Abstract
The present study has evaluated the association of growth differentiation factor9 (GDF9) and bone morphogenetic protein15 (BMP15) mRNA expression in cumulus-oocyte complexes (COCs) of buffalo ovary during in vitro maturation (IVM). GDF9 and BMP15 are expressed specifically in mammalian oocytes and also participate in cumulus-oocyte crosstalk. Quantitative real-time polymerase chain reaction (qRT-PCR) technique was applied to investigate the relative abundance (RA) of GDF9 and BMP15 mRNA transcripts throughout the IVM process. Relative mRNA expression pattern of these specific genes were assessed in oocytes and cumulus cells at 0, 6, 12 and 24 h of in vitro culture. Our results revealed that RA of GDF9 during different hours of IVM showed significant reduction between 0 h and 24 h of maturation in oocytes and BMP15 transcript increased significantly (P<0.05) between 6 h and 12 h and decreased again between 12 h and 24. In cumulus cells, GDF9 remained stable during IVM upto 12 h of maturation and decreased significantly between 12 h and 24 h of maturation. Conversely, significant reduction of BMP15 was observed between 0 h and 6 h, stayed stable upto 12 h and became undetectable at 24 h of maturation. In conclusion, these two genes were differentially expressed during the period of oocyte maturation process and notably, BMP15 expression pattern is associated specifically with the period of cumulus cell expansion.
Similar content being viewed by others
Introduction
Growth factors synthesized from mammalian oocytes popularly known as oocyte secreted factors (OSFs) play numerous role in ovarian folliculogenesis. In particular, a growing body of evidence in recent years indicated that two famous members of Transforming growth factor-β superfamily, GDF9 and BMP15 have been involved in the control of ovulation rate in mammals (.Moore et al. 2004). They act synergistically to affect development of the cumulus–oocyte complexes in mice (Yan et al. 2001). GDF9 plays critical roles in granulosa and theca cells growth, as well as in differentiation and maturation of the oocyte (Hreinsson et al. 2002). Further, the level of GDF9 in human follicular fluid has been found significantly correlated with the nuclear maturation of the oocytes and embryo quality (Gode et al. 2011). Moreover, BMP15 has also been thought to be involved in oocyte maturation and follicular development alone or along with the related protein, GDF9. Both the proteins are progressively expressed by the oocytes of growing follicles throughout folliculogenesis (Dube et al. 1998; Laitinen et al. 1998). This stage specific expression through intra-follicular cascade at the correct time is vital for follicular growth in order to reach the ovulatory phase (Campbell 2009). In this regard, the expression of GDF9 and BMP15 has been studied extensively in follicular compartment cells in the ovaries of various species, while poorly explored in the COCs within culture systems.
The domestic Asian water buffalo (Bubalus bubalis) is a multipurpose livestock animal. It is of particular important production animal due to the source of high quality animal protein in developing countries (Zicarelli 1994). IVM is an essential technology under assisted reproductive technologies (ARTs), which enables oocytes to achieve maturation and acquire the competence for subsequent embryonic division leading to blastocyst formation (Lonergan et al. 2003; Somfai et al. 2011). This technique is valuable not only because they allow the production of large numbers of oocytes, but also because they provide a valuable in vitro model to study the gene expression at different maturational stages of oocytes. During the maturation process, oocytes accumulate large amount of maternal mRNAs to dictate the developmental competence of oocytes (Watson 2007). Quantification of RNA developmentally important oocyte secreted factors in oocytes during maturation might be valuable when setting up IVM medium that lack the normal hormonal interplay found in vivo. Considering the importance of buffalo and the necessity of designing potent ART protocols for this species, the current study aimed for analysing expression pattern of two oogenesis specific genes during maturation. For this purpose, we used quantitative real-time PCR (qRT-PCR) technique for mRNA expression of GDF9 and BMP15 genes. Accordingly, the current work will provide important leads for understanding the mRNA transcriptional level of GDF9 and BMP15 genes in oocytes during IVM in case of buffalo.
Materials and methods
All chemicals and culture media were obtained from Sigma Chemical Co. (USA) unless otherwise stated. Disposable plastic wares used were purchased from Nunc, Denmark.
Collection of cumulus oocyte complexes and IVM
Buffalo ovaries were collected from an abattoir and transported in thermos containing pre-warmed (37°C) normal saline to the laboratory within 2-3 h. In the laboratory, ovaries were washed five times in modified Dulbecco’s phosphate buffered saline (mDPBS) containing gentamicin. Oocytes were aspirated with a 10 cc syringe using 20 gauge needle into 50 ml conical tubes from follicles (>3 mm in diameter) of buffalo ovaries. The aspirated COCs were graded based on morphological appearance of the cumulus cell investments and homogeneity of ooplasm under stereo zoom microscope as described by Nandi et al. (1998) and the different grades are, Grade A (Excellent): COCs with more than 4 layers of compact cumulus cells with evenly granular homogenous ooplasm; Grade B (Good): COCs with 2–4 layers of compact cumulus cells with evenly granular homogenous ooplasm; Grade C (Poor): COCs with one layer of cumulus cells with irregular dark ooplasm; Grade D (Poor): Naked oocytes with irregular dark ooplasm. The aspirated oocytes of grade A and B alone were chosen for in vitro maturation studies. Average of 20–30 oocytes were cultured in maturation media and each experiment was replicated on eight separate occasions. From 851 ovaries, total of 749 oocytes were used for this study. Maturation medium consisted of TCM-199 supplemented with 10% FBS, 10 IU/ml LH, 0.5 μg/ml FSH, 1 μg/ml estradiol-17β, 100 IU/ml penicillin and 100 μg/ml streptomycin. The oocytes were subjected to mature in the maturation medium in 35×10 mm petridishes for 24 h in a CO2 incubator maintained at 39°C, 5% CO2 with maximum relative humidity.
Assessment of oocyte maturation
The maturation of oocytes was evaluated under stereomicroscope based on the degree of cumulus expansion (Kobayashi et al. 1994) and further evaluated by identifying the first polar body in the perivitelline space when the oocytes were denuded after maturation. For this purpose, a proportion of COCs were recovered from the culture and transferred to 1.5 ml tube containing 400 μl of 0.1% trypsin-EDTA solution. Then vortex-agitated for 1 min to remove cumulus cells and thus denuded oocytes were visualised for extrusion of polar body.
Collection of samples for expression analysis
Immature oocytes (0 h) were treated with 0.1 per cent trypsin-EDTA for 2–3 min and mechanically denuded of surrounding cumulus cells by repeated pipetting. The cumulus-free oocytes were observed under stereo zoom microscope to ensure that they were free of cumulus cells. The oocytes were washed twice in PBS and transferred to a fresh 1.5 ml micro centrifuge tube with minimum volume of PBS and then frozen at −80°C. After the removal of cumulus free oocytes, the remaining cumulus cells in PBS were transferred to micro centrifuge tube and centrifuged at 300 g for 2 min according to Caixeta et al. (2009). The supernatant was removed and the cell pellet was resuspended with minimum volume of PBS and stored at −80°C for RNA isolation. During different hours of IVM (6, 12 and 24 h), the cumulus denuded from matured oocytes were collected as mentioned above and the oocytes with evenly granular ooplasm after cumulus removal with a visible polar body in the peri-vitelline space were also stored at −80°C for RNA isolation.
RNA isolation and cDNA preparation
Total RNA was extracted from pools of immature (IM 75), in vitro matured oocytes (MO 50), cumulus cells (cells removed from 50–75 COCs) using RNeasy Plus Mini Kit (Qiagen, USA) and eluted in 30 μl RNase free-water. The purity and concentration of isolated RNA was determined by ND1000 spectrophotometer (Nanodrop technologies, USA) and purity was assessed using the A260/A280 nm ratio with expected values between 1.8 and 2.0. The cDNA synthesis was carried out with 6 μl of RNA using Superscript III first-strand synthesis system (Invitrogen, USA) and random hexamer primer in a final reaction volume of 25 μl. The cDNA synthesis reactions were carried at 25°C for 10 min for annealing, 50°C for 60 min for extension, followed by heat inactivation of the enzyme at 85°C for 5 min. Reactions without Reverse transcriptase served as controls. After termination of cDNA synthesis, each RT reactions were diluted with nuclease-free water to a final volume of 50 μl and stored at −80°C till further use.
Primer designing and validation
Oligonucleotide primers for qRT-PCR analysis were designed from Bubalus bubalis complete mRNA sequence of GDF-9 (GenBank: FJ529501.2), BMP-15 (GenBank: EF375880.1) and GAPDH (GenBank: GU324291.1). Oligo Analyzer (1.1.2 Version) software was used to determine the Tm, GC per cent, primer loops, primer dimers and primer-primer compatibility. The FastPCR Professional (6.2.45 beta 4 version) has been used for Real time PCR primer designing and the oligonucleotide primers were synthesized from Sigma-Aldrich (USA). The designed primers were optimized prior to quantification experiments using polymerase chain reaction. For these genes, the expected sizes of the products were confirmed by gel electrophoresis on a 2% agarose gel. The primer sequences, expected fragment size of amplified products and genbank accession numbers are shown in Table 1.
Real-time polymerase chain reaction
Real-time PCR was performed to quantify the mRNA transcripts of GDF-9, BMP-15 in oocytes and cumulus cells using SYBR Green JumpStart Taq ReadyMix kit from Sigma-Aldrich, USA. Each cDNA sample was analysed in duplicate using real-time thermal cycler (Applied Biosystems, USA, ABI 7500). A non template control (NTC) was prepared using DEPC water without cDNA. Reactions were performed in 20 μl volumes and loaded in the real-time qPCR 96-well plate (10 μl/well) and the plate was centrifuged in cooling (4°C) plate at 560 rpm for 2 min to remove the air bubbles and kept for reaction in real time thermal cycler.
The PCR cyclic conditions were 95°C for 10 min followed by 40 cycles of denaturing at 95°C for 15 seconds and then annealing (60°C for GDF-9, 56°C for BMP-15, 58°C for GAPDH) for 1 min. The standard curve was obtained for GAPDH, GDF9 and BMP15 genes using 10-fold dilutions of the sample and resultant slope of the curve was utilized for the calculation of the quantitative expression of both the target genes. The Ct values were recorded for both the target (GDF-9, BMP-15) and endogenous control genes (GAPDH). The data were accepted only if the NTC had no amplification.
Interpretation of qRT-PCR
Data were quantified using the method of relative quantification in qPCR (Pfaffl 2001). The threshold cycle value of reference gene was used to normalize the target gene signals in each sample. The amount of target transcripts relative to the calibrator was calculated by subtracting the Ct value of sample reference gene from the Ct value of the sample target gene. All samples were run in duplicate and the mean value of each duplicate was used for all further calculations. The 40-ΔCt value gives the relative quantification of the transcripts.
Statistical analysis
Statistical analysis of the data was done as per the standard procedures of Snedecor and Cochran (1994). The gene expression status of all the genes analyzed was subjected to ANOVA using a computer-aided statistical software package SPSS 16.0 for windows. All the results are expressed as the mean ± SE and statistical significance was accepted for P < 0.05.
Results
In vitro maturation of buffalo oocytes and cumulus expansion
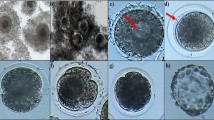
Total of 692 culturable grade oocytes were collected in a batch of experiments and 20–25 oocytes were cultured in 100 μl medium at each time-point to observe the progress of IVM. The COCs morphological changes were found at different time periods of culture (Figure 1). Maturation is assessed based on cumulus expansion and polarbody extrusion. Cumulus expansion was markedly increased at 12 h after IVM cultivation and continuously increased up to 24 h. When preparing samples for expression analysis at different maturation stages, special attention was paid to selecting COCs with a uniform ooplasm and appropriate cumulus expansion to ensure an accurate status of oocytes and cumulus cells.
Relative expression of GDF9 and BMP15 in buffalo oocytes during IVM
The mRNA expression of GDF9 and BMP15 in oocytes during different hours of IVM showed differential changes (Figure 2A). Specifically, the RA of GDF9 mRNA gradually declined throughout the time period upto 24 h of maturation and showed significant (P<0.05) variation in the RA of GDF9 transcripts in cumulus free oocytes at between 0 h and 24 h of maturation. A significant (P<0.05) variation in the RA of BMP15 transcript was observed in cumulus free oocytes of different maturation times. It increased between 6 h and 12 h of maturation and reduced between 12 h and 24 h.
Relative expression of GDF9 and BMP15 in buffalo COCs during IVM. (A) Relative expression of GDF9 and BMP15 mRNA transcripts from oocytes. (B) Relative expression of GDF9 and BMP15 mRNA transcripts from cumulus cells. Data are presented as (mean ± SE). For each gene (GDF9 or BMP15), bars with different superscripts show significant difference (P < 0.05).
Relative expression of GDF9 and BMP15 in buffalo cumulus cells during IVM
The differential expression pattern of GDF9 and BMP15 mRNA transcripts in cumulus cells during different hours of IVM were presented in Figure 2B. The RA of GDF9 remained stable upto 12 h of maturation, but decreased significantly at 24 h of maturation. However, BMP15 showed a significant reduction in the RA between 0 h and 6 h, stayed stable upto 12 h and become undetectable at 24 h of maturation.
Discussion
Ovarian folliculogenesis is a complex process depends upon numerous growth factors that regulate the growth and differentiation of oocytes, granulosa cells and theca cells. Oocyte-secreted GDF9 and BMP15 play essential roles during follicle maturation through actions on granulosa cells. GDF9 and BMP15 expression are less well characterized especially during IVM of COCs. Oocyte maturation is one of the important stages for the successful production of embryos in vitro. Moreover, it is well known that oocyte developmental potential is a reflection of proper maturation. So understanding the molecular mechanisms regulated during maturation is of great importance to optimize the IVM conditions. Since GDF9 and BMP15 are considered as important oocyte maturation factors during folliculogenesis (Gilchrist et al. 2004), this study investigated the potential relationship between GDF9 and BMP15 mRNA expression from in vitro matured oocytes and cumulus cells during the course of in vitro maturation.
In the present study significant difference in the RA of GDF9 transcripts in cumulus free oocytes, which showed gradual decline during IVM between 0h and 24 h of maturation. This is in agreement with earlier findings that suggested the GDF9 expression to remain stable in oocyte during IVM (Prochazka et al. 2004), yet showed gradual decline during oocyte maturation (Li et al. 2008). Zhu et al. (2008) studied the temporal and spatial expression patterns of GDF9 gene in porcine COCs throughout in vitro maturation and reported that GDF9 highly transcribed in oocytes from fresh COCs, whereas expression was gradually decreased during IVM. Relative abundance of BMP15 from denuded oocytes during different hours of IVM increased significantly (P<0.05) between 6 h and 12 h and decreased again between 12 h and 24. This increase in BMP15 transcript may be correlated with the cumulus expansion after 12 h of maturation in vitro. Similar to our finding, Li et al. (2008) observed a specific increase in porcine BMP15 transcript at 18 h of cumulus expansion during maturation. Zhu et al. (2008) reported that BMP15 was highly transcribed in oocytes from fresh COCs and the expression gradually declined during IVM.
It was also found that GDF9 RA in cumulus cells remained stable during IVM upto 12 h of maturation and decreased significantly between 12 and 24 h of maturation. This is in accordance with earlier studies, where RA of GDF9 mRNA was remained stable during the first 16 h of culture and significantly reduced during later stages of culture period in cumulus cells (Prochazka et al. 2004). This drastic change in the expression pattern of GDF9 during later stages of IVM could specify any probable role of this gene in selection of dominant follicle, which warrants further investigation. Whereas, BMP15 transcript was significantly reduced between 0 h and 6 h, stayed stable upto 12 h and undetectable at 24 h of maturation. Likewise, Zhu et al. (2008) reported undetectable BMP15 expression pattern in cumulus cells during maturation. The absence of RA might be related to the degradation of transcripts after maturation of oocytes.
Conclusion
To conclude, during maturation of COCs in vitro, GDF9 mRNA showed significant reduction between 0 h and 24 h of maturation in oocytes. On the other hand, the RA of BMP15 varied significantly (P<0.05) during IVM; It showed gradual increase between 6 h and 12 h and decreased again between 12 h and 24. However in cumulus cells, GDF9 remained stable during IVM upto 12 h of maturation and decreased significantly between 12 h and 24 h of maturation. In case of BMP15 significant reduction was observed between 0 h and 6 h, stayed stable upto 12 h and become undetectable at 24 h of maturation. In future, further characterization of these genes from in vivo matured buffalo COCs will help in better understanding of the GDF9 and BMP15 role in oocyte maturation of this species.
Abbreviations
- GDF9:
-
Growth differentiation factor9
- BMP15:
-
Bone morphogenetic protein15
- mRNA:
-
Messenger Ribonucleic acid
- COCs:
-
Cumulus-oocyte complexes
- IVM:
-
In vitro maturation
- qRT-PCR:
-
Quantitative real-time PCR
- RA:
-
Relative abundance.
References
Caixeta ES, Ripamonte P, Franco MM, Junior JB, Dode MAN: Effect of follicle size on mRNA expression in cumulus cells and oocytes of Bos indicus : an approach to identify marker genes for developmental competence. Reprod Fertil Dev 2009, 21: 655-664. 10.1071/RD08201
Campbell BK: The endocrine and local control of ovarian follicle development in the ewe. Anim Reprod 2009, 6: 159-171.
Dube JL, Wang P, Elvin J, Lyons KM, Celeste AJ, Matzuk MM: The bone morphogenetic protein-15 gene is X-Linked and expressed in oocytes. Mol Endocrinol 1998, 12: 1809-1817. 10.1210/me.12.12.1809
Gilchrist RB, Ritter LJ, Armstrong DT: Oocyte-somatic cell interactions during follicle development in mammals. Anim Reprod Sci 2004, 82–83: 431-446.
Gode F, Gulekli B, Dogan E, Korhan P, Dogan S, Bige O, Cimrin D, Atabey N: Influence of follicular fluid GDF9 and BMP15 on embryo quality. Fertil Steril 2011, 95: 2274-2278. 10.1016/j.fertnstert.2011.03.045
Hreinsson J, Scott J, Rasmussen C, Swahn M, Hsueh A, Hovatta O: Growth differentiation factor-9 promotes the growth, development, and survival of human ovarian follicles in organ culture. J Clin Endocrinol Metab 2002, 87(1):316-321. 10.1210/jc.87.1.316
Kobayashi K, Yamashita S, Hoshi H: Influence of EGF and TGF-α on in vitro maturation of cumulus cell enclosed bovine oocytes in a defined medium. J Reprod Fertil 1994, 100: 439-446. 10.1530/jrf.0.1000439
Laitinen M, Vuojolainen K, Jaatinen R, Ketola I, Aaltonen J, Lehtonen E, Heikinheimo M, Ritvos O: A novel growth differentiation factor-9 (GDF-9) related factor is co-expressed with GDF-9 in mouse oocytes during folliculogenesis. Mech Dev 1998, 78: 135-140. 10.1016/S0925-4773(98)00161-0
Li HK, Kuo TK, Yang HS, Chen LR, Li SSL, Huang HW: Differential gene expression of bone morphogenetic protein-15 and growth differentiation factor-9 during in vitro maturation of porcine oocytes and early embryos. Anim Reprod Sci 2008, 103: 312-322. 10.1016/j.anireprosci.2006.12.017
Lonergan P, Gutierrez-Adan A, Rizos D, Pintado B, de la Fuente J, Boland MP: Relative messenger RNA abundance in bovine oocytes collected in vitro or in vivo before and 20 hr after the pre-ovulatory luteinizing hormone surge. Mol Reprod Dev 2003, 66: 297-305. 10.1002/mrd.10357
Moore RK, Erickson GF, Shimasaki S: Are BMP-15 and GDF-9 primary determinants of ovulation quota in mammals. Trends Endocrinol Metab 2004, 15: 356-361.
Nandi S, Chauhan MS, Palta P: Influence of cumulus cells and sperm concentration on cleavage rate and subsequent embryonic development of buffalo ( Bubalus bubalis ) oocytes matured and fertilized in vitro . Theriogenology 1998, 50: 1251-1262. 10.1016/S0093-691X(98)00224-6
Pfaffl MW: A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001, 29: e45. 10.1093/nar/29.9.e45
Prochazka R, Nemcova L, Nagyova E, Kanka J: Expression of growth differentiation factor 9 messenger RNA in porcine growing and preovulatory ovarian follicles. Biol Reprod 2004, 71: 1290-1295. 10.1095/biolreprod.104.027912
Snedecor GW, Cochran WG: Statistical methods. 8th edition. USA: Iowa State Univeristy Press; 1994.
Somfai T, Imai K, Kaneda M, Akagi S, Watanabe S, Haraguchi S, Mizutani E, Dang-Nguyen TQ, Inaba Y, Geshi M, Nagai T: The effect of ovary storage and in vitro maturation on mRNA levels in bovine oocytes; a possible impact of maternal ATP1A1 on blastocyst development in slaughterhouse-derived oocytes. J Reprod Dev 2011, 57: 723-730. 10.1262/jrd.11-020H
Watson AJ: Oocyte cytoplasmic maturation: a key mediator of oocyte and embryo developmental competence. J Anim Sci 2007, 85: E1-E3. 10.2527/jas.2006-432
Yan C, Wang P, DeMayo J, Elvin JA, Carino C, Prasad SV, Skinner SS, Dunbar BS, Dube JL, Celeste AJ, Matzuk MM: Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol 2001, 15: 854-866. 10.1210/me.15.6.854
Zhu G, Guo B, Pan D, Mu Y, Feng S: Expression of bone morphogenetic proteins and receptors in porcine cumulus oocyte complexes during in vitro maturation. Anim Reprod Sci 2008, 104: 275-283. 10.1016/j.anireprosci.2007.02.011
Zicarelli L: Management under different environmental condition. Buffalo Journal 1994, Suppl. 2: 17-38.
Acknowledgement
The authors are thankful to the Dean, Madras Veterinary College and Director of Research, TANUVAS for the facilities provided to carry out the research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MK carried out the in vitro maturation and real time experiments, participated in data analysis and drafted the manuscript. ES participated in manuscript correction and helped in statistical analysis of data and VK participated in realtime data analysis. All the authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kathirvel, M., Soundian, E. & Kumanan, V. Differential expression dynamics of Growth differentiation factor9 (GDF9) and Bone morphogenetic factor15 (BMP15) mRNA transcripts during in vitro maturation of buffalo (Bubalus bubalis) cumulus–oocyte complexes. SpringerPlus 2, 206 (2013). https://doi.org/10.1186/2193-1801-2-206
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/2193-1801-2-206