies concerning these polymorphisms and cancer risk are warranted to validate our findings.
It is a very useful method for checking the gene-disease correlation to investigate candidate genes, but the shortcomings are inevitable. The true relationship between cancer risk and single polymorphism may be interpreted wrongly if multiple genetic variants (polymorphisms) are responsible for cancer susceptibility. The previous studies found that the combination (multiple-variant haplotype) of multiple AhR polymorphisms contributed to the susceptibility of lung cancer [13, 24]. Jorge-Nebert et al. constructed multiple-variant haplotypes of AhR gene to investigate the association of theses haplotypes with head-and-neck squamous cell carcinoma, but no AhR haplotypes displayed statistically significant correlation with the risk of that cancer [36]. Haplotypic analysis which has bigger statistical power than single polymorphism analysis was carried out by some original studies, but it was impossible to perform the corresponding meta-analysis of AhR haplotypes attributing to inadequate haplotype data. More investigations are required to evaluate the effect of AhR haplotypes on cancer susceptibility, which may supply the data basis for the meta-analysis of haplotypes.
Heterogeneity should be taken into account when interpreting the pooled results of AhR rs2066853 polymorphism in meta-analysis. Despite meticulous literature retrieval and precise data extraction, the low to moderate between-study heterogeneity was observed among the overall population (Table 3). After performing subgroup analysis by ethnicity and cancer type, the heterogeneity was not substantially reduced or eliminated apart from among the subpopulation of Caucasians. The relatively limited studies (n = 3) of rs2066853 polymorphism may be one reason of no obvious between-study heterogeneity in the Caucasian subgroup. Noticeably, there was low to high heterogeneity in Asian and Chinese subgroups (Table 3). Hence, we infer that the ethnicity and cancer type might not be the source of heterogeneity for rs2066853 polymorphism. The obvious between-study heterogeneity, along with the stable negative results in cumulative meta-analysis and sensitivity analysis, possibly reflects the obscure association of rs2066853 polymorphism with cancer susceptibility.
Apart from the heterogeneity, several limitations of this current meta-analysis must be addressed. First, only the published studies in limitative databases were included, so we may have missed some reports in other databases or unpublished negative investigations; additionally, the observational studies themselves were prone to publication bias. Therefore, potential publication bias could not be ruled out, even if no statistical evidence of publication bias was found in this meta-analysis (Table 4). Second, it is important to take some clinical parameters and environmental exposures into account when exploring the gene-disease association, but we failed to stratify data by age, gender, smoking status, and environmental factors due to insufficient corresponding information in the included studies. Third, the limited cases and controls in the overall or subgroup analysis may lead to relatively low statistical power and inability in detecting the low or moderate correlation. Fourth, the data of our meta-analysis were from Caucasians, Asians, and Chinese, so our findings were merely applicable to these ethnic populations. For these reasons, caution should be adopted when explaining our meta-analysis results. In order to minimize the likelihood of bias from the above limitations, we created a detailed protocol of data extraction and inclusion, conducted a comprehensive literature searching, and performed meticulous statistical analysis.
Conclusions
In summary, this present study is the most comprehensive and latest meta-analysis concerning multiple AhR polymorphisms and cancer risk to date. Evidence from our meta-analysis suggests that AhR rs2066853 polymorphism is not a risk factor for cancer either in the overall population, Caucasians, Asians, or Chinese, and this polymorphism is not a risk factor for lung cancer or breast cancer. Our results demonstrate that AhR rs7796976 or rs2074113 polymorphism does not confer susceptibility to cancer. Considering the deep involvement of AhR gene in immune response, the influence of immune on oncogenesis, and the undeniable limitations of our meta-analysis, our findings should be viewed with caution. Future researches focusing on biological mechanisms and clinical phenotypes are required to clarify the exact role of AhR polymorphisms in cancer development, which may provide a sophisticated understanding of the link between AhR polymorphisms and cancer susceptibility.
Availability of data and materials
All data generated or analyzed in this study are included in this published article and its supplementary information files.
Abbreviations
- PAHs:
-
Polycyclic aromatic hydrocarbons
- AhR:
-
Aryl hydrocarbon receptor
- ARNT:
-
AhR nuclear translocator protein
- TCDD:
-
2,3,7,8-Tetrachlorodibenzo-p-dioxin
- HWE:
-
Hardy–Weinberg equilibrium
- OR:
-
Odds ratio
- 95%CI:
-
95% Confidence interval
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Davie-Martin CL, Stratton KG, Teeguarden JG, Waters KM, Simonich SLM. Implications of Bioremediation of Polycyclic Aromatic Hydrocarbon-Contaminated Soils for Human Health and Cancer Risk. Environ Sci Technol. 2017;51:9458–68.
Lewandowska AM, Rudzki M, Rudzki S, Lewandowski T, Laskowska B. Environmental risk factors for cancer - review paper. Ann Agric Environ Med. 2019;26:1–7.
Yu H, Frank C, Sundquist J, Hemminki A, Hemminki K. Common cancers share familial susceptibility: implications for cancer genetics and counselling. J Med Genet. 2017;54:248–53.
Zhang DS, Lin GF, Ma QW, Shen JH. Nonassociation of aryl hydrocarbon receptor genotypes with susceptibility to bladder cancer in Shanghai population. Acta Pharmacol Sin. 2002;23:188–92.
Guarnieri T, Abruzzo PM, Bolotta A. More than a cell biosensor: aryl hydrocarbon receptor at the intersection of physiology and inflammation. Am J Physiol Cell Physiol. 2020;318:C1078–c82.
Kawajiri K, Fujii-Kuriyama Y. The aryl hydrocarbon receptor: a multifunctional chemical sensor for host defense and homeostatic maintenance. Exp Anim. 2017;66:75–89.
Yakkundi P, Gonsalves E, Galou-Lameyer M, Selby MJ, Chan WK. Aryl hydrocarbon receptor acts as a tumor suppressor in a syngeneic MC38 colon carcinoma tumor model. Hypoxia (Auckl). 2019;7:1–16.
Murray IA, Patterson AD, Perdew GH. Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat Rev Cancer. 2014;14:801–14.
Xue P, Fu J, Zhou Y. The Aryl Hydrocarbon Receptor and Tumor Immunity. Front Immunol. 2018;9:286.
Kolluri SK, Jin UH, Safe S. Role of the aryl hydrocarbon receptor in carcinogenesis and potential as an anti-cancer drug target. Arch Toxicol. 2017;91:2497–513.
Gu A, Ji G, Jiang T, Lu A, You Y, Liu N, et al. Contributions of aryl hydrocarbon receptor genetic variants to the risk of glioma and PAH-DNA adducts. Toxicol Sci. 2012;128:357–64.
Chen D, Tian T, Wang H, Liu H, Hu Z, Wang Y, et al. Association of human aryl hydrocarbon receptor gene polymorphisms with risk of lung cancer among cigarette smokers in a Chinese population. Pharmacogenet Genomics. 2009;19:25–34.
Kawajiri K, Watanabe J, Eguchi H, Nakachi K, Kiyohara C, Hayashi S. Polymorphisms of human Ah receptor gene are not involved in lung cancer. Pharmacogenetics. 1995;5:151–8.
Li Y, Qin HZ, Song Q, Wu XD, Zhu JH. Lack of association between the aryl hydrocarbon receptor rs2066853 polymorphism and breast cancer: A meta-analysis on Ahr polymorphism and breast cancer. Genet Mol Res. 2015;14:16162–8.
Luo C, Zou P, Ji G, Gu A, Zhao P, Zhao C. The aryl hydrocarbon receptor (AhR) 1661G>A polymorphism in human cancer: a meta-analysis. Gene. 2013;513:225–30.
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327:557–60.
Sangrajrang S, Sato Y, Sakamoto H, Ohnami S, Laird NM, Khuhaprema T, et al. Genetic polymorphisms of estrogen metabolizing enzyme and breast cancer risk in Thai women. Int J Cancer. 2009;125:837–43.
Sierra-Martinez M, Hernandez-Cadena L, Garcia-Sanchez JR, Acosta-Altamirano G, Palacios-Reyes C, Alonso-Themann PG, et al. Predictive polymorphisms for breast cancer in postmenopausal Mexican women. J Cancer Res Ther. 2018;14:640–6.
Long JR, Egan KM, Dunning L, Shu XO, Cai Q, Cai H, et al. Population-based case-control study of AhR (aryl hydrocarbon receptor) and CYP1A2 polymorphisms and breast cancer risk. Pharmacogenet Genomics. 2006;16:237–43.
Le Marchand L, Donlon T, Kolonel LN, Henderson BE, Wilkens LR. Estrogen metabolism-related genes and breast cancer risk: the multiethnic cohort study. Cancer Epidemiol Biomarkers Prev. 2005;14:1998–2003.
Budhwar S, Bahl C, Sharma S, Singh N, Behera D. Role of Sequence Variations in AhR Gene Towards Modulating Smoking Induced Lung Cancer Susceptibility in North Indian Population: A Multiple Interaction Analysis. Curr Genomics. 2018;19:313–26.
Cauchi S, Stucker I, Solas C, Laurent-Puig P, Cenee S, Hemon D, et al. Polymorphisms of human aryl hydrocarbon receptor (AhR) gene in a French population: relationship with CYP1A1 inducibility and lung cancer. Carcinogenesis. 2001;22:1819–24.
Kim JH, Kim H, Lee KY, Kang JW, Lee KH, Park SY, et al. Aryl hydrocarbon receptor gene polymorphisms affect lung cancer risk. Lung Cancer. 2007;56:9–15.
Pérez-Morales R, Méndez-Ramírez I, Moreno-Macias H, Mendoza-Posadas AD, Martínez-Ramírez OC, Castro-Hernández C, et al. Genetic susceptibility to lung cancer based on candidate genes in a sample from the Mexican Mestizo population: a case-control study. Lung. 2014;192:167–73.
Figueroa JD, Malats N, Garcia-Closas M, Real FX, Silverman D, Kogevinas M, et al. Bladder cancer risk and genetic variation in AKR1C3 and other metabolizing genes. Carcinogenesis. 2008;29:1955–62.
Cotterchio M, Boucher BA, Manno M, Gallinger S, Okey AB, Harper PA. Red meat intake, doneness, polymorphisms in genes that encode carcinogen-metabolizing enzymes, and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:3098–107.
De Roos AJ, Gold LS, Wang S, Hartge P, Cerhan JR, Cozen W, et al. Metabolic gene variants and risk of non-Hodgkin's lymphoma. Cancer Epidemiol Biomarkers Prev. 2006;15:1647–53.
Ng CH, Janoo-Gilani R, Sipahimalani P, Gallagher RP, Gascoyne RD, Connors JM, et al. Interaction between organochlorines and the AHR gene, and risk of non-Hodgkin lymphoma. Cancer Causes Control. 2010;21:11–22.
Guo J, Zhang J, Yin L, Pu Y. A Case-Control Study on the Relation of Aryl Hydrocarbon Receptor(Ahr)Polymorphisms with Susceptibility to Adult Acute Leukemia in Chinese HAN Population. J Environ Occup Med. 2005;22:388–91.
Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–30.
Wang GZ, Zhang L, Zhao XC, Gao SH, Qu LW, Yu H, et al. The Aryl hydrocarbon receptor mediates tobacco-induced PD-L1 expression and is associated with response to immunotherapy. Nat Commun. 2019;10:1125.
Gao M, Li Y, Xue X, Long J, Chen L, Shah W, et al. Impact of AhR, CYP1A1 and GSTM1 genetic polymorphisms on TP53 R273G mutations in individuals exposed to polycyclic aromatic hydrocarbons. Asian Pac J Cancer Prev. 2014;15:2699–705.
Smart J, Daly AK. Variation in induced CYP1A1 levels: relationship to CYP1A1, Ah receptor and GSTM1 polymorphisms. Pharmacogenetics. 2000;10:11–24.
Celius T, Matthews J. Functional analysis of six human aryl hydrocarbon receptor variants in human breast cancer and mouse hepatoma cell lines. Toxicology. 2010;277:59–65.
Jorge-Nebert LF, Zhang G, Wilson KM, Jiang Z, Butler R, Gluckman JL, et al. Head-and-neck squamous cell carcinoma risk in smokers: no association detected between phenotype and AHR, CYP1A1, CYP1A2, or CYP1B1 genotype. Hum Genomics. 2016;10:39.
Acknowledgements
Not applicable
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 81700015).
Author information
Authors and Affiliations
Contributions
RZ conceived and designed this study. HL and LL searched for literature and collected the data. HL, LL, and DW analyzed the data. RZ, HL, and LL wrote the paper. RZ, HL, LL, DW, and JD revised the whole paper. DW and JD contributed to the analysis tools and performed the statistical analysis. The authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors proclaim that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Fig. S1.
Forest plot for AhR rs2066853 polymorphism and the overall cancer risk in the recessive model.
Additional file 2: Fig. S2.
Forest plot for AhR rs2066853 polymorphism and the overall cancer risk in the codominant model.
Additional file 3: Fig. S3.
Forest plot for AhR rs2066853 polymorphism and the overall cancer risk in the allelic model.
Additional file 4: Fig. S4.
Forest plot for AhR rs2066853 polymorphism and the stratified cancer risk in the recessive model.
Additional file 5: Fig. S5.
Forest plot for AhR rs2066853 polymorphism and the stratified cancer risk in the codominant model. Asians (a), Caucasians (a), Chinese (a), breast cancer (b), and lung cancer (b).
Additional file 6: Fig. S6.
Forest plot for AhR rs2066853 polymorphism and the stratified cancer risk in the allelic model.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, H., Luo, L., Wang, D. et al. Lack of association between multiple polymorphisms in aryl hydrocarbon receptor (AhR) gene and cancer susceptibility. Environ Health Prev Med 25, 79 (2020). https://doi.org/10.1186/s12199-020-00907-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12199-020-00907-z