Abstract
Nanoparticles of molecularly imprinted polymers (MIPs) were prepared by precipitation polymerization method. Glucose was used as a template molecule. The impact of different process parameters on the preparation of nanoparticles was investigated in order to reach the maximum binding capacity of MIPs. Experimental data based on uniform design were analyzed using artificial neural network to find the optimal condition. The results showed that the binding ability of nanoparticles of MIPs prepared under optimum condition was much higher than that of the corresponding non-imprinted nanoparticles (NIPs). The findings also demonstrated high glucose selectivity of imprinted nanoparticles. The results exhibited that the particle size for MIP nanoparticles was about 557.6 nm, and the Brunauer-Emmett-Teller analysis also confirmed that the particle pores were mesopores and macropores around 40 nm and possessed higher volume, surface area, and uniform size compared to the corresponding NIPs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Molecularly imprinted polymers (MIPs) are smart and tailor-made polymers owing high selectivity for the predetermined template molecule. Selectivity of these materials originates from the synthetic procedure which is followed for their preparation. Indeed, molecular imprinting technique is a method of inducing receptor-like binding sites in synthetic polymers. Recently, these materials have attracted an increasing interest for many applications, most notably for pharmaceutical and analytical applications [1–4].
Conventional MIPs have been prepared in the form of bulk monolith. The polymers are then ground and sieved to obtain appropriate size particles with irregularly shape for further use. This procedure is time-consuming and yields only moderate amounts of useful product owing low capacity and poor site accessibility for the template molecules because the grinding process may be detrimental to some of the binding sites. In order to overcome the above shortcomings, recent efforts have been made to prepare MIPs with desired shape and suitable for wide applications. Since decreasing the size of the particles would have led to a larger increase in the surface area, it seems the ideal shape of MIPs for many applications may be the nanoparticles with regular size and shape.
In addition, successful applications of MIPs have been limited due to their low binding capacity. It is known that the binding capacity relies on an adequate selection of the process parameters that influence the success of imprinting process. Different researchers have shown that several factors including functional monomer [5–7], cross-linker [8–10], porogenic solvent [11], initiator [12], polymerization method [13, 14], type of solvent [15], time [16], and temperature [17, 18] of polymerization process are concerned with the imprinting process.
Optimization of such parameters has been traditionally carried out using a univariate method, i.e., changing each factor at a time over the investigated range while the others are held constant at a selected level (one-factor-at-a-time method). The main disadvantages of this approach are that it needs high number of experiments and it does not consider any possible interaction between the studied variables. To overcome this shortcoming, multivariate optimization methods are considered as promising techniques which use experimental design in conjunction with multivariate analysis techniques to optimize different processes [19–22]. Zhu et al. [19] investigated the influence of process parameters on the preparation of vinblastine-imprinted polymers. The optimal conditions were achieved by the regression model, and the results demonstrated that the performance of the imprinted polymer improved by optimizing the polymerization parameters. Experimental design and partial least square model have been employed in order to optimize the synthesis of bisphenol A selective MIPs [20]. The findings revealed that the photoinitiated polymers yield the highest specific binding capacity.
There are different methods to search for the optimization condition in the multidimensional space [19–22]. Simplex method is one of the most simple and general optimization methods [23]. In this method, to get the numerical value of the evaluation function for a defined system, response variables have to be measured or a mathematical model which predicts the properties of system has to be found and incorporated into the optimization process.
Artificial neural networks (ANNs) have been introduced to model various chemical processes owing non-linear relationships between variables [24, 25]. Radial basis function (RBF) neural network is one type of artificial neural networks. The theory of RBF has been addressed in detail in the literature [26].
Uniform design as an experimental design method presents the attractive advantages to evaluate multiple parameters and their interactions with significantly reduced number of experiments [27]. The uniform design principle is to replace the complete combination of experimental parameters by using relatively fewer experiment trials uniformly distributed within the parameter space.
This paper describes a novel approach for the preparation and optimization of nanoparticles of MIPs through simplex technique in conjunction with RBF in order to obtain MIPs with high binding capacity. Glucose was used as a template molecule, and imprinted particles were prepared by non-covalent imprinting method. The glucose-imprinted nanoparticles under optimized condition were also synthesized and characterized.
Methods
Materials
2,2-Azobisisobutyronitrile (AIBN) and hexadecane were provided from Kemikalieimport (Lyngby, Denmark) and Meryer Chemical Technology Co., Ltd. (Shenzhen, China), respectively. Phenol was purchased from Scharlau Chemie (Gillman, South Australia, Australia). All other materials were obtained from Merck (Darmstadt, Germany), purchased locally and used as received. MATLAB version 7.0 and MultiSimplx.1 were used for construction of RBF and simplex analysis, respectively.
Preparation of molecularly imprinted nanoparticles
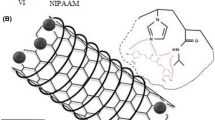
Nanoparticles of MIPs were prepared by precipitation polymerization. Briefly, 0.09 g of glucose was dissolved in the mixture of acetonitrile and dimethylformamide (DMF) (20/10) and then 4 mmol of methacrylic acid (MAA), 6 mmol of ethyleneglycol dimethacrylate (EGDMA), and 0.2 ml of hexadecane were added and stirred at 2 rpm for 24 h. The prepolymerization mixture was ultrasonicated for 1 min and then 30 mg of AIBN was added and sparged with nitrogen for 5 min. Polymerization was carried out for 24 h at 60°C in an oil bath. Nanoparticles were centrifuged (Sigma 3K30; Sigma-Aldrich Corp., St. Louis, MO, USA) at 20,000 rpm and washed two times with the mixture of water and acetonitril to remove unreacted monomers and free glucose molecules. The particles were lyophilized and stored for future experiments. The template was extracted by batch method with water at 2 rpm for 24 h. The extent recovery of glucose from MIPs was analyzed by spectrophotometery of collected extracts at the wavelength of 490 nm using the procedure reported elsewhere [28]. Scheme 1 illustrates the preparation process of nanoparticles of MIPs. Non-imprinted nanoparticles (NIPs) were prepared following the same procedure in the absence of the template in the polymerization mixture.
Experimental design
An experimental design approach based on uniform design was taken in which factors that have significant impact on the extent of imprinted glucose were considered. In this study, seven factors at six levels were included in an 18-trial setup. The selected factors were (1) the amount of the functional monomer (MAA), (2) the amount of the cross-linker (EGDMA), (3) the amount of initiator (AIBN), (4) the amount of progen (hexadecane), (5) the polymerization solvent (DMF), (6) time (t), and (7) temperature (T). The table U18(67) of uniform design was employed to arrange selected parameters (Table 1). The range of variables was decided after some preliminary work and literature data for MIP compositions.
Artificial neural network
RBF was used to build predictive artificial neural network model. The network consists of three layers, namely an input layer with seven inputs representing the selected parameters, one hidden layer in which the number of nodes was determined during training and testing, and one output layer with a single output node, A, corresponding to the glucose absorption in the extracted solution. A bias was used to calculate the net input of a neuron from all the neurons connected to it. The error function sum square error (SSE) was used as criterion for finalizing the learning process. It can be obtained from the following equation:
where t j is the experimental result, and out j indicates the output of RBF.
There are two adaptive parameters (the spread and the number of nodes in the hidden layer) to be adjusted in order to achieve the best predictive model. Training and testing of the network is a process of determination of the neural network's topology and optimization of its adjustable parameters, where we seek the minimum of an error surface in a multidimensional space. The experiments which are used for training and testing the network were selected randomly. In order to determine the optimum number of hidden nodes, a series of different topologies was used, in which the nodes were varied from 1 to 8. The RBF of the lowest error constructed in this manner was then used as the best model for optimization of nanoparticle preparation process.
Optimum condition for MIP preparation
To achieve the highest glucose recovery, simplex technique was employed. The extent of glucose absorption of extracted solution of MIPs prepared at predetermined condition by simplex technique was predicted by the optimized RBF and was used to search for optimum condition. In order to validate the empirical model, MIPs and the corresponding NIPs were prepared under optimum conditions and were evaluated for glucose recovery.
MIP characterization
Particle size
The size and zeta potential of the particles were determined by photon correlation spectroscopy (PCS) on a Malvern Instruments' Nano/Zeta (Malvern Instruments Ltd., Worcestershire, UK) in water.
Binding experiments
The binding capacities of the particles were evaluated by equilibrium binding experiments. The binding capacity of the imprinted nanoparticles was determined using 10 mg of imprinted and NIPs added into ten tubes with 2.5 ml of glucose solutions at concentrations ranging from 10 to 90 mg l−1 at room temperature for 20 h with shaking. The polymer particles were then separated by centrifugation at 20,000 rpm for 15 min. The concentration of free glucose in the supernatant was assayed by spectrophotometry. The amount of glucose bound to the MIPs was calculated by subtracting the amount of free substrate from the initial concentration.
The distribution coefficient K was utilized to evaluate the molecular selectivity of MIPs [21]. K is defined as follows:
where CP (mmol g−1) is the amount of adsorbed glucose on MIPs, and CS (mmol l−1) is the equilibrium concentration of substrates in the solution.
Selectivity of MIPs
Selectivity of the MIPs was determined by incubating 10 mg of MIPs and NIPs with 2.5 ml of different concentrations of fructose under similar condition used for glucose binding experiments.Two different selectivity factors were also calculated as indicated in Equations 3 and 4, the selectivity factor (α) as the ratio between the template and template analogue rebinding and specific selectivity factor (α′) which corresponds to the specific glucose and fructose binding:
Pore size distribution and surface area analysis
Pore size distribution and surface areas of the washed polymers were analyzed by Brunauer-Emmett-Teller (BET) analysis. Relevant information was obtained as follows: A plot of pore size versus incremental pore volume gave pore size distribution. A plot of pore size versus pore volume gave total pore volume. The surface areas and total pore volumes of the polymers were also obtained.
Results and discussion
Data analysis using ANN and developing predictive model
Optimization of nanoparticles of MIP preparation condition to obtain a global optimum via experimental approach would be a significant and time-consuming challenge. Therefore, alternative approach is to model process using limited experimental points representing all variable space. RBF with logistic sigmoidal function was used to model the process. Since there are no theoretical principles for choosing the proper network topology and spread, several different structures were tested. Neural networks were trained using different numbers of hidden nodes (1 to 8) and spread (1 to 10). At the start of training run, all weights and all biases were initialized with random values. The data partitioning as training and test set was done to avoid over-training and over parameterization. The lowest testing SSE is obtained with 7 hidden neurons and at a spread of 1 (Figure 1). The optimized RBF resulted in the least value for the test set SSE, i.e., Etst = 0.233, and the corresponding SSE value for training data was Etrn = 0.134. These results indicate high predictive capability of developed model for the preparation process of nanoparticles of MIPs.
Optimization of experimental conditions
The aim of nanoparticle preparation process was to obtain MIPs with high recovery which in turn leads to achieve high selectivity and binding capacity while keeping non-selective interactions as low as possible. The simplex technique was used to optimize the input space of RBF model with the objective of maximizing glucose recovery. Since when all the adjustable parameters had been optimized, the neural network showed high ability of generalization; therefore, the optimized neural network model put forward to predict glucose recovery at predetermined condition by the simplex method. Based on the simplex method, the optimal conditions were found to be as follows: 0.5 ml of MAA, 1.7 ml of EGDMA, and 0.15 ml of hexadecane, 40 mg of AIBN, and 9 ml of DMF, at 65°C for 4 h which correspond to the maximum value of A (A = 0.568).
In order to verify the accuracy of the simplex method in conjunction with RBF in optimizing the MIP preparation process parameters, the glucose imprinted polymer was prepared under the optimal conditions. Glucose recovery for optimized MIPs was obtained to be 0.524 (n = 4, RSD 6.94%). As the results show, the prediction error in optimum recovery by RBF was 8.40%. From the results obtained, it can be concluded that the measured recovery was in good agreement with that predicted by the model which subsequently confirms the significance of the simplex method and RBF to optimize and represent this system, respectively. This can be explained by this fact that the neural network method scanned the whole range of experimental parameters intensively.
Influence of polymerization conditions on adsorption properties of MIP
For the better view of given results and parameter interaction, the graphs of the response surface were drawn (Figure 2). As a matter of fact, the response surface plots can visualize the effect and interaction between dependent variables. Each surface response plot represents a combination of two test variables with the other ones maintained at their optimum levels. Several important conclusions can be drawn from these graphs. Figure 2a shows that a maximum glucose recovery can be obtained at 75°C using 30 mg of AIBN which is far from the optimum value obtained by the simplex method. This can be an evidence for the existence of interaction between temperature and AIBN with the other variables. A similar behavior was observed for time (maximum 4 h) and AIBN (30 mg), too (Figure 2g). The maximum response for other variables was in agreement with the results obtained by the simplex method which indicates lack of interaction between these variables.
Influence of the variation of two input factors on glucose recovery. The remaining input factors were kept constant at optimal level. 3D plot of glucose recovery predicted by the ANN model for (a) Temperature and AIBN, (b) DMF and Time, (c) AIBN and Hexadecane, (d) Hexadecane and DMF, (e) MAA and DMF, (f) Temperature and Hexadecane, (h) Temperature and MAA, (g) Time and AIBN, (i) Time and EGDMA, (j) Time and Hexadecane, (l) Time and MAA, (m) AIBN and DMF, (n) EGDMA and MAA, (o) EGDMA and DMF, (r) Temperature and Time, (s) Temperature and DMF, (q) EGDMA and AIBN, (w) EGDMA and Hexadecane, (y) Hexadecane and MAA, (z) Temperature and EGDMA.
The response surface plots can also be employed to show the extent of impact of each variable on the glucose imprinting. As Figure 2 illustrates among investigated variable time, temperature and the amount of AIBN show the most significant impact on imprinting performance. This finding is consistent with the results reported in the literature [29]. The effect of temperature is not surprising; it arises from the negative effect of high temperature on the complex formation between the template and functional monomers. In case of AIBN amount, it can be related to the heat generated during the polymerization reaction. Thus, it can be postulated that large concentrations of initiator added to the monomer mixture should increase the polymerization rate and increase the heat of reaction. The increased temperature would disrupt the complex formed and reduce the affinity and selectivity of MIPs. The effect of time on imprinting performance can be related to the formation of large particles which reduces the surface area and subsequently decreases the available imprinted sites for the template molecule.
Optimized MIP nanoparticle characterization
The particle size and pore distribution
The hydrodynamic diameter and zeta potential of nanoparticles were determined by PCS (Figure 3). As it can be seen, almost narrow particle size distribution with a z-average about 557.6 nm (polydispersity index = 0.465) in aqueous dispersant was obtained, and the zeta potential was determined to be -11.2 mV.
The pore volume and pore diameter are considered as two of the most important properties of MIPs. Typically, pore sizes have been separated into three size categories, micropores (<2 nm), mesopores (2 to 50 nm), and macropores (over 50 nm). Table 2 shows the surface areas, total pore volumes, and average pore sizes (diameter) for the MIP nanoparticles along with their corresponding NIP nanoparticles.
In terms of the surface area, the MIPs produced a surface area of 78.59 m2 g− 1, whereas NIP produced a surface area of 0.8859 m2 g− 1. From this data, it can be concluded that imprinting template molecule significantly can improve the surface area. Additionally, Barrett-Joyner-Halenda calculation for MIP nanoparticles is indicative of its better formed and uniform pore distribution compared to the NIP nanoparticles. It would be expected that increasing the size of the pores would have led to a larger decrease in the surface area. The BET plot shows that MIP nanoparticles possess a relatively large population of pores in the micro- and mesoporous range having size around 40 nm, while in the case of NIPs, the isotherm corresponds to the type III pores, which is related to the nonporous materials with weak interaction.
Binding property and selectivity of MIP nanoparticles
The binding properties of MIPs were measured with initial concentrations of glucose ranging from 20 to 90 mg l−1. MIPs exhibited a high distribution coefficient (0.19) for glucose. This indicates that the higher recognizing cavities have been created in the resultant MIP nanoparticles.
The substrate selectivity of MIPs was also investigated using glucose and its structurally similar compound, fructose, with the uniform molecular weight and the chemical structures except for the different replacement site of hydroxy groups. The αtemplate/analogue and α′template/analogue values for MIPs prepared at optimized condition were calculated to be 2.025 and 4.66, respectively, which are indicative of high selectivity of prepared MIPs.
Binding profile of MIPs and the corresponding NIPs in the presence of glucose and its analogue, fructose, at concentration ranges from 10 to 90 mg l−1 is shown in Figure 4. Curves (a) and (b) in Figure 4 show the binding profile of glucose on MIPs and NIPs, respectively. It is obvious that the binding amount of template molecules to MIP nanoparticles were much higher than that of NIPs. Curves (c) and (d) in Figure 4 show the adsorption isotherms of fructose to MIP and NIP nanoparticles, respectively. The binding ability of this compound and glucose to MIPs is speculated to be based on the same interaction due to their similar chemical structure. When comparing curves (a) with curve (c), it is obvious that the binding amounts of fructose to MIP nanoparticles are much lower than that of glucose. This observation can be explained by the fact that since the structure of the analogue molecule is not complementary to the imprinted cavities in the MIPs produced by the glucose molecular imprinting, the ability of fructose interaction with the binding sites is expected to be weaker than that of glucose which leads to low binding capacity. Therefore, the higher binding of glucose MIPs may be attributed to the shape selective fitting of glucose into complementary cavities created on the MIP nanoparticles during the imprinting procedure.
Conclusion
In the present study, nanoparticles of MIPs with narrow particle size distribution were prepared by precipitation polymerization. Simplex technique in conjunction with radial basis function neural network was demonstrated to be an effective and reliable approach in finding the optimal conditions for the preparation of MIPs for glucose. The optimal conditions were found to be as follows: 0.5 ml of MAA, 1.7 ml of EGDMA, 0.15 ml of hexadecane, 40 mg of AIBN, and 9 ml of DMF, at 65°C for 4 h. The results showed that among the studied variables, time, temperature, and the amount of initiator showed the most significant impact on MIP nanoparticle performance. Comparison of experimental and predicted recoveries by RBF revealed that the results were closely in agreement with each other, indicating high capability of designed RBF to model process and subsequently simplex technique usefulness in optimizing MIP nanoparticle preparation parameters. The binding ability was also evaluated, and the findings demonstrated high glucose binding and selectivity of imprinted particles compared to NIPs. BET analysis also confirmed that the particle pores were mesopores and macropores around 40 nm and possessed higher volume, surface area, and uniform size compared to the NIPs. Hence, it is worthy to employ simplex technique in conjunction with RBF to optimize the parameters of polymerization process and obtain high efficiency MIPs for different purposes.
References
Allender CJ, Richardson C, Woodhouse B, Heard CM, Brain KR: Pharmaceutical applications for molecularly imprinted polymers. Int. J. Pharm 2000,195(1–2):39–43.
Lachová M, Lehotay J, Cizmárik J: Applications of molecularly imprinted polymers in analytical and pharmaceutical chemistry. Ceska Slov. Farm. 2007,56(4):159–164.
Alvarez-Lorenzo C, Concheiro A: Molecularly imprinted materials as advanced excipients for drug delivery systems. Biotechnol. Annu. Rev. 2006, 12: 225–268.
Yoshimi Y, Narimatsu A, Nakayama K, Sekine S, Hattori K, Sakai K: Development of an enzyme-free glucose sensor using the gate effect of a molecularly imprinted polymer. J. Artif. Organs. 2009,12(4):264–270. 10.1007/s10047-009-0473-4
Sellergren B, Lepisto M, Mosbach K: Highly enantioselective and substrateselective polymers obtained by molecular imprinting utilizing noncovalent interactions. NMR and chromatographic studies on the nature of recognition. J. Am. Chem. Soc. 1988, 110: 5853–5860.
Haginaka J, Sanbe H: Uniformly sized molecularly imprinted polymer for (S)-naproxen retention and molecular recognition properties in aqueous mobile phase. J. Chromatogr. A. 2001, 913: 141–146. 10.1016/S0021-9673(01)00555-6
Karlsson JG, Karlsson B, Andersson LI, Nichollsa IA: The roles of template complexation and ligand binding conditions on recognition in bupivacaine molecularly imprinted polymers. Analyst. 2004, 129: 456–462. 10.1039/b316716j
Sibrian-Vazquez M, Spivak DA: Characterization of molecularly imprinted polymers employing crosslinkers with non-symmetric polymerizable groups. J. Polym. Sci. A: Polym. Chem. 2004, 42: 3668–3675. 10.1002/pola.20186
Syu MJ, Nian YM, Chang YS, Lin XZ, Shiesh SC, Chou TC: Ionic effect on the binding of bilirubin to the imprinted poly(methacrylic acid-co-ethylene glycol dimethylacrylate). J. Chromatogr. A. 2006, 1122: 54–62. 10.1016/j.chroma.2006.04.024
Cummins W, Duggan P, McLoughlin P: Systematic cross-selectivity study of the factors influencing template receptor interactions in molecularly imprinted nitrogen heterocycles. Biosens. Bioelectron. 2006, 22: 372–380. 10.1016/j.bios.2006.05.003
Perez-Moral N, Mayes AG: Non-covalent imprinting in the shell of core-shell nanoparticles. Langmuir. 2004, 20: 3775–3779. 10.1021/la0356755
Vaughan AD, Sizemore SP, Byrne ME: Enhancing molecularly imprinted polymer binding properties via controlled/living radical polymerization and reaction analysis. Polymer. 2007, 48: 74–81. 10.1016/j.polymer.2006.11.013
O'Shannessy DJ, Ekberg B, Mosbach K: Molecular imprinting of amino-acid derivatives at low-temperature (0-degrees-C) using photolytic homolysis of azobisnitriles. Anal. Biochem. 1989, 177: 144–149. 10.1016/0003-2697(89)90029-8
Pérez-Moral N, Mayes AG: Comparative study of imprinted polymer particles prepared by different polymerisation methods. Anal. Chim. Acta. 2004, 504: 15–21. 10.1016/S0003-2670(03)00533-6
Nomachi M, Kubo T, Hosoya K, Kaya K: Solvent effects in the preparation of molecularly imprinted polymers for melatonin using N-propionyl-5-methoxytryptamine as the pseudo template. Anal. Bioanal. Chem. 2006, 384: 1291–1296. 10.1007/s00216-006-0310-z
Piletsky SA, Mijangos I, Guerreiro A, Piletska EV, Chianella I, Karim K, Turner APF: Polymer cookery: influence of polymerization time and different initiation conditions on performance of molecularly imprinted polymers. Macromolecules 2005, 38: 1410–1414. 10.1021/ma048021r
Hosoya K, Iwakoshi Y, Yoshizako K, Kimata K, Tanaka N, Takehira H, Haginaka J: An unexpected molecular imprinting effect for a polyaromatic hydrocarbon, anthracene, using uniform size ethylene dimethacrylate particles. J. High Resolut. Chromatogr. 1999, 22: 256–260. 10.1002/(SICI)1521-4168(19990501)22:5<256::AID-JHRC256>3.0.CO;2-B
Lu Y, Li CX, Wang XD, Sun PC, Xing XH: Influence of polymerization temperature on the molecular recognition of imprinted polymers. J. Chromatogr. B. 2004, 804: 53–59. 10.1016/j.jchromb.2003.10.013
Zhu QH, He JF, Feng JY: Optimization of the process parameters of synthesis of vinblastine imprinted polymer. Eur. Polym. J. 2007, 43: 4043–4051. 10.1016/j.eurpolymj.2007.06.036
Villoslada FN, Vicente BS, Moreno-Bondi MC: Application of multivariate analysis to the screening of molecularly imprinted polymers for bisphenol A. Anal. Chim. Acta. 2004, 504: 149–162. 10.1016/S0003-2670(03)00766-9
Davies MP, Biasi VD, Perrett D: Approaches to the rational design of molecularly imprinted polymers. Anal. Chim. Acta. 2004, 504: 7–14. 10.1016/S0003-2670(03)00812-2
Rossi C, Haupt K: Application of the Doehlert experimental design to molecularly imprinted polymers: surface response optimization of specific template recognition as a function of the type and degree of cross-linking. Anal. Bioanal. Chem. 2007, 389: 455–460. 10.1007/s00216-007-1484-8
Bostyn S, Cagnon B, Fauduet H: Optimization by the simplex method of the separation of phenolic acids by high-performance liquid chromatography in wastewater olive and sugar beet vinasse. Talanta 2009,80(1):1–7. 10.1016/j.talanta.2009.06.040
Agatonovic-Kustrin S, Beresford R: Basic concepts of artificial neural network (ANN) modeling and its application in pharmaceutical research. J. Pharm. Biomed. Anal. 2000,22(5):717–27. 10.1016/S0731-7085(99)00272-1
Asadi H, Rostamizadeh K, Salari D, Hamidi M: Preparation of biodegradable nanoparticles of tri-block PLA–PEG–PLA copolymer and determination of factors controlling the particle size using artificial neural network. J. Microencapsul. 2011,28(5):406–416. 10.3109/02652048.2011.576784
Matera F: Radial basis function neural network. Subst. Use Misuse. 1998,33(2):317–334. 10.3109/10826089809115868
Liu RS, Tang YJ: Quantitative analysis for the effect of plant oil and fatty acid on Tuber melanosporum culture by uniform design combined with partial least squares regression. Appl. Microbiol. Biotechnol. 2010,87(5):1689–1697. 10.1007/s00253-010-2602-7
Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F: Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956,28(3):350–356. 10.1021/ac60111a017
Mijangos I, Navarro-Villoslada F, Guerreiro A, Piletska E, Chianella I, Karim K, Turner A, Piletsky S: Influence of initiator and different polymerisation conditions on performance of molecularly imprinted polymers. Biosens. Bioelectron. 2006, 22: 381–387. 10.1016/j.bios.2006.05.012
Acknowledgments
The authors are most grateful for the continuing financial support for this research project by Zanjan Pharmaceutical Nanotechnology Research Center, Research Center for Pharmaceutical Nanotechnology of Tabriz University of Medical Sciences, and Institute for Advanced Studies in Basic Sciences.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
KR participated in the synthesis and characterization of nanoparticles and drafted the manuscript. HA was responsible for the optimization session and analysis of the data. CP carried out the experiments. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Rostamizadeh, K., Abdollahi, H. & Parsajoo, C. Synthesis, optimization, and characterization of molecularly imprinted nanoparticles. Int Nano Lett 3, 20 (2013). https://doi.org/10.1186/2228-5326-3-20
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/2228-5326-3-20