Abstract
Introduction
Biomarkers can be useful for identifying or ruling out sepsis, identifying patients who may benefit from specific therapies or assessing the response to therapy.
Methods
We used an electronic search of the PubMed database using the key words "sepsis" and "biomarker" to identify clinical and experimental studies which evaluated a biomarker in sepsis.
Results
The search retrieved 3370 references covering 178 different biomarkers.
Conclusions
Many biomarkers have been evaluated for use in sepsis. Most of the biomarkers had been tested clinically, primarily as prognostic markers in sepsis; relatively few have been used for diagnosis. None has sufficient specificity or sensitivity to be routinely employed in clinical practice. PCT and CRP have been most widely used, but even these have limited ability to distinguish sepsis from other inflammatory conditions or to predict outcome.
Similar content being viewed by others
Introduction
Sepsis is a leading cause of death in critically ill patients despite the use of modern antibiotics and resuscitation therapies [1]. The septic response is an extremely complex chain of events involving inflammatory and anti-inflammatory processes, humoral and cellular reactions and circulatory abnormalities [2, 3]. The diagnosis of sepsis and evaluation of its severity is complicated by the highly variable and non-specific nature of the signs and symptoms of sepsis [4]. However, the early diagnosis and stratification of the severity of sepsis is very important, increasing the possibility of starting timely and specific treatment [5, 6].
Biomarkers can have an important place in this process because they can indicate the presence or absence or severity of sepsis [7, 8], and can differentiate bacterial from viral and fungal infection, and systemic sepsis from local infection. Other potential uses of biomarkers include roles in prognostication, guiding antibiotic therapy, evaluating the response to therapy and recovery from sepsis, differentiating Gram-positive from Gram-negative microorganisms as the cause of sepsis, predicting sepsis complications and the development of organ dysfunction (heart, kidneys, liver or multiple organ dysfunction). However, the exact role of biomarkers in the management of septic patients remains undefined [9]. C-reactive protein (CRP) has been used for many years [10, 11] but its specificity has been challenged [12]. Procalcitonin (PCT) has been proposed as a more specific [13] and better prognostic [14] marker than CRP, although its value has also been challenged [15]. It remains difficult to differentiate sepsis from other non-infectious causes of systemic inflammatory response syndrome [16], and there is a continuous search for better biomarkers of sepsis.
With this background in mind, we reviewed the literature on sepsis biomarkers that have been used in clinical or experimental studies to help better evaluate their utility.
Materials and methods
The entire Medline database was searched in February 2009 using the key words 'sepsis' and 'biomarker'. All studies, both clinical and experimental, which evaluated a biomarker were included. For each identified biomarker, the Medline database was searched again using the biomarker name and the key word 'biomarker'.
Results
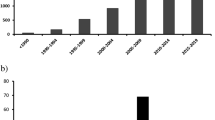
A total of 3370 studies that assessed a biomarker in sepsis were retrieved; 178 different biomarkers were evaluated in the 3370 studies. The retrieved biomarkers and the major findings from key studies using these biomarkers are listed in Tables 1, 2, 3, 4, 5, 6, 7, 8 and 9. Of the 178 biomarkers, 18 had been evaluated in experimental studies only, 58 in both experimental and clinical studies, and 101 in clinical studies only. Thirty-four biomarkers were identified that have been assessed for use specifically in the diagnosis of sepsis (Table 10); of these just five reported sensitivity and specificity values greater than 90%.
Discussion
A multitude of biomarkers has been proposed in the field of sepsis, many more than in other disease processes; for example, a study of patients with myocardial infarction revealed 14 biomarkers suitable for diagnosis and determination of prognosis [17] and in patients with Alzheimer's disease, just 8 biomarkers were identified [18]. This large difference in the numbers of biomarkers for sepsis is likely to be related to the very complex pathophysiology of sepsis, which involves many mediators of inflammation [19], but also other pathophysiological mechanisms. Coagulation, complement, contact system activation, inflammation, and apoptosis are all involved in the sepsis process, and separate markers for each (part of each) system have been proposed (Tables 1 to 9). Additionally, the systemic nature of sepsis and the large numbers of cell types, tissues and organs involved expand the number of potential biomarker candidates, compared with disease processes that involve individual organs or are more localized.
It is interesting to note that most of the biomarkers we identified have been tested clinically and not experimentally. This is likely to be in part related to difficulties creating an experimental model that accurately reflects all aspects of human sepsis, problems with species differences, and problems in determining end-points in animal studies. Additionally, as the sepsis response varies with time, the exact time period during which any specific biomarker may be useful varies, and this is difficult to assess reliably in experimental models. Moreover, as there is no 'gold standard' for the diagnosis of sepsis, the effectiveness of a biomarker needs to be compared with current methods used to diagnose and monitor sepsis in everyday clinical practice, i.e., by the combination of clinical signs and available laboratory variables [20]; experimental models cannot be used for this purpose.
Our study revealed that there are many more potential biomarkers for sepsis than are currently used in clinical studies. Some of these markers may require considerable time, effort and costs to measure. Some are already routinely used for other purposes and easily obtained, such as coagulation tests or cholesterol concentrations. In many cases, the reliability and validity of the proposed biomarker have not been tested properly [8]. Of the many proposed markers for sepsis, acute phase proteins have perhaps been most widely assessed. PCT has been used particularly extensively in recent years. The specificity and sensitivity of PCT for the diagnosis of sepsis is relatively low (typically below 90%), regardless of the cut-off value [21, 22]. Raised PCT levels have also been reported in other conditions associated with inflammatory response, such as trauma [23], major surgery [24] and cardiac surgery [25]. Although CRP is often reported as inferior compared with PCT in terms of sepsis diagnosis, it is frequently used in clinical practice because of its greater availability. Elevated concentrations of serum CRP are correlated with an increased risk of organ failure and death [26], and the study of its time course may be helpful to evaluate the response to therapy in septic patients [11].
Another group of compounds that has been widely assessed as potential biomarkers are the cytokines. These are important mediators in the pathophysiology of sepsis, and most are produced fairly rapidly after sepsis onset. In a clinical study, levels of TNF and IL-10 were increased within the first 24 hours after admission of the patient [27]. However, blood cytokine concentrations are rather erratic and their time course is not clearly in concert with the course of sepsis [27, 28], making interpretation difficult.
The diagnosis of sepsis is a challenge. Clinical and standard laboratory tests are not very helpful because most critically ill patients develop some degree of inflammatory response, whether or not they have sepsis. Even microbiological assessment is unreliable because many culture samples do not yield microorganisms in these patients. However, biomarkers have also not been shown to be a great asset in the diagnosis of sepsis. Indeed, relatively few biomarkers have been evaluated as diagnostic markers (Table 10). Our search retrieved only 10 biomarkers that have been assessed for their ability to distinguish septic patients from non-septic patients with systemic immune response syndrome. However, none of these biomarkers has been tested for both sensitivity and specificity, and there is therefore no biomarker clearly identified as being able to differentiate sepsis syndrome from an inflammatory response due to other causes.
Early diagnosis of sepsis is also an important issue as early institution of appropriate therapy, including antibiotics, is associated with improved outcomes. We identified 16 factors that have been evaluated specifically for the early diagnosis of sepsis; five of these had reported sensitivity and specificity of more than 90%. IL-12 was measured in newborns at the time when sepsis was first suspected clinically and was higher in patients with sepsis than in those without [29]. Interferon-induced protein 10 (IP-10) was higher in neonates with sepsis and necrotizing enterocolitis than in neonates who had only necrotizing enterocolitis [30]. These two biomarkers have not been evaluated for this purpose in adults. Group II phospholipase 2 (PLA2-II) was reported to have high sensitivity and specificity for the diagnosis of bacteremia in critically ill adult patients within 24 hours after admission [31]. CD64 had high sensitivity and specificity for the early diagnosis of sepsis in adults, but could not reliably distinguish viral from bacterial infections, or local infection from systemic sepsis [32]. Neutrophil CD11b could distinguish septic pediatric patients from those with possible infection with good sensitivity and specificity [33]. The sensitivity and specificity of the other 11 biomarkers used to diagnose early sepsis were not reported or were less than 90%.
Biomarkers can be more useful to rule out sepsis than to rule it in. We identified three biomarkers with high negative predictive value to rule out sepsis: PCT (99% at a cut-off value of 0.2 ng/ml) [34]; activated partial thromboplastin time (aPTT) waveform (96%) [35]; and fibrin degradation products (100% for Gram-negative sepsis by ELISA assay) [36]. It is important to emphasize that culture-positive sepsis was generally used as the gold standard in all these studies, although cultures may remain negative in many patients with sepsis.
The majority of the biomarkers that we identified in our search were assessed for their ability to differentiate patients likely to survive from those likely to die. Indeed, any biomarker is expected to have some prognostic value and sepsis biomarkers are no exception; however, this is not an absolute rule because some sepsis biomarkers failed to have prognostic value [37–39]. Moreover, sensitivity and specificity were tested in only some of the proposed prognostic markers, and none had sufficient (more than 90%) sensitivity and specificity to predict which patients were at greater risk of dying due to sepsis. Other biomarkers were assessed for their ability to predict the development of multiple organ failure and to evaluate response to therapy. It is known that the extent of infection and the severity of organ failure has a significant impact on the prognosis of patients with sepsis. Additionally, the response to therapy varies among patients. Recently, the PIRO model has been proposed as a way of stratifying septic patients according to their Predisposing condition, the severity of Infection, the Response to therapy and the degree of Organ dysfunction [20]. In the future, sepsis biomarkers may contribute to this model of classification rather than just being used as prognostic markers.
No biomarker has, therefore, established itself sufficiently to be of great help to clinicians in everyday clinical practice. As each biomarker has limited sensitivity and specificity, it may be interesting to combine several biomarkers [40, 41]; however, this hypothesis requires further study. A clinical study showed that the combination of aPTT waveform with PCT increased the specificity of the aPTT waveform in the diagnosis of sepsis [35]. Studies using panels of sepsis biomarkers have also provided encouraging results [42–44]. The cost-effectiveness of all these methods must also be evaluated.
In this study, we tried to categorize the sepsis biomarkers according to their pathophysiological role in sepsis. A useful sepsis marker must not only help to identify or rule out sepsis, but it should also be able to be used to guide therapy. It has been shown that using PCT levels to guide therapy reduces antibiotic use and may be associated with improved outcomes [45, 46]. The use of novel therapies that modify the pathophysiological process of sepsis may also be guided by biomarkers [47, 48]. A study is underway to evaluate the value of protein C levels to guide the administration of activated protein C (clinicaltrials.gov identifier NCT00386425). In the future, sepsis biomarkers may help us administer these therapies to the right patient at the right time.
Conclusions
Our literature review indicates that there are many biomarkers that can be used in sepsis, but none has sufficient specificity or sensitivity to be routinely employed in clinical practice. PCT and CRP have been most widely used, but even these have limited abilities to distinguish sepsis from other inflammatory conditions or to predict outcome. In view of the complexity of the sepsis response, it is unlikely that a single ideal biomarker will ever be found. A combination of several sepsis biomarkers may be more effective, but this requires further evaluation.
Key messages
• More than 170 different biomarkers have been assessed for potential use in sepsis, more for prognosis than for diagnosis.
• None has sufficient specificity or sensitivity to be routinely employed in clinical practice.
• Combinations of several biomarkers may be more effective than single biomarkers, but this requires further evaluation.
Abbreviations
- aPTT:
-
activated partial thromboplastin time
- CRP:
-
C-reactive protein
- ELISA:
-
enzyme-linked immunosorbent assay
- IL:
-
interleukin
- IP-10:
-
interferon-induced protein 10
- PCT:
-
procalcitonin
- PLA2-II:
-
group II phospholipase 2
- TNF:
-
tumor necrosis factor.
References
Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR: Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med 2001, 29: 1303-1310. 10.1097/00003246-200107000-00002
Hotchkiss RS, Karl IE: The pathophysiology and treatment of sepsis. N Engl J Med 2003, 348: 138-150. 10.1056/NEJMra021333
Gullo A, Bianco N, Berlot G: Management of severe sepsis and septic shock: challenges and recommendations. Crit Care Clin 2006, 22: 489-501. 10.1016/j.ccc.2006.03.006
Lever A, Mackenzie I: Sepsis: definition, epidemiology, and diagnosis. BMJ 2007, 335: 879-883. 10.1136/bmj.39346.495880.AE
Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A, Cheang M: Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 2006, 34: 1589-1596. 10.1097/01.CCM.0000217961.75225.E9
Zambon M, Ceola M, Almeida-de-Castro R, Gullo A, Vincent JL: Implementation of the Surviving Sepsis Campaign guidelines for severe sepsis and septic shock: we could go faster. J Crit Care 2008, 23: 455-460. 10.1016/j.jcrc.2007.08.003
Biomarkers Definitions Working Group: Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther 2001, 69: 89-95. 10.1067/mcp.2001.113989
Marshall JC, Reinhart K: Biomarkers of sepsis. Crit Care Med 2009, 37: 2290-2298. 10.1097/CCM.0b013e3181a02afc
Dellinger RP, Levy MM, Carlet JM, Bion J, Parker MM, Jaeschke R, Reinhart K, Angus DC, Brun-Buisson C, Beale R, Calandra T, Dhainaut JF, Gerlach H, Harvey M, Marini JJ, Marshall J, Ranieri M, Ramsay G, Sevransky J, Thompson BT, Townsend S, Vender JS, Zimmerman JL, Vincent JL: Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock: 2008. Crit Care Med 2008, 36: 296-327. 10.1097/01.CCM.0000298158.12101.41
Povoa P, Coelho L, Almeida E, Fernandes A, Mealha R, Moreira P, Sabino H: C-reactive protein as a marker of infection in critically ill patients. Clin Microbiol Infect 2005, 11: 101-108. 10.1111/j.1469-0691.2004.01044.x
Schmit X, Vincent JL: The time course of blood C-reactive protein concentrations in relation to the response to initial antimicrobial therapy in patients with sepsis. Infection 2008, 36: 213-219. 10.1007/s15010-007-7077-9
Clyne B, Olshaker JS: The C-reactive protein. J Emerg Med 1999, 17: 1019-1025. 10.1016/S0736-4679(99)00135-3
Nakamura A, Wada H, Ikejiri M, Hatada T, Sakurai H, Matsushima Y, Nishioka J, Maruyama K, Isaji S, Takeda T, Nobori T: Efficacy of procalcitonin in the early diagnosis of bacterial infections in a critical care unit. Shock 2009, 31: 586-591.
Luzzani A, Polati E, Dorizzi R, Rungatscher A, Pavan R, Merlini A: Comparison of procalcitonin and C-reactive protein as markers of sepsis. Crit Care Med 2003, 31: 1737-1741. 10.1097/01.CCM.0000063440.19188.ED
Tang BM, Eslick GD, Craig JC, McLean AS: Accuracy of procalcitonin for sepsis diagnosis in critically ill patients: systematic review and meta-analysis. Lancet Infect Dis 2007, 7: 210-217. 10.1016/S1473-3099(07)70052-X
Giamarellos-Bourboulis EJ, Giannopoulou P, Grecka P, Voros D, Mandragos K, Giamarellou H: Should procalcitonin be introduced in the diagnostic criteria for the systemic inflammatory response syndrome and sepsis? J Crit Care 2004, 19: 152-157. 10.1016/j.jcrc.2004.07.001
Penttila I, Penttila K, Rantanen T: Laboratory diagnosis of patients with acute chest pain. Clin Chem Lab Med 2000, 38: 187-197. 10.1515/CCLM.2000.027
Tang BL, Kumar R: Biomarkers of mild cognitive impairment and Alzheimer's disease. Ann Acad Med Singapore 2008, 37: 406-410.
Marshall JC, Vincent JL, Fink MP, Cook DJ, Rubenfeld G, Foster D, Fisher CJ Jr, Faist E, Reinhart K: Measures, markers, and mediators: toward a staging system for clinical sepsis. A report of the Fifth Toronto Sepsis Roundtable, Toronto, Ontario, Canada, October 25-26, 2000. Crit Care Med 2003, 31: 1560-1567. 10.1097/01.CCM.0000065186.67848.3A
Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D, Cohen J, Opal SM, Vincent JL, Ramsay G: 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit Care Med 2003, 31: 1250-1256. 10.1097/01.CCM.0000050454.01978.3B
Ugarte H, Silva E, Mercan D, de Mendonca A, Vincent JL: Procalcitonin used as a marker of infection in the intensive care unit. Crit Care Med 1999, 27: 498-504. 10.1097/00003246-199903000-00024
Suprin E, Camus C, Gacouin A, Le Tulzo Y, Lavoue S, Feuillu A, Thomas R: Procalcitonin: a valuable indicator of infection in a medical ICU? Intensive Care Med 2000, 26: 1232-1238. 10.1007/s001340000580
Mimoz O, Benoist JF, Edouard AR, Assicot M, Bohuon C, Samii K: Procalcitonin and C-reactive protein during the early posttraumatic systemic inflammatory response syndrome. Intensive Care Med 1998, 24: 185-188. 10.1007/s001340050543
Meisner M, Tschaikowsky K, Hutzler A, Schick C, Schuttler J: Postoperative plasma concentrations of procalcitonin after different types of surgery. Intensive Care Med 1998, 24: 680-684. 10.1007/s001340050644
Hensel M, Volk T, Docke WD, Kern F, Tschirna D, Egerer K, Konertz W, Kox WJ: Hyperprocalcitonemia in patients with noninfectious SIRS and pulmonary dysfunction associated with cardiopulmonary bypass. Anesthesiology 1998, 89: 93-104. 10.1097/00000542-199807000-00016
Lobo SM, Lobo FR, Bota DP, Lopes-Ferreira F, Soliman HM, Melot C, Vincent JL: C-reactive protein levels correlate with mortality and organ failure in critically ill patients. Chest 2003, 123: 2043-2049. 10.1378/chest.123.6.2043
Pinsky MR, Vincent JL, Deviere J, Alegre M, Kahn RJ, Dupont E: Serum cytokine levels in human septic shock. Relation to multiple-system organ failure and mortality. Chest 1993, 103: 565-575. 10.1378/chest.103.2.565
Wu HP, Chen CK, Chung K, Tseng JC, Hua CC, Liu YC, Chuang DY, Yang CH: Serial cytokine levels in patients with severe sepsis. Inflamm Res, in press.
Sherwin C, Broadbent R, Young S, Worth J, McCaffrey F, Medlicott NJ, Reith D: Utility of interleukin-12 and interleukin-10 in comparison with other cytokines and acute-phase reactants in the diagnosis of neonatal sepsis. Am J Perinatol 2008, 25: 629-636. 10.1055/s-0028-1090585
Ng PC, Li K, Chui KM, Leung TF, Wong RP, Chu WC, Wong E, Fok TF: IP-10 is an early diagnostic marker for identification of late-onset bacterial infection in preterm infants. Pediatr Res 2007, 61: 93-98. 10.1203/01.pdr.0000250207.95723.96
Rintala EM, Aittoniemi J, Laine S, Nevalainen TJ, Nikoskelainen J: Early identification of bacteremia by biochemical markers of systemic inflammation. Scand J Clin Lab Invest 2001, 61: 523-530. 10.1080/003655101753218283
Nuutila J, Hohenthal U, Laitinen I, Kotilainen P, Rajamaki A, Nikoskelainen J, Lilius EM: Simultaneous quantitative analysis of FcgammaRI (CD64) expression on neutrophils and monocytes: a new, improved way to detect infections. J Immunol Methods 2007, 328: 189-200. 10.1016/j.jim.2007.09.002
Nupponen I, Andersson S, Jarvenpaa AL, Kautiainen H, Repo H: Neutrophil CD11b expression and circulating interleukin-8 as diagnostic markers for early-onset neonatal sepsis. Pediatrics 2001, 108: E12. 10.1542/peds.108.1.e12
Liaudat S, Dayer E, Praz G, Bille J, Troillet N: Usefulness of procalcitonin serum level for the diagnosis of bacteremia. Eur J Clin Microbiol Infect Dis 2001, 20: 524-527. 10.1007/s100960100548
Zakariah AN, Cozzi SM, Van Nuffelen M, Clausi CM, Pradier O, Vincent JL: Combination of biphasic transmittance waveform with blood procalcitonin levels for diagnosis of sepsis in acutely ill patients. Crit Care Med 2008, 36: 1507-1512. 10.1097/CCM.0b013e3181709f19
Deitcher SR, Eisenberg PR: Elevated concentrations of cross-linked fibrin degradation products in plasma. An early marker of gram-negative bacteremia. Chest 1993, 103: 1107-1112. 10.1378/chest.103.4.1107
Emmanuilidis K, Weighardt H, Matevossian E, Heidecke CD, Ulm K, Bartels H, Siewert JR, Holzmann B: Differential regulation of systemic IL-18 and IL-12 release during postoperative sepsis: high serum IL-18 as an early predictive indicator of lethal outcome. Shock 2002, 18: 301-305. 10.1097/00024382-200210000-00002
Figueras-Aloy J, Gomez-Lopez L, Rodriguez-Miguelez JM, Salvia-Roiges MD, Jordan-Garcia I, Ferrer-Codina I, Carbonell-Estrany X, Jimenez-Gonzalez R: Serum soluble ICAM-1, VCAM-1, L-selectin, and P-selectin levels as markers of infection and their relation to clinical severity in neonatal sepsis. Am J Perinatol 2007, 24: 331-338. 10.1055/s-2007-981851
Sakr Y, Burgett U, Nacul FE, Reinhart K, Brunkhorst F: Lipopolysaccharide binding protein in a surgical intensive care unit: a marker of sepsis? Crit Care Med 2008, 36: 2014-2022. 10.1097/CCM.0b013e31817b86e3
Carrigan SD, Scott G, Tabrizian M: Toward resolving the challenges of sepsis diagnosis. Clin Chem 2004, 50: 1301-1314. 10.1373/clinchem.2004.032144
Ng PC, Lam HS: Diagnostic markers for neonatal sepsis. Curr Opin Pediatr 2006, 18: 125-131. 10.1097/01.mop.0000193293.87022.4c
Bozza FA, Salluh JI, Japiassu AM, Soares M, Assis EF, Gomes RN, Bozza MT, Castro-Faria-Neto HC, Bozza PT: Cytokine profiles as markers of disease severity in sepsis: a multiplex analysis. Crit Care 2007, 11: R49. 10.1186/cc5783
Kofoed K, Schneider UV, Scheel T, Andersen O, Eugen-Olsen J: Development and validation of a multiplex add-on assay for sepsis biomarkers using xMAP technology. Clin Chem 2006, 52: 1284-1293. 10.1373/clinchem.2006.067595
Shapiro NI, Trzeciak S, Hollander JE, Birkhahn R, Otero R, Osborn TM, Moretti E, Nguyen HB, Gunnerson KJ, Milzman D, Gaieski DF, Goyal M, Cairns CB, Ngo L, Rivers EP: A prospective, multicenter derivation of a biomarker panel to assess risk of organ dysfunction, shock, and death in emergency department patients with suspected sepsis. Crit Care Med 2009, 37: 96-104. 10.1097/CCM.0b013e318192fd9d
Nobre V, Harbarth S, Graf JD, Rohner P, Pugin J: Use of procalcitonin to shorten antibiotic treatment duration in septic patients: a randomized trial. Am J Respir Crit Care Med 2008, 177: 498-505. 10.1164/rccm.200708-1238OC
Briel M, Schuetz P, Mueller B, Young J, Schild U, Nusbaumer C, Periat P, Bucher HC, Christ-Crain M: Procalcitonin-guided antibiotic use vs a standard approach for acute respiratory tract infections in primary care. Arch Intern Med 2008, 168: 2000-2007. 10.1001/archinte.168.18.2000
Bernard GR, Vincent JL, Laterre PF, LaRosa SP, Dhainaut JF, Lopez-Rodriguez A, Steingrub JS, Garber GE, Helterbrand JD, Ely EW, Fisher CJ Jr: Efficacy and safety of recombinant human activated protein C for severe sepsis. N Engl J Med 2001, 344: 699-709. 10.1056/NEJM200103083441001
Panacek EA, Marshall JC, Albertson TE, Johnson DH, Johnson S, MacArthur RD, Miller M, Barchuk WT, Fischkoff S, Kaul M, Teoh L, Van Meter L, Daum L, Lemeshow S, Hicklin G, Doig C: Efficacy and safety of the monoclonal anti-tumor necrosis factor antibody F(ab')2 fragment afelimomab in patients with severe sepsis and elevated interleukin-6 levels. Crit Care Med 2004, 32: 2173-2182.
Vermont CL, Hazelzet JA, de Kleijn ED, Dobbelsteen GP, de Groot R: CC and CXC chemokine levels in children with meningococcal sepsis accurately predict mortality and disease severity. Crit Care 2006, 10: R33. 10.1186/cc4836
Rodriguez-Wilhelmi P, Montes R, Matsukawa A, Hurtado V, Montes M, Hermida J, Rocha E: Interleukin (IL)-8 and growth related oncogene-alpha in severe endotoxemia and the effects of a tumor necrosis factor-alpha/IL-1beta inhibitor on these chemokines. Exp Mol Pathol 2002, 73: 220-229. 10.1006/exmp.2002.2467
Karlsson S, Pettila V, Tenhunen J, Laru-Sompa R, Hynninen M, Ruokonen E: HMGB1 as a predictor of organ dysfunction and outcome in patients with severe sepsis. Intensive Care Med 2008, 34: 1046-1053. 10.1007/s00134-008-1032-9
Shao YM, Yao HG, Liang XZ, Xia YH: [Relation between level of expression of high mobility group protein B1 in hepatic tissue with the severity and prognosis of sepsis in rat]. Zhongguo Wei Zhong Bing Ji Jiu Yi Xue 2006, 18: 668-672.
Schmidhammer R, Wassermann E, Germann P, Redl H, Ullrich R: Infusion of increasing doses of endotoxin induces progressive acute lung injury but prevents early pulmonary hypertension in pigs. Shock 2006, 25: 389-394. 10.1097/01.shk.0000209529.43367.00
Hynninen M, Valtonen M, Markkanen H, Vaara M, Kuusela P, Jousela I, Piilonen A, Takkunen O: Interleukin 1 receptor antagonist and E-selectin concentrations: a comparison in patients with severe acute pancreatitis and severe sepsis. J Crit Care 1999, 14: 63-68. 10.1016/S0883-9441(99)90015-1
Kuster H, Weiss M, Willeitner AE, Detlefsen S, Jeremias I, Zbojan J, Geiger R, Lipowsky G, Simbruner G: Interleukin-1 receptor antagonist and interleukin-6 for early diagnosis of neonatal sepsis 2 days before clinical manifestation. Lancet 1998, 352: 1271-1277. 10.1016/S0140-6736(98)08148-3
Murch O, Collin M, Sepodes B, Foster SJ, Mota-Filipe H, Thiemermann C: Lysophosphatidylcholine reduces the organ injury and dysfunction in rodent models of gram-negative and gram-positive shock. Br J Pharmacol 2006, 148: 769-777. 10.1038/sj.bjp.0706788
Kurt AN, Aygun AD, Godekmerdan A, Kurt A, Dogan Y, Yilmaz E: Serum IL-1beta, IL-6, IL-8, and TNF-alpha levels in early diagnosis and management of neonatal sepsis. Mediators Inflamm 2007, 2007: 31397. 10.1155/2007/31397
BalcI C, Sungurtekin H, Gurses E, Sungurtekin U, Kaptanoglu B: Usefulness of procalcitonin for diagnosis of sepsis in the intensive care unit. Crit Care 2003, 7: 85-90. 10.1186/cc1843
DiPiro JT, Howdieshell TR, Goddard JK, Callaway DB, Hamilton RG, Mansberger AR Jr: Association of interleukin-4 plasma levels with traumatic injury and clinical course. Arch Surg 1995, 130: 1159-1162.
Patel RT, Deen KI, Youngs D, Warwick J, Keighley MR: Interleukin 6 is a prognostic indicator of outcome in severe intra-abdominal sepsis. Br J Surg 1994, 81: 1306-1308. 10.1002/bjs.1800810914
El Maghraby SM, Moneer MM, Ismail MM, Shalaby LM, El Mahallawy HA: The diagnostic value of C-reactive protein, interleukin-8, and monocyte chemotactic protein in risk stratification of febrile neutropenic children with hematologic malignancies. J Pediatr Hematol Oncol 2007, 29: 131-136. 10.1097/MPH.0b013e3180308770
Fujishima S, Sasaki J, Shinozawa Y, Takuma K, Kimura H, Suzuki M, Kanazawa M, Hori S, Aikawa N: Serum MIP-1 alpha and IL-8 in septic patients. Intensive Care Med 1996, 22: 1169-1175. 10.1007/BF01709331
Heper Y, Akalin EH, Mistik R, Akgoz S, Tore O, Goral G, Oral B, Budak F, Helvaci S: Evaluation of serum C-reactive protein, procalcitonin, tumor necrosis factor alpha, and interleukin-10 levels as diagnostic and prognostic parameters in patients with community-acquired sepsis, severe sepsis, and septic shock. Eur J Clin Microbiol Infect Dis 2006, 25: 481-491. 10.1007/s10096-006-0168-1
Wang CH, Gee MJ, Yang C, Su YC: A new model for outcome prediction in intra-abdominal sepsis by the linear discriminant function analysis of IL-6 and IL-10 at different heart rates. J Surg Res 2006, 132: 46-51. 10.1016/j.jss.2005.08.030
Marchant A, Alegre ML, Hakim A, Pierard G, Marecaux G, Friedman G, De Groote D, Kahn RJ, Vincent JL, Goldman M: Clinical and biological significance of interleukin-10 plasma levels in patients with septic shock. J Clin Immunol 1995, 15: 266-273. 10.1007/BF01540884
Castellheim A, Thorgersen EB, Hellerud BC, Pharo A, Johansen HT, Brosstad F, Gaustad P, Brun H, Fosse E, Tonnessen TI, Nielsen EW, Mollnes TE: New biomarkers in an acute model of live Escherichia coli-induced sepsis in pigs. Scand J Immunol 2008, 68: 75-84. 10.1111/j.1365-3083.2008.02122.x
Weighardt H, Heidecke CD, Westerholt A, Emmanuilidis K, Maier S, Veit M, Gerauer K, Matevossian E, Ulm K, Siewert JR, Holzmann B: Impaired monocyte IL-12 production before surgery as a predictive factor for the lethal outcome of postoperative sepsis. Ann Surg 2002, 235: 560-567. 10.1097/00000658-200204000-00015
Collighan N, Giannoudis PV, Kourgeraki O, Perry SL, Guillou PJ, Bellamy MC: Interleukin 13 and inflammatory markers in human sepsis. Br J Surg 2004, 91: 762-768. 10.1002/bjs.4521
Matsukawa A, Hogaboam CM, Lukacs NW, Lincoln PM, Evanoff HL, Strieter RM, Kunkel SL: Expression and contribution of endogenous IL-13 in an experimental model of sepsis. J Immunol 2000, 164: 2738-2744.
Oberholzer A, Steckholzer U, Kurimoto M, Trentz O, Ertel W: Interleukin-18 plasma levels are increased in patients with sepsis compared to severely injured patients. Shock 2001, 16: 411-414. 10.1097/00024382-200116060-00001
Tsujimoto H, Ono S, Majima T, Kawarabayashi N, Takayama E, Kinoshita M, Seki S, Hiraide H, Moldawer LL, Mochizuki H: Neutrophil elastase, MIP-2, and TLR-4 expression during human and experimental sepsis. Shock 2005, 23: 39-44. 10.1097/01.shk.0000145936.31967.d7
O'Grady NP, Tropea M, Preas HL, Reda D, Vandivier RW, Banks SM, Suffredini AF: Detection of macrophage inflammatory protein (MIP)-1alpha and MIP-1beta during experimental endotoxemia and human sepsis. J Infect Dis 1999, 179: 136-141. 10.1086/314559
Bacher M, Meinhardt A, Lan HY, Mu W, Metz CN, Chesney JA, Calandra T, Gemsa D, Donnelly T, Atkins RC, Bucala R: Migration inhibitory factor expression in experimentally induced endotoxemia. Am J Pathol 1997, 150: 235-246.
Jansen PM, Van Damme J, Put W, de Jong IW, Taylor FB Jr, Hack CE: Monocyte chemotactic protein 1 is released during lethal and sublethal bacteremia in baboons. J Infect Dis 1995, 171: 1640-1642.
Vaschetto R, Nicola S, Olivieri C, Boggio E, Piccolella F, Mesturini R, Damnotti F, Colombo D, Navalesi P, Della CF, Dianzani U, Chiocchetti A: Serum levels of osteopontin are increased in SIRS and sepsis. Intensive Care Med 2008, 34: 2176-2184. 10.1007/s00134-008-1268-4
Moller AS, Bjerre A, Brusletto B, Joo GB, Brandtzaeg P, Kierulf P: Chemokine patterns in meningococcal disease. J Infect Dis 2005, 191: 768-775. 10.1086/427514
VanOtteren GM, Strieter RM, Kunkel SL, Paine R III, Greenberger MJ, Danforth JM, Burdick MD, Standiford TJ: Compartmentalized expression of RANTES in a murine model of endotoxemia. J Immunol 1995, 154: 1900-1908.
Calandra T, Baumgartner JD, Grau GE, Wu MM, Lambert PH, Schellekens J, Verhoef J, Glauser MP: Prognostic values of tumor necrosis factor/cachectin, interleukin-1, interferon-alpha, and interferon-gamma in the serum of patients with septic shock. Swiss-Dutch J5 Immunoglobulin Study Group. J Infect Dis 1990, 161: 982-987.
Riche F, Panis Y, Laisne MJ, Briard C, Cholley B, Bernard-Poenaru O, Graulet AM, Gueris J, Valleur P: High tumor necrosis factor serum level is associated with increased survival in patients with abdominal septic shock: a prospective study in 59 patients. Surgery 1996, 120: 801-807. 10.1016/S0039-6060(96)80087-0
Kaneko T, Stearns-Kurosawa DJ, Taylor F Jr, Twigg M, Osaki K, Kinasewitz GT, Peer G, Kurosawa S: Reduced neutrophil CD10 expression in nonhuman primates and humans after in vivo challenge with E. coli or lipopolysaccharide. Shock 2003, 20: 130-137. 10.1097/01.shk.0000068326.68761.34
Martens A, Eppink GJ, Woittiez AJ, Eidhof H, de Leij LF: Neutrophil function capacity to express CD10 is decreased in patients with septic shock. Crit Care Med 1999, 27: 549-553. 10.1097/00003246-199903000-00034
Weiss DJ, Welle M, Mortiz A, Walcheck B: Evaluation of leukocyte cell surface markers in dogs with septic and nonseptic inflammatory diseases. Am J Vet Res 2004, 65: 59-63. 10.2460/ajvr.2004.65.59
Russwurm S, Vickers J, Meier-Hellmann A, Spangenberg P, Bredle D, Reinhart K, Losche W: Platelet and leukocyte activation correlate with the severity of septic organ dysfunction. Shock 2002, 17: 263-268. 10.1097/00024382-200204000-00004
Williams MA, White SA, Miller JJ, Toner C, Withington S, Newland AC, Kelsey SM: Granulocyte-macrophage colony-stimulating factor induces activation and restores respiratory burst activity in monocytes from septic patients. J Infect Dis 1998, 177: 107-115. 10.1086/513802
Aalto H, Takala A, Kautiainen H, Siitonen S, Repo H: Monocyte CD14 and soluble CD14 in predicting mortality of patients with severe community acquired infection. Scand J Infect Dis 2007, 39: 596-603. 10.1080/00365540701199808
Ebdrup L, Krog J, Granfeldt A, Tonnesen E, Hokland M: Dynamic expression of the signal regulatory protein alpha and CD18 on porcine PBMC during acute endotoxaemia. Scand J Immunol 2008, 68: 430-437. 10.1111/j.1365-3083.2008.02157.x
Saito K, Wagatsuma T, Toyama H, Ejima Y, Hoshi K, Shibusawa M, Kato M, Kurosawa S: Sepsis is characterized by the increases in percentages of circulating CD4+CD25+ regulatory T cells and plasma levels of soluble CD25. Tohoku J Exp Med 2008, 216: 61-68. 10.1620/tjem.216.61
Nolan A, Weiden M, Kelly A, Hoshino Y, Hoshino S, Mehta N, Gold JA: CD40 and CD80/86 act synergistically to regulate inflammation and mortality in polymicrobial sepsis. Am J Respir Crit Care Med 2008, 177: 301-308. 10.1164/rccm.200703-515OC
Sugimoto K, Galle C, Preiser JC, Creteur J, Vincent JL, Pradier O: Monocyte CD40 expression in severe sepsis. Shock 2003, 19: 24-27. 10.1097/00024382-200301000-00005
Katsuura M, Shimizu Y, Akiba K, Kanazawa C, Mitsui T, Sendo D, Kawakami T, Hayasaka K, Yokoyama S: CD48 expression on leukocytes in infectious diseases: flow cytometric analysis of surface antigen. Acta Paediatr Jpn 1998, 40: 580-585.
Livaditi O, Kotanidou A, Psarra A, Dimopoulou I, Sotiropoulou C, Augustatou K, Papasteriades C, Armaganidis A, Roussos C, Orfanos SE, Douzinas EE: Neutrophil CD64 expression and serum IL-8: sensitive early markers of severity and outcome in sepsis. Cytokine 2006, 36: 283-290. 10.1016/j.cyto.2007.02.007
Schwulst SJ, Muenzer JT, Chang KC, Brahmbhatt TS, Coopersmith CM, Hotchkiss RS: Lymphocyte phenotyping to distinguish septic from nonseptic critical illness. J Am Coll Surg 2008, 206: 335-342. 10.1016/j.jamcollsurg.2007.07.038
Moller HJ, Moestrup SK, Weis N, Wejse C, Nielsen H, Pedersen SS, Attermann J, Nexo E, Kronborg G: Macrophage serum markers in pneumococcal bacteremia: Prediction of survival by soluble CD163. Crit Care Med 2006, 34: 2561-2566. 10.1097/01.CCM.0000239120.32490.AB
Monneret G, Lepape A, Voirin N, Bohe J, Venet F, Debard AL, Thizy H, Bienvenu J, Gueyffier F, Vanhems P: Persisting low monocyte human leukocyte antigen-DR expression predicts mortality in septic shock. Intensive Care Med 2006, 32: 1175-1183. 10.1007/s00134-006-0204-8
Zhou Y, Yang Y, Warr G, Bravo R: LPS down-regulates the expression of chemokine receptor CCR2 in mice and abolishes macrophage infiltration in acute inflammation. J Leukoc Biol 1999, 65: 265-269.
Drouin SM, Kildsgaard J, Haviland J, Zabner J, Jia HP, McCray PB Jr, Tack BF, Wetsel RA: Expression of the complement anaphylatoxin C3a and C5a receptors on bronchial epithelial and smooth muscle cells in models of sepsis and asthma. J Immunol 2001, 166: 2025-2032.
Huber-Lang M, Sarma JV, Rittirsch D, Schreiber H, Weiss M, Flierl M, Younkin E, Schneider M, Suger-Wiedeck H, Gebhard F, McClintock SD, Neff T, Zetoune F, Bruckner U, Guo RF, Monk PN, Ward PA: Changes in the novel orphan, C5a receptor (C5L2), during experimental sepsis and sepsis in humans. J Immunol 2005, 174: 1104-1110.
Venet F, Lepape A, Debard AL, Bienvenu J, Bohe J, Monneret G: The Th2 response as monitored by CRTH2 or CCR3 expression is severely decreased during septic shock. Clin Immunol 2004, 113: 278-284. 10.1016/j.clim.2004.07.005
De Freitas I, Fernandez-Somoza M, Essenfeld-Sekler E, Cardier JE: Serum levels of the apoptosis-associated molecules, tumor necrosis factor-alpha/tumor necrosis factor type-I receptor and Fas/FasL, in sepsis. Chest 2004, 125: 2238-2246. 10.1378/chest.125.6.2238
Muller Kobold AC, Zijlstra JG, Koene HR, de Haas M, Kallenberg CG, Tervaert JW: Levels of soluble Fc gammaRIII correlate with disease severity in sepsis. Clin Exp Immunol 1998, 114: 220-227. 10.1046/j.1365-2249.1998.00727.x
Ebihara I, Hirayama K, Kaneko S, Nagai M, Ogawa Y, Fujita S, Usui J, Mase K, Yamagata K, Kobayashi M: Vascular endothelial growth factor and soluble fms-like tyrosine kinase-1 in septic shock patients treated with direct hemoperfusion with a polymyxin B-immobilized fiber column. Ther Apher Dial 2008, 12: 285-291. 10.1111/j.1744-9987.2008.00589.x
Tsao PN, Chan FT, Wei SC, Hsieh WS, Chou HC, Su YN, Chen CY, Hsu WM, Hsieh FJ, Hsu SM: Soluble vascular endothelial growth factor receptor-1 protects mice in sepsis. Crit Care Med 2007, 35: 1955-1960. 10.1097/01.CCM.0000275273.56547.B8
Marsik C, Halama T, Cardona F, Schlifke I, Mittermayer F, Jilma B: Endotoxemia enhances expression of the signaling receptor (GP130) on protein and molecular level. Clin Immunol 2005, 114: 293-298. 10.1016/j.clim.2004.11.003
Delogu G, Casula MA, Mancini P, Tellan G, Signore L: Serum neopterin and soluble interleukin-2 receptor for prediction of a shock state in gram-negative sepsis. J Crit Care 1995, 10: 64-71. 10.1016/0883-9441(95)90018-7
Yokota Y, Ikeda M, Higashino K, Nakano K, Fujii N, Arita H, Hanasaki K: Enhanced tissue expression and elevated circulating level of phospholipase A(2) receptor during murine endotoxic shock. Arch Biochem Biophys 2000, 379: 7-17. 10.1006/abbi.2000.1849
Endo S, Inada K, Nakae H, Takakuwa T, Yamada Y, Suzuki T, Taniguchi S, Yoshida M, Ogawa M, Teraoka H: Plasma levels of type II phospholipase A2 and cytokines in patients with sepsis. Res Commun Mol Pathol Pharmacol 1995, 90: 413-421.
Bopp C, Hofer S, Weitz J, Bierhaus A, Nawroth PP, Martin E, Buchler MW, Weigand MA: sRAGE is elevated in septic patients and associated with patients outcome. J Surg Res 2008, 147: 79-83. 10.1016/j.jss.2007.07.014
Brunner M, Krenn C, Roth G, Moser B, Dworschak M, Jensen-Jarolim E, Spittler A, Sautner T, Bonaros N, Wolner E, Boltz-Nitulescu G, Ankersmit HJ: Increased levels of soluble ST2 protein and IgG1 production in patients with sepsis and trauma. Intensive Care Med 2004, 30: 1468-1473. 10.1007/s00134-004-2184-x
Williams DL, Ha T, Li C, Kalbfleisch JH, Schweitzer J, Vogt W, Browder IW: Modulation of tissue Toll-like receptor 2 and 4 during the early phases of polymicrobial sepsis correlates with mortality. Crit Care Med 2003, 31: 1808-1818. 10.1097/01.CCM.0000069343.27691.F3
Orliac ML, Peroni RN, Abramoff T, Neuman I, Podesta EJ, Adler-Graschinsky E: Increases in vanilloid TRPV1 receptor protein and CGRP content during endotoxemia in rats. Eur J Pharmacol 2007, 566: 145-152. 10.1016/j.ejphar.2007.03.032
Gibot S, Massin F, Le Renard P, Bene MC, Faure GC, Bollaert PE, Levy B: Surface and soluble triggering receptor expressed on myeloid cells-1: expression patterns in murine sepsis. Crit Care Med 2005, 33: 1787-1793. 10.1097/01.CCM.0000172614.36571.75
Gibot S, Cravoisy A, Kolopp-Sarda MN, Bene MC, Faure G, Bollaert PE, Levy B: Time-course of sTREM (soluble triggering receptor expressed on myeloid cells)-1, procalcitonin, and C-reactive protein plasma concentrations during sepsis. Crit Care Med 2005, 33: 792-796. 10.1097/01.CCM.0000159089.16462.4A
Ertel W, Scholl FA, Gallati H, Bonaccio M, Schildberg FW, Trentz O: Increased release of soluble tumor necrosis factor receptors into blood during clinical sepsis. Arch Surg 1994, 129: 1330-1336.
Wittenhagen P, Kronborg G, Weis N, Nielsen H, Obel N, Pedersen SS, Eugen-Olsen J: The plasma level of soluble urokinase receptor is elevated in patients with Streptococcus pneumoniae bacteraemia and predicts mortality. Clin Microbiol Infect 2004, 10: 409-415. 10.1111/j.1469-0691.2004.00850.x
Pettila V, Pentti J, Pettila M, Takkunen O, Jousela I: Predictive value of antithrombin III and serum C-reactive protein concentration in critically ill patients with suspected sepsis. Crit Care Med 2002, 30: 271-275. 10.1097/00003246-200202000-00001
Kinasewitz GT, Yan SB, Basson B, Comp P, Russell JA, Cariou A, Um SL, Utterback B, Laterre PF, Dhainaut JF: Universal changes in biomarkers of coagulation and inflammation occur in patients with severe sepsis, regardless of causative micro-organism [ISRCTN74215569]. Crit Care 2004, 8: R82-R90. 10.1186/cc2459
Lorenz R, Brauer M: Platelet factor 4 (PF4) in septicaemia. Infection 1988, 16: 273-276. 10.1007/BF01645070
Madoiwa S, Nunomiya S, Ono T, Shintani Y, Ohmori T, Mimuro J, Sakata Y: Plasminogen activator inhibitor 1 promotes a poor prognosis in sepsis-induced disseminated intravascular coagulation. Int J Hematol 2006, 84: 398-405. 10.1532/IJH97.05190
Pralong G, Calandra T, Glauser MP, Schellekens J, Verhoef J, Bachmann F, Kruithof EK: Plasminogen activator inhibitor 1: a new prognostic marker in septic shock. Thromb Haemost 1989, 61: 459-462.
Fisher CJ Jr, Yan SB: Protein C levels as a prognostic indicator of outcome in sepsis and related diseases. Crit Care Med 2000, 28: S49-S56. 10.1097/00003246-200009001-00011
Heuer JG, Sharma GR, Gerlitz B, Zhang T, Bailey DL, Ding C, Berg DT, Perkins D, Stephens EJ, Holmes KC, Grubbs RL, Fynboe KA, Chen YF, Grinnell B, Jakubowski JA: Evaluation of protein C and other biomarkers as predictors of mortality in a rat cecal ligation and puncture model of sepsis. Crit Care Med 2004, 32: 1570-1578. 10.1097/01.CCM.0000129488.54282.1A
Lin SM, Wang YM, Lin HC, Lee KY, Huang CD, Liu CY, Wang CH, Kuo HP: Serum thrombomodulin level relates to the clinical course of disseminated intravascular coagulation, multiorgan dysfunction syndrome, and mortality in patients with sepsis. Crit Care Med 2008, 36: 683-689. 10.1097/CCM.0B013E31816537D8
Drake TA, Cheng J, Chang A, Taylor FB Jr: Expression of tissue factor, thrombomodulin, and E-selectin in baboons with lethal Escherichia coli sepsis. Am J Pathol 1993, 142: 1458-1470.
Mimuro J, Niimura M, Kashiwakura Y, Ishiwata A, Ono T, Ohmori T, Madoiwa S, Okada K, Matsuo O, Sakata Y: Unbalanced expression of ADAMTS13 and von Willebrand factor in mouse endotoxinemia. Thromb Res 2008, 122: 91-97. 10.1016/j.thromres.2007.09.011
Ono T, Mimuro J, Madoiwa S, Soejima K, Kashiwakura Y, Ishiwata A, Takano K, Ohmori T, Sakata Y: Severe secondary deficiency of von Willebrand factor-cleaving protease (ADAMTS13) in patients with sepsis-induced disseminated intravascular coagulation: its correlation with development of renal failure. Blood 2006, 107: 528-534. 10.1182/blood-2005-03-1087
Orfanos SE, Kotanidou A, Glynos C, Athanasiou C, Tsigkos S, Dimopoulou I, Sotiropoulou C, Zakynthinos S, Armaganidis A, Papapetropoulos A, Roussos C: Angiopoietin-2 is increased in severe sepsis: correlation with inflammatory mediators. Crit Care Med 2007, 35: 199-206. 10.1097/01.CCM.0000251640.77679.D7
Scherpereel A, Depontieu F, Grigoriu B, Cavestri B, Tsicopoulos A, Gentina T, Jourdain M, Pugin J, Tonnel AB, Lassalle P: Endocan, a new endothelial marker in human sepsis. Crit Care Med 2006, 34: 532-537. 10.1097/01.CCM.0000198525.82124.74
Tissier S, Lancel S, Marechal X, Mordon S, Depontieu F, Scherpereel A, Chopin C, Neviere R: Calpain inhibitors improve myocardial dysfunction and inflammation induced by endotoxin in rats. Shock 2004, 21: 352-357. 10.1097/00024382-200404000-00010
Boldt J, Muller M, Kuhn D, Linke LC, Hempelmann G: Circulating adhesion molecules in the critically ill: a comparison between trauma and sepsis patients. Intensive Care Med 1996, 22: 122-128. 10.1007/BF01720718
Redl H, Dinges HP, Buurman WA, Linden CJ, Pober JS, Cotran RS, Schlag G: Expression of endothelial leukocyte adhesion molecule-1 in septic but not traumatic/hypovolemic shock in the baboon. Am J Pathol 1991, 139: 461-466.
Rafat N, Hanusch C, Brinkkoetter PT, Schulte J, Brade J, Zijlstra JG, Woude FJ, van Ackern K, Yard BA, Beck GC: Increased circulating endothelial progenitor cells in septic patients: correlation with survival. Crit Care Med 2007, 35: 1677-1684. 10.1097/01.CCM.0000269034.86817.59
Presterl E, Lassnigg A, Mueller-Uri P, Wenisch C, El Menyawi I, Graninger W: High serum laminin concentrations in patients with Candida sepsis. Eur J Clin Invest 1999, 29: 992-996. 10.1046/j.1365-2362.1999.00571.x
Ruokonen E, Ilkka L, Niskanen M, Takala J: Procalcitonin and neopterin as indicators of infection in critically ill patients. Acta Anaesthesiol Scand 2002, 46: 398-404. 10.1034/j.1399-6576.2002.460412.x
Redl H, Schlag G, Togel E, Assicot M, Bohuon C: Procalcitonin release patterns in a baboon model of trauma and sepsis: relationship to cytokines and neopterin. Crit Care Med 2000, 28: 3659-3663. 10.1097/00003246-200011000-00021
Brueckmann M, Hoffmann U, Engelhardt C, Lang S, Fukudome K, Haase KK, Liebe V, Kaden JJ, Putensen C, Borggrefe M, Huhle G: Prognostic value of platelet-derived growth factor in patients with severe sepsis. Growth Factors 2007, 25: 15-24. 10.1080/08977190701272784
Cummings CJ, Sessler CN, Beall LD, Fisher BJ, Best AM, Fowler AA III: Soluble E-selectin levels in sepsis and critical illness. Correlation with infection and hemodynamic dysfunction. Am J Respir Crit Care Med 1997, 156: 431-437.
Seidelin JB, Nielsen OH, Strom J: Soluble L-selectin levels predict survival in sepsis. Intensive Care Med 2002, 28: 1613-1618. 10.1007/s00134-002-1501-5
Lopez S, Prats N, Marco AJ: Expression of E-selectin, P-selectin, and intercellular adhesion molecule-1 during experimental murine listeriosis. Am J Pathol 1999, 155: 1391-1397.
Whalen MJ, Doughty LA, Carlos TM, Wisniewski SR, Kochanek PM, Carcillo JA: Intercellular adhesion molecule-1 and vascular cell adhesion molecule-1 are increased in the plasma of children with sepsis-induced multiple organ failure. Crit Care Med 2000, 28: 2600-2607. 10.1097/00003246-200007000-00070
van Oosten M, Bilt E, de Vries HE, van Berkel TJ, Kuiper J: Vascular adhesion molecule-1 and intercellular adhesion molecule-1 expression on rat liver cells after lipopolysaccharide administration in vivo. Hepatology 1995, 22: 1538-1546. 10.1002/hep.1840220529
Flier M, van Leeuwen HJ, van Kessel KP, Kimpen JL, Hoepelman AI, Geelen SP: Plasma vascular endothelial growth factor in severe sepsis. Shock 2005, 23: 35-38. 10.1097/01.shk.0000150728.91155.41
Kim CO, Huh AJ, Kim MS, Chin BS, Han SH, Choi SH, Jeong SJ, Choi HK, Choi JY, Song YG, Kim JM: LPS-induced vascular endothelial growth factor expression in rat lung pericytes. Shock 2008, 30: 92-97. 10.1097/SHK.0b013e31815d19ad
Rubin DB, Wiener-Kronish JP, Murray JF, Green DR, Turner J, Luce JM, Montgomery AB, Marks JD, Matthay MA: Elevated von Willebrand factor antigen is an early plasma predictor of acute lung injury in nonpulmonary sepsis syndrome. J Clin Invest 1990, 86: 474-480. 10.1172/JCI114733
Novotny MJ, Turrentine MA, Johnson GS, Adams HR: Experimental endotoxemia increases plasma von Willebrand factor antigen concentrations in dogs with and without free-radical scavenger therapy. Circ Shock 1987, 23: 205-213.
Christ-Crain M, Morgenthaler NG, Struck J, Harbarth S, Bergmann A, Muller B: Mid-regional pro-adrenomedullin as a prognostic marker in sepsis: an observational study. Crit Care 2005, 9: R816-R824. 10.1186/cc3885
Jiang W, Jiang HF, Cai DY, Pan CS, Qi YF, Pang YZ, Tang CS: Relationship between contents of adrenomedullin and distributions of neutral endopeptidase in blood and tissues of rats in septic shock. Regul Pept 2004, 118: 199-208. 10.1016/j.regpep.2003.12.013
Maccarrone M, De Petrocellis L, Bari M, Fezza F, Salvati S, Di MV, Finazzi-Agro A: Lipopolysaccharide downregulates fatty acid amide hydrolase expression and increases anandamide levels in human peripheral lymphocytes. Arch Biochem Biophys 2001, 393: 321-328. 10.1006/abbi.2001.2500
Deitz DM, Swartz KR, Wright M, Murphy E, Connell RS, Harrison MW: Effects of E. coli endotoxin on rat plasma angiotensin converting enzyme activity in vitro and in vivo. Circ Shock 1987, 21: 23-29.
Casey L, Krieger B, Kohler J, Rice C, Oparil S, Szidon P: Decreased serum angiotensin converting enzyme in adult respiratory distress syndrome associated with sepsis: a preliminary report. Crit Care Med 1981, 9: 651-654. 10.1097/00003246-198109000-00008
Wang Y, Liu Y, Ito Y, Hashiguchi T, Kitajima I, Yamakuchi M, Shimizu H, Matsuo S, Imaizumi H, Maruyama I: Simultaneous measurement of anandamide and 2-arachidonoylglycerol by polymyxin B-selective adsorption and subsequent high-performance liquid chromatography analysis: increase in endogenous cannabinoids in the sera of patients with endotoxic shock. Anal Biochem 2001, 294: 73-82. 10.1006/abio.2001.5015
Seligman R, Papassotiriou J, Morgenthaler NG, Meisner M, Teixeira PJ: Copeptin, a novel prognostic biomarker in ventilator-associated pneumonia. Crit Care 2008, 12: R11. 10.1186/cc6780
Hama N, Itoh H, Shirakami G, Suga S, Komatsu Y, Yoshimasa T, Tanaka I, Mori K, Nakao K: Detection of C-type natriuretic peptide in human circulation and marked increase of plasma CNP level in septic shock patients. Biochem Biophys Res Commun 1994, 198: 1177-1182. 10.1006/bbrc.1994.1166
Broner CW, O'Dorisio MS, Rosenberg RB, O'Dorisio TM: Cyclic nucleotides and vasoactive intestinal peptide production in a rabbit model of Escherichia coli septicemia. Am J Med Sci 1995, 309: 267-277.
Rosenberg RB, Broner CW, O'Dorisio MS: Modulation of cyclic guanosine monophosphate production during Escherichia coli septic shock. Biochem Med Metab Biol 1994, 51: 149-155. 10.1006/bmmb.1994.1019
Faury G, Wynnychenko TM, Cand F, Leone M, Jacob MP, Verdetti J, Boyle WA: Decreased circulating elastin peptide levels in humans with sepsis. Pathol Biol (Paris) 2005, 53: 443-447.
Beer S, Weighardt H, Emmanuilidis K, Harzenetter MD, Matevossian E, Heidecke CD, Bartels H, Siewert JR, Holzmann B: Systemic neuropeptide levels as predictive indicators for lethal outcome in patients with postoperative sepsis. Crit Care Med 2002, 30: 1794-1798. 10.1097/00003246-200208000-00020
Arden WA, Fiscus RR, Wang X, Yang L, Maley R, Nielsen M, Lanzo S, Gross DR: Elevations in circulating calcitonin gene-related peptide correlate with hemodynamic deterioration during endotoxic shock in pigs. Circ Shock 1994, 42: 147-153.
Asmis LM, Asmis R, Sulzer I, Furlan M, Lammle B: Contact system activation in human sepsis - 47 kD HK, a marker of sepsis severity? Swiss Med Wkly 2008, 138: 142-149.
Arnalich F, Sanchez JF, Martinez M, Jimenez M, Lopez J, Vazquez JJ, Hernanz A: Changes in plasma concentrations of vasoactive neuropeptides in patients with sepsis and septic shock. Life Sci 1995, 56: 75-81. 10.1016/0024-3205(94)00416-P
Wang X, Jones SB, Zhou Z, Han C, Fiscus RR: Calcitonin gene-related peptide (CGRP) and neuropeptide Y (NPY) levels are elevated in plasma and decreased in vena cava during endotoxin shock in the rat. Circ Shock 1992, 36: 21-30.
de Werra I, Jaccard C, Corradin SB, Chiolero R, Yersin B, Gallati H, Assicot M, Bohuon C, Baumgartner JD, Glauser MP, Heumann D: Cytokines, nitrite/nitrate, soluble tumor necrosis factor receptors, and procalcitonin concentrations: comparisons in patients with septic shock, cardiogenic shock, and bacterial pneumonia. Crit Care Med 1997, 25: 607-613. 10.1097/00003246-199704000-00009
van Amsterdam JG, Berg C, Zuidema J, te Biesebeek JD, Rokos H: Effect of septicaemia on the plasma levels of biopterin and nitric oxide metabolites in rats and rabbits. Biochem Pharmacol 1996, 52: 1447-1451. 10.1016/S0006-2952(96)00511-4
Zamir O, Hasselgren PO, Higashiguchi T, Frederick JA, Fischer JE: Effect of sepsis or cytokine administration on release of gut peptides. Am J Surg 1992, 163: 181-184. 10.1016/0002-9610(92)90273-T
Galley HF, Le Cras AE, Yassen K, Grant IS, Webster NR: Circulating tetrahydrobiopterin concentrations in patients with septic shock. Br J Anaesth 2001, 86: 578-580. 10.1093/bja/86.4.578
Hattori Y, Nakanishi N, Kasai K, Murakami Y, Shimoda S: Tetrahydrobiopterin and GTP cyclohydrolase I in a rat model of endotoxic shock: relation to nitric oxide synthesis. Exp Physiol 1996, 81: 665-671.
Jacob P, Mueller MH, Hahn J, Wolk I, Mayer P, Nagele U, Hennenlotter J, Stenzl A, Konigsrainer A, Glatzle J: Alterations of neuropeptides in the human gut during peritonitis. Langenbecks Arch Surg 2007, 392: 267-271. 10.1007/s00423-007-0168-3
Brandtzaeg P, Oktedalen O, Kierulf P, Opstad PK: Elevated VIP and endotoxin plasma levels in human gram-negative septic shock. Regul Pept 1989, 24: 37-44. 10.1016/0167-0115(89)90209-7
Morgenthaler NG, Struck J, Christ-Crain M, Bergmann A, Muller B: Pro-atrial natriuretic peptide is a prognostic marker in sepsis, similar to the APACHE II score: an observational study. Crit Care 2005, 9: R37-R45. 10.1186/cc3015
Hartemink KJ, Groeneveld AB, de Groot MC, Strack van Schijndel RJ, van Kamp G, Thijs LG: alpha-atrial natriuretic peptide, cyclic guanosine monophosphate, and endothelin in plasma as markers of myocardial depression in human septic shock. Crit Care Med 2001, 29: 80-87. 10.1097/00003246-200101000-00019
Post F, Weilemann LS, Messow CM, Sinning C, Munzel T: B-type natriuretic peptide as a marker for sepsis-induced myocardial depression in intensive care patients. Crit Care Med 2008, 36: 3030-3037. 10.1097/CCM.0b013e31818b9153
Kandil E, Burack J, Sawas A, Bibawy H, Schwartzman A, Zenilman ME, Bluth MH: B-type natriuretic peptide: a biomarker for the diagnosis and risk stratification of patients with septic shock. Arch Surg 2008, 143: 242-246. 10.1001/archsurg.2007.69
Rivers EP, McCord J, Otero R, Jacobsen G, Loomba M: Clinical utility of B-type natriuretic peptide in early severe sepsis and septic shock. J Intensive Care Med 2007, 22: 363-373. 10.1177/0885066607307523
Struck J, Uhlein M, Morgenthaler NG, Furst W, Hoflich C, Bahrami S, Bergmann A, Volk HD, Redl H: Release of the mitochondrial enzyme carbamoyl phosphate synthase under septic conditions. Shock 2005, 23: 533-538.
Piechota M, Banach M, Irzmanski R, Barylski M, Piechota-Urbanska M, Kowalski J, Pawlicki L: Plasma endothelin-1 levels in septic patients. J Intensive Care Med 2007, 22: 232-239. 10.1177/0885066607301444
Nakamura T, Kasai K, Sekiguchi Y, Banba N, Takahashi K, Emoto T, Hattori Y, Shimoda S: Elevation of plasma endothelin concentrations during endotoxin shock in dogs. Eur J Pharmacol 1991, 205: 277-282. 10.1016/0014-2999(91)90910-I
Brauner JS, Rohde LE, Clausell N: Circulating endothelin-1 and tumor necrosis factor-alpha: early predictors of mortality in patients with septic shock. Intensive Care Med 2000, 26: 305-313. 10.1007/s001340051154
Schuetz P, Stolz D, Mueller B, Morgenthaler NG, Struck J, Mueller C, Bingisser R, Tamm M, Christ-Crain M: Endothelin-1 precursor peptides correlate with severity of disease and outcome in patients with community acquired pneumonia. BMC Infect Dis 2008, 8: 22. 10.1186/1471-2334-8-22
Jha P, Jacobs H, Bose D, Wang R, Yang J, Light RB, Mink S: Effects of E. coli sepsis and myocardial depressant factor on interval-force relations in dog ventricle. Am J Physiol 1993, 264: H1402-H1410.
Dahl B, Schiodt FV, Ott P, Wians F, Lee WM, Balko J, O'Keefe GE: Plasma concentration of Gc-globulin is associated with organ dysfunction and sepsis after injury. Crit Care Med 2003, 31: 152-156. 10.1097/00003246-200301000-00024
Hsu AA, Fenton K, Weinstein S, Carpenter J, Dalton H, Bell MJ: Neurological injury markers in children with septic shock. Pediatr Crit Care Med 2008, 9: 245-251. 10.1097/PCC.0b013e3181727b22
Koo DJ, Zhou M, Chaudry IH, Wang P: Plasma alpha-glutathione S-transferase: a sensitive indicator of hepatocellular damage during polymicrobial sepsis. Arch Surg 2000, 135: 198-203. 10.1001/archsurg.135.2.198
Nayeri F, Nilsson I, Brudin L, Fryden A, Soderstrom C, Forsberg P: High serum hepatocyte growth factor levels in the acute stage of community-acquired infectious diseases. Scand J Infect Dis 2002, 34: 127-130. 10.1080/00365540110077236
Masson S, Daveau M, Francois A, Bodenant C, Hiron M, Teniere P, Salier JP, Scotte M: Up-regulated expression of HGF in rat liver cells after experimental endotoxemia: a potential pathway for enhancement of liver regeneration. Growth Factors 2001, 18: 237-250. 10.3109/08977190109029113
Igonin AA, Armstrong VW, Shipkova M, Kukes VG, Oellerich M: The monoethylglycinexylidide (MEGX) test as a marker of hepatic dysfunction in septic patients with pneumonia. Clin Chem Lab Med 2000, 38: 1125-1128. 10.1515/CCLM.2000.169
McKindley DS, Chichester C, Raymond R: Effect of endotoxin shock on the clearance of lidocaine and indocyanine green in the perfused rat liver. Shock 1999, 12: 468-472. 10.1097/00024382-199912000-00009
Ji Y, Ren X, Zhao Y, Dong L, Wu L, Su J: Role of intracardiac angiotensin II in cardiac dysfunction of rat during septic shock. Chin Med J (Engl) 1996, 109: 864-867.
Nguyen DN, Spapen H, Su F, Schiettecatte J, Shi L, Hachimi-Idrissi S, Huyghens L: Elevated serum levels of S-100beta protein and neuron-specific enolase are associated with brain injury in patients with severe sepsis and septic shock. Crit Care Med 2006, 34: 1967-1974. 10.1097/01.CCM.0000217218.51381.49
Tribl B, Filipp D, Bodeker H, Yu P, Hammerrmuller I, McKerlie C, Keim V, Sibbald WJ: Pseudomonas pneumonia-mediated sepsis induces expression of pancreatitis-associated protein-I in rat pancreas. Pancreas 2004, 29: 33-40. 10.1097/00006676-200407000-00053
Ye SQ, Simon BA, Maloney JP, Zambelli-Weiner A, Gao L, Grant A, Easley RB, McVerry BJ, Tuder RM, Standiford T, Brower RG, Barnes KC, Garcia JG: Pre-B-cell colony-enhancing factor as a potential novel biomarker in acute lung injury. Am J Respir Crit Care Med 2005, 171: 361-370. 10.1164/rccm.200404-563OC
Larsson A, Lipcsey M, Sjolin J, Hansson LO, Eriksson MB: Slight increase of serum S-100B during porcine endotoxemic shock may indicate blood-brain barrier damage. Anesth Analg 2005, 101: 1465-1469. 10.1213/01.ANE.0000180193.29655.6A
Lewis JF, Veldhuizen R, Possmayer F, Sibbald W, Whitsett J, Qanbar R, McCaig L: Altered alveolar surfactant is an early marker of acute lung injury in septic adult sheep. Am J Respir Crit Care Med 1994, 150: 123-130.
Endo S, Sato N, Nakae H, Yamada Y, Makabe H, Abe H, Imai S, Wakabayashi G, Inada K, Sato S: Surfactant protein A and D (SP-A, AP-D) levels in patients with septic ARDS. Res Commun Mol Pathol Pharmacol 2002, 111: 245-251.
Mehta NJ, Khan IA, Gupta V, Jani K, Gowda RM, Smith PR: Cardiac troponin I predicts myocardial dysfunction and adverse outcome in septic shock. Int J Cardiol 2004, 95: 13-17. 10.1016/j.ijcard.2003.02.005
Cicarelli DD, Vieira JE, Bensenor FE: Comparison of C-reactive protein and serum amyloid a protein in septic shock patients. Mediators Inflamm 2008, 2008: 631414. 10.1155/2008/631414
Orro T, Sankari S, Pudas T, Oksanen A, Soveri T: Acute phase response in reindeer after challenge with Escherichia coli endotoxin. Comp Immunol Microbiol Infect Dis 2004, 27: 413-422. 10.1016/j.cimid.2004.01.005
Chiarla C, Giovannini I, Siegel JH: Patterns of correlation of plasma ceruloplasmin in sepsis. J Surg Res 2008, 144: 107-110. 10.1016/j.jss.2007.03.024
Suri M, Sharma VK, Thirupuram S: Evaluation of ceruloplasmin in neonatal septicemia. Indian Pediatr 1991, 28: 489-493.
Couto RC, Barbosa JA, Pedrosa TM, Biscione FM: C-reactive protein-guided approach may shorten length of antimicrobial treatment of culture-proven late-onset sepsis: an intervention study. Braz J Infect Dis 2007, 11: 240-245. 10.1590/S1413-86702007000200015
Seller-Perez G, Herrera-Gutierrez ME, Lebron-Gallardo M, Toro-Peinado I, Martin-Hita L, Porras-Ballesteros JA: [Serum C-reactive protein as a marker of outcome and infection in critical care patients]. Med Clin (Barc) 2005, 125: 761-765. 10.1016/S0025-7753(05)72184-9
Garcia PC, Longhi F, Branco RG, Piva JP, Lacks D, Tasker RC: Ferritin levels in children with severe sepsis and septic shock. Acta Paediatr 2007, 96: 1829-1831. 10.1111/j.1651-2227.2007.00564.x
Linden EC, van Ommen EC, van Dijk W: Glycosylation of alpha 1-acid glycoprotein in septic shock: changes in degree of branching and in expression of sialyl Lewis(x) groups. Glycoconj J 1996, 13: 27-31. 10.1007/BF01049676
Scotte M, Daveau M, Hiron M, Delers F, Lemeland JF, Teniere P, Lebreton JP: Interleukin-6 (IL-6) and acute-phase proteins in rats with biliary sepsis. Eur Cytokine Netw 1991, 2: 177-182.
Li H, Rose MJ, Tran L, Zhang J, Miranda LP, James CA, Sasu BJ: Development of a method for the sensitive and quantitative determination of hepcidin in human serum using LC-MS/MS. J Pharmacol Toxicol Methods 2009,59(3):171-180. 10.1016/j.vascn.2009.02.004
Oude Nijhuis CS, Vellenga E, Daenen SM, Graaf WT, Gietema JA, Groen HJ, Kamps WA, De Bont ES: Lipopolysaccharide-binding protein: a possible diagnostic marker for Gram-negative bacteremia in neutropenic cancer patients. Intensive Care Med 2003, 29: 2157-2161. 10.1007/s00134-003-2026-2
Becker KL, Snider R, Nylen ES: Procalcitonin assay in systemic inflammation, infection, and sepsis: clinical utility and limitations. Crit Care Med 2008, 36: 941-952. 10.1097/CCM.0B013E318165BABB
Muller B, Peri G, Doni A, Torri V, Landmann R, Bottazzi B, Mantovani A: Circulating levels of the long pentraxin PTX3 correlate with severity of infection in critically ill patients. Crit Care Med 2001, 29: 1404-1407. 10.1097/00003246-200107000-00017
al Ramadi BK, Ellis M, Pasqualini F, Mantovani A: Selective induction of pentraxin 3, a soluble innate immune pattern recognition receptor, in infectious episodes in patients with haematological malignancy. Clin Immunol 2004, 112: 221-224. 10.1016/j.clim.2004.03.012
Andrejko KM, Chen J, Deutschman CS: Intrahepatic STAT-3 activation and acute phase gene expression predict outcome after CLP sepsis in the rat. Am J Physiol 1998, 275: G1423-G1429.
Wang XY, Li WQ, Lu J, Li N, Li JS: Lipopolysaccharide decreasing albumin expression in rat hepatocytes. Hepatobiliary Pancreat Dis Int 2005, 4: 410-415.
Maury E, Blanchard HS, Chauvin P, Guglielminotti J, Alzieu M, Guidet B, Offenstadt G: Circulating endotoxin and antiendotoxin antibodies during severe sepsis and septic shock. J Crit Care 2003, 18: 115-120. 10.1053/jcrc.2003.50004
Schippers EF, Berbee JF, van Disseldorp IM, Versteegh MI, Havekes LM, Rensen PC, van Dissel JT: Preoperative apolipoprotein CI levels correlate positively with the proinflammatory response in patients experiencing endotoxemia following elective cardiac surgery. Intensive Care Med 2008, 34: 1492-1497. 10.1007/s00134-008-1077-9
Berbee JF, Mooijaart SP, de Craen AJ, Havekes LM, van Heemst D, Rensen PC, Westendorp RG: Plasma apolipoprotein CI protects against mortality from infection in old age. J Gerontol A Biol Sci Med Sci 2008, 63: 122-126.
Berbee JF, Hoogt CC, de Haas CJ, van Kessel KP, Dallinga-Thie GM, Romijn JA, Havekes LM, van Leeuwen HJ, Rensen PC: Plasma apolipoprotein CI correlates with increased survival in patients with severe sepsis. Intensive Care Med 2008, 34: 907-911. 10.1007/s00134-008-1006-y
Adrie C, Bachelet M, Vayssier-Taussat M, Russo-Marie F, Bouchaert I, Adib-Conquy M, Cavaillon JM, Pinsky MR, Dhainaut JF, Polla BS: Mitochondrial membrane potential and apoptosis peripheral blood monocytes in severe human sepsis. Am J Respir Crit Care Med 2001, 164: 389-395.
Nakamura T, Ebihara I, Shoji H, Ushiyama C, Suzuki S, Koide H: Treatment with polymyxin B-immobilized fiber reduces platelet activation in septic shock patients: decrease in plasma levels of soluble P-selectin, platelet factor 4 and beta-thromboglobulin. Inflamm Res 1999, 48: 171-175. 10.1007/s000110050442
Delogu G, Famularo G, Tellan G, Marandola M, Antonucci A, Signore M, Marcellini S, Moretti S: Lymphocyte apoptosis, caspase activation and inflammatory response in septic shock. Infection 2008, 36: 485-487. 10.1007/s15010-008-7070-y
Delogu G, Famularo G, Amati F, Signore L, Antonucci A, Trinchieri V, Di Marzio L, Cifone MG: Ceramide concentrations in septic patients: a possible marker of multiple organ dysfunction syndrome. Crit Care Med 1999, 27: 2413-2417. 10.1097/00003246-199911000-00015
Memis D, Gursoy O, Tasdogan M, Sut N, Kurt I, Ture M, Karamanlioglu B: High C-reactive protein and low cholesterol levels are prognostic markers of survival in severe sepsis. J Clin Anesth 2007, 19: 186-191. 10.1016/j.jclinane.2006.10.008
Stove S, Welte T, Wagner TO, Kola A, Klos A, Bautsch W, Kohl J: Circulating complement proteins in patients with sepsis or systemic inflammatory response syndrome. Clin Diagn Lab Immunol 1996, 3: 175-183.
Gressner OA, Koch A, Sanson E, Trautwein C, Tacke F: High C5a levels are associated with increased mortality in sepsis patients--no enhancing effect by actin-free Gc-globulin. Clin Biochem 2008, 41: 974-980. 10.1016/j.clinbiochem.2008.05.005
Schuerholz T, Leuwer M, Cobas-Meyer M, Vangerow B, Kube F, Kirschfink M, Marx G: Terminal complement complex in septic shock with capillary leakage: marker of complement activation? Eur J Anaesthesiol 2005, 22: 541-547. 10.1017/S0265021505000931
Guisset O, Dilhuydy MS, Thiebaut R, Lefevre J, Camou F, Sarrat A, Gabinski C, Moreau JF, Blanco P: Decrease in circulating dendritic cells predicts fatal outcome in septic shock. Intensive Care Med 2007, 33: 148-152. 10.1007/s00134-006-0436-7
Efron PA, Martins A, Minnich D, Tinsley K, Ungaro R, Bahjat FR, Hotchkiss R, Clare-Salzler M, Moldawer LL: Characterization of the systemic loss of dendritic cells in murine lymph nodes during polymicrobial sepsis. J Immunol 2004, 173: 3035-3043.
Bergmann A, Bohuon C: Decrease of serum dipeptidylpeptidase activity in severe sepsis patients: relationship to procalcitonin. Clin Chim Acta 2002, 321: 123-126. 10.1016/S0009-8981(02)00042-6
Meinhold H, Gramm HJ, Meissner W, Zimmermann J, Schwander J, Dennhardt R, Voigt K: Elevated serum diiodotyrosine (DIT) in severe infections and sepsis: DIT, a possible new marker of leukocyte activity. J Clin Endocrinol Metab 1991, 72: 945-953. 10.1210/jcem-72-4-945
Morlion BJ, Torwesten E, Kuhn KS, Puchstein C, Furst P: Cysteinyl-leukotriene generation as a biomarker for survival in the critically ill. Crit Care Med 2000, 28: 3655-3658. 10.1097/00003246-200011000-00020
Uozumi N, Kita Y, Shimizu T: Modulation of lipid and protein mediators of inflammation by cytosolic phospholipase A2alpha during experimental sepsis. J Immunol 2008, 181: 3558-3566.
Peters KM, Koberg K, Rosendahl T, Haubeck HD: PMN elastase in bone and joint infections. Int Orthop 1994, 18: 352-355. 10.1007/BF00187079
Geiger R, Sokal S, Trefz G, Siebeck M, Hoffmann H: PMN elastase and leukocyte neutral proteinase inhibitor (LNPI) from granulocytes as inflammation markers in experimental-septicemia. Adv Exp Med Biol 1988, 240: 465-471.
Tegtmeyer FK, Horn C, Richter A, van Wees J: Elastase alpha 1 proteinase inhibitor complex, granulocyte count, ratio of immature to total granulocyte count, and C-reactive protein in neonatal septicaemia. Eur J Pediatr 1992, 151: 353-356. 10.1007/BF02113257
Duswald KH, Jochum M, Schramm W, Fritz H: Released granulocytic elastase: an indicator of pathobiochemical alterations in septicemia after abdominal surgery. Surgery 1985, 98: 892-899.
Tamion F, Cam-Duchez V, Menard JF, Girault C, Coquerel A, Bonmarchand G: Serum erythropoietin levels in septic shock. Anaesth Intensive Care 2005, 33: 578-584.
Puthucheary SD, Nathan SA: Comparison of serum F2 isoprostane levels in diabetic patients and diabetic patients infected with Burkholderia pseudomallei. Singapore Med J 2008, 49: 117-120.
Tanaka M, Yanagihara I, Takahashi H, Hamaguchi M, Nakahira K, Sakata I: The mRNA expression of fatty acid amide hydrolase in human whole blood correlates with sepsis. J Endotoxin Res 2007, 13: 35-38. 10.1177/0968051907078607
Rhodes A, Wort SJ, Thomas H, Collinson P, Bennett ED: Plasma DNA concentration as a predictor of mortality and sepsis in critically ill patients. Crit Care 2006, 10: R60. 10.1186/cc4894
Perry SE, Mostafa SM, Wenstone R, McLaughlin PJ: Low plasma granulocyte-macrophage colony stimulating factor is an indicator of poor prognosis in sepsis. Intensive Care Med 2002, 28: 981-984. 10.1007/s00134-002-1335-1
Kennon C, Overturf G, Bessman S, Sierra E, Smith KJ, Brann B: Granulocyte colony-stimulating factor as a marker for bacterial infection in neonates. J Pediatr 1996, 128: 765-769. 10.1016/S0022-3476(96)70327-X
Wang H, Cheng B, Chen Q, Wu S, Lv C, Xie G, Jin Y, Fang X: Time course of plasma gelsolin concentrations during severe sepsis in critically ill surgical patients. Crit Care 2008, 12: R106. 10.1186/cc6988
Hataya Y, Akamizu T, Hosoda H, Kanamoto N, Moriyama K, Kangawa K, Takaya K, Nakao K: Alterations of plasma ghrelin levels in rats with lipopolysaccharide-induced wasting syndrome and effects of ghrelin treatment on the syndrome. Endocrinology 2003, 144: 5365-5371. 10.1210/en.2003-0427
Yilmaz Z, Ilcol YO, Ulus IH: Endotoxin increases plasma leptin and ghrelin levels in dogs. Crit Care Med 2008, 36: 828-833. 10.1097/01.CCM.0B013E3181611F5AA
Borgel D, Clauser S, Bornstain C, Bieche I, Bissery A, Remones V, Fagon JY, Aiach M, Diehl JL: Elevated growth-arrest-specific protein 6 plasma levels in patients with severe sepsis. Crit Care Med 2006, 34: 219-222. 10.1097/01.CCM.0000195014.56254.8A
Ofenstein JP, Heidemann S, Juett-Wilstermann A, Sarnaik A: Expression of stress proteins HSP 72 & HSP 32 in response to endotoxemia. Ann Clin Lab Sci 2000, 30: 92-98.
Delogu G, Lo BL, Marandola M, Famularo G, Lenti L, Ippoliti F, Signore L: Heat shock protein (HSP70) expression in septic patients. J Crit Care 1997, 12: 188-192. 10.1016/S0883-9441(97)90031-9
Weiss YG, Bouwman A, Gehan B, Schears G, Raj N, Deutschman CS: Cecal ligation and double puncture impairs heat shock protein 70 (HSP-70) expression in the lungs of rats. Shock 2000, 13: 19-23. 10.1097/00024382-200013010-00004
Ramaglia V, Harapa GM, White N, Buck LT: Bacterial infection and tissue-specific Hsp72, -73 and -90 expression in western painted turtles. Comp Biochem Physiol C Toxicol Pharmacol 2004, 138: 139-148. 10.1016/j.cca.2004.06.007
Monneret G, Voirin N, Krawice-Radanne I, Bohe J, Lepape A, Rouas-Freiss N, Carosella ED: Soluble human leukocyte antigen-G5 in septic shock: marked and persisting elevation as a predictor of survival. Crit Care Med 2007, 35: 1942-1947. 10.1097/01.CCM.0000277039.84372.1C
Li L, Bhatia M, Zhu YZ, Zhu YC, Ramnath RD, Wang ZJ, Anuar FB, Whiteman M, Salto-Tellez M, Moore PK: Hydrogen sulfide is a novel mediator of lipopolysaccharide-induced inflammation in the mouse. FASEB J 2005, 19: 1196-1198.
Berg S, Brodin B, Hesselvik F, Laurent TC, Maller R: Elevated levels of plasma hyaluronan in septicaemia. Scand J Clin Lab Invest 1988, 48: 727-732. 10.3109/00365518809088752
Berg S, Jansson I, Hesselvik FJ, Laurent TC, Lennquist S, Walther S: Hyaluronan: relationship to hemodynamics and survival in porcine injury and sepsis. Crit Care Med 1992, 20: 1315-1321. 10.1097/00003246-199209000-00020
Lacroix-Desmazes S, Bayry J, Kaveri SV, Hayon-Sonsino D, Thorenoor N, Charpentier J, Luyt CE, Mira JP, Nagaraja V, Kazatchkine MD, Dhainaut JF, Mallet VO: High levels of catalytic antibodies correlate with favorable outcome in sepsis. Proc Natl Acad Sci USA 2005, 102: 4109-4113. 10.1073/pnas.0500586102
Opal SM, Lim YP, Siryaporn E, Moldawer LL, Pribble JP, Palardy JE, Souza S: Longitudinal studies of inter-alpha inhibitor proteins in severely septic patients: a potential clinical marker and mediator of severe sepsis. Crit Care Med 2007, 35: 387-392. 10.1097/01.CCM.0000253810.08230.83
Soriano AO, Jy W, Chirinos JA, Valdivia MA, Velasquez HS, Jimenez JJ, Horstman LL, Kett DH, Schein RM, Ahn YS: Levels of endothelial and platelet microparticles and their interactions with leukocytes negatively correlate with organ dysfunction and predict mortality in severe sepsis. Crit Care Med 2005, 33: 2540-2546. 10.1097/01.CCM.0000186414.86162.03
Nguyen HB, Rivers EP, Knoblich BP, Jacobsen G, Muzzin A, Ressler JA, Tomlanovich MC: Early lactate clearance is associated with improved outcome in severe sepsis and septic shock. Crit Care Med 2004, 32: 1637-1642. 10.1097/01.CCM.0000132904.35713.A7
Haji-Michael PG, Ladriere L, Sener A, Vincent JL, Malaisse WJ: Leukocyte glycolysis and lactate output in animal sepsis and ex vivo human blood. Metabolism 1999, 48: 779-785. 10.1016/S0026-0495(99)90179-8
Gutteberg TJ, Rokke O, Jorgensen T, Andersen O: Lactoferrin as an indicator of septicemia and endotoxemia in pigs. Scand J Infect Dis 1988, 20: 659-666. 10.3109/00365548809035667
Soderquist B, Sundqvist KG, Jones I, Holmberg H, Vikerfors T: Interleukin-6, C-reactive protein, lactoferrin and white blood cell count in patients with S. aureus septicemia. Scand J Infect Dis 1995, 27: 375-380. 10.3109/00365549509032734
Tzanela M, Orfanos SE, Tsirantonaki M, Kotanidou A, Sotiropoulou C, Christophoraki M, Vassiliadi D, Thalassinos NC, Roussos C: Leptin alterations in the course of sepsis in humans. In Vivo 2006, 20: 565-570.
Jones JW Jr, Su S, Jones MB, Heniford BT, McIntyre K, Granger DK: Serum lysozyme activity can differentiate infection from rejection in organ transplant recipients. J Surg Res 1999, 84: 134-137. 10.1006/jsre.1999.5628
Hoffmann U, Bertsch T, Dvortsak E, Liebetrau C, Lang S, Liebe V, Huhle G, Borggrefe M, Brueckmann M: Matrix-metalloproteinases and their inhibitors are elevated in severe sepsis: prognostic value of TIMP-1 in severe sepsis. Scand J Infect Dis 2006, 38: 867-872. 10.1080/00365540600702058
Piliponsky AM, Chen CC, Nishimura T, Metz M, Rios EJ, Dobner PR, Wada E, Wada K, Zacharias S, Mohanasundaram UM, Faix JD, Abrink M, Pejler G, Pearl RG, Tsai M, Galli SJ: Neurotensin increases mortality and mast cells reduce neurotensin levels in a mouse model of sepsis. Nat Med 2008, 14: 392-398. 10.1038/nm1738
Oudenhoven IM, Klaasen HL, Lapre JA, Weerkamp AH, Van der MR: Nitric oxide-derived urinary nitrate as a marker of intestinal bacterial translocation in rats. Gastroenterology 1994, 107: 47-53.
Williams JP, Thompson JP, Young SP, Gold SJ, McDonald J, Rowbotham DJ, Lambert DG: Nociceptin and urotensin-II concentrations in critically ill patients with sepsis. Br J Anaesth 2008, 100: 810-814. 10.1093/bja/aen093
Arnalich F, Garcia-Palomero E, Lopez J, Jimenez M, Madero R, Renart J, Vazquez JJ, Montiel C: Predictive value of nuclear factor kappaB activity and plasma cytokine levels in patients with sepsis. Infect Immun 2000, 68: 1942-1945. 10.1128/IAI.68.4.1942-1945.2000
Zeerleder S, Zwart B, Wuillemin WA, Aarden LA, Groeneveld AB, Caliezi C, van Nieuwenhuijze AE, van Mierlo GJ, Eerenberg AJ, Lammle B, Hack CE: Elevated nucleosome levels in systemic inflammation and sepsis. Crit Care Med 2003, 31: 1947-1951. 10.1097/01.CCM.0000074719.40109.95
Shimizu T, Endo Y, Tabata T, Mori T, Hanasawa K, Tsuchiya M, Tani T: Diagnostic and predictive value of the silkworm larvae plasma test for postoperative infection following gastrointestinal surgery. Crit Care Med 2005, 33: 1288-1295. 10.1097/01.CCM.0000165810.97971.DD
Yano K, Liaw PC, Mullington JM, Shih SC, Okada H, Bodyak N, Kang PM, Toltl L, Belikoff B, Buras J, Simms BT, Mizgerd JP, Carmeliet P, Karumanchi SA, Aird WC: Vascular endothelial growth factor is an important determinant of sepsis morbidity and mortality. J Exp Med 2006, 203: 1447-1458. 10.1084/jem.20060375
Chiarla C, Giovannini I, Siegel JH, Boldrini G, Castagneto M: The relationship between plasma taurine and other amino acid levels in human sepsis. J Nutr 2000, 130: 2222-2227.
Poeze M, Luiking YC, Breedveld P, Manders S, Deutz NE: Decreased plasma glutamate in early phases of septic shock with acute liver dysfunction is an independent predictor of survival. Clin Nutr 2008, 27: 523-530. 10.1016/j.clnu.2008.04.006
Basler T, Meier-Hellmann A, Bredle D, Reinhart K: Amino acid imbalance early in septic encephalopathy. Intensive Care Med 2002, 28: 293-298. 10.1007/s00134-002-1217-6
Ruiz Martin G, Prieto PJ, Veiga dC, Gomez LL, Barberan J, Gonzalez Landa JM, Fernandez C: Plasma fibronectin as a marker of sepsis. Int J Infect Dis 2004, 8: 236-243. 10.1016/j.ijid.2003.10.005
Mavrommatis AC, Theodoridis T, Economou M, Kotanidou A, El Ali M, Christopoulou-Kokkinou V, Zakynthinos SG: Activation of the fibrinolytic system and utilization of the coagulation inhibitors in sepsis: comparison with severe sepsis and septic shock. Intensive Care Med 2001, 27: 1853-1859. 10.1007/s00134-001-1139-8
Tamion F, Cam-Duchez V, Menard JF, Girault C, Coquerel A, Bonmarchand G: Erythropoietin and renin as biological markers in critically ill patients. Crit Care 2004, 8: R328-R335. 10.1186/cc2902
Sunden-Cullberg J, Nystrom T, Lee ML, Mullins GE, Tokics L, Andersson J, Norrby-Teglund A, Treutiger CJ: Pronounced elevation of resistin correlates with severity of disease in severe sepsis and septic shock. Crit Care Med 2007, 35: 1536-1542. 10.1097/01.CCM.0000266536.14736.03
Sakr Y, Reinhart K, Bloos F, Marx G, Russwurm S, Bauer M, Brunkhorst F: Time course and relationship between plasma selenium concentrations, systemic inflammatory response, sepsis, and multiorgan failure. Br J Anaesth 2007, 98: 775-784. 10.1093/bja/aem091
Hollenbach B, Morgenthaler NG, Struck J, Alonso C, Bergmann A, Kohrle J, Schomburg L: New assay for the measurement of selenoprotein P as a sepsis biomarker from serum. J Trace Elem Med Biol 2008, 22: 24-32. 10.1016/j.jtemb.2007.11.003
Ramzi J, Mohamed Z, Yosr B, Karima K, Raihane B, Lamia A, Hela BA, Zaher B, Balkis M: Predictive factors of septic shock and mortality in neutropenic patients. Hematology 2007, 12: 543-548. 10.1080/10245330701384237
Claus RA, Bunck AC, Bockmeyer CL, Brunkhorst FM, Losche W, Kinscherf R, Deigner HP: Role of increased sphingomyelinase activity in apoptosis and organ failure of patients with severe sepsis. FASEB J 2005, 19: 1719-1721.
Mitsuhashi H, Ikeuchi H, Yamashita S, Kuroiwa T, Kaneko Y, Hiromura K, Ueki K, Nojima Y: Increased levels of serum sulfite in patients with acute pneumonia. Shock 2004, 21: 99-102. 10.1097/01.shk.0000105501.75189.85
Briassoulis G, Papassotiriou I, Mavrikiou M, Lazaropoulou C, Margeli A: Longitudinal course and clinical significance of TGF-beta1, sL- and sE-Selectins and sICAM-1 levels during severe acute stress in children. Clin Biochem 2007, 40: 299-304. 10.1016/j.clinbiochem.2006.11.015
Hafez HM, Berwanger CS, Lintott P, Delis K, Wolfe JH, Mansfield AO, Stansby G: Endotoxemia during supraceliac aortic crossclamping is associated with suppression of the monocyte CD14 mechanism: possible role of transforming growth factor-beta1. J Vasc Surg 2000, 31: 520-531. 10.1016/S0741-5214(00)90314-8
Wang H, Yu XY, Sun M, Pan JK, Gao H: [Effects of glutamine on matrix metalloproteinase-3 and tissue inhibitor of metalloproteinase-3 expressions in myocardium of rats with sepsis]. Zhonghua Er Ke Za Zhi 2006, 44: 587-591.
Giovannini I, Chiarla C, Giuliante F, Vellone M, Ardito F, Pallavicini F, Nuzzo G: Biochemical and clinical correlates of hypouricemia in surgical and critically ill patients. Clin Chem Lab Med 2007, 45: 1207-1210. 10.1515/CCLM.2007.502
Cheng WE, Shih CM, Hang LW, Wu KY, Yang HL, Hsu WH, Hsia TC: Urinary biomarker of oxidative stress correlating with outcome in critically septic patients. Intensive Care Med 2007, 33: 1187-1190. 10.1007/s00134-007-0628-9
Otani K, Shimizu S, Chijiiwa K, Yamaguchi K, Kuroki S, Tanaka M: Increased urinary excretion of bilirubin oxidative metabolites in septic patients: a new marker for oxidative stress in vivo. J Surg Res 2001, 96: 44-49. 10.1006/jsre.2000.6036
Weiss M, Elsharkawi M, Welt K, Schneider EM: Transient leukocytosis, granulocyte colony-stimulating factor plasma concentrations, and apoptosis determined by binding of annexin V by peripheral leukocytes in patients with severe sepsis. Ann N Y Acad Sci 2003, 1010: 742-747. 10.1196/annals.1299.134
Luchtemberg MN, Petronilho F, Constantino L, Gelain DP, Andrades M, Ritter C, Moreira JC, Streck EL, Dal Pizzol F: Xanthine oxidase activity in patients with sepsis. Clin Biochem 2008, 41: 1186-1190. 10.1016/j.clinbiochem.2008.07.015
Cardelli P, Ferraironi M, Amodeo R, Tabacco F, De Blasi RA, Nicoletti M, Sessa R, Petrucca A, Costante A, Cipriani P: Evaluation of neutrophil CD64 expression and procalcitonin as useful markers in early diagnosis of sepsis. Int J Immunopathol Pharmacol 2008, 21: 43-49.
Gibot S, Cravoisy A, Levy B, Bene MC, Faure G, Bollaert PE: Soluble triggering receptor expressed on myeloid cells and the diagnosis of pneumonia. N Engl J Med 2004, 350: 451-458. 10.1056/NEJMoa031544
Gibot S, Kolopp-Sarda MN, Bene MC, Cravoisy A, Levy B, Faure GC, Bollaert PE: Plasma level of a triggering receptor expressed on myeloid cells-1: its diagnostic accuracy in patients with suspected sepsis. Ann Intern Med 2004, 141: 9-15.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
CP and JLV conceived the study. CP conducted the literature search. CP and JLV wrote the manuscript.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Pierrakos, C., Vincent, JL. Sepsis biomarkers: a review. Crit Care 14, R15 (2010). https://doi.org/10.1186/cc8872
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/cc8872