Abstract
Background
RNAs for embryo patterning and for germ cell specification are localized to the vegetal cortex of the oocyte of Xenopus laevis. In oocytes of the direct developing frog Eleutherodactylus coqui, orthologous RNAs for patterning are not localized, raising the question as to whether RNAs and other components of germ plasm are localized in this species.
Methods
To identify germ plasm, E. coqui embryos were stained with DiOC6(3) or examined by in situ hybridization for dazl and DEADSouth RNAs. The cDNAs for the E. coqui orthologues were cloned by RT-PCR using degenerate primers. To examine activity of the E. coqui orthologues, RNAs, made from constructs of their 3'UTRs with mCherry, were injected into X. laevis embryos.
Results
Both DiOC6(3) and dazl and DEADSouth in situs identified many small islands at the vegetal surface of cleaving E. coqui embryos, indicative of germ plasm. Dazl was also expressed in primordial germ cells in the genital ridge. The 3'UTRs of E. coqui dazl and DEADSouth directed primordial germ cell specific protein synthesis in X. laevis.
Conclusions
E. coqui utilizes germ plasm with RNAs localized to the vegetal cortex to specify primordial germ cells. The large number of germ plasm islands suggests that an increase in the amount of germ plasm was important in the evolution of the large E. coqui egg.
Similar content being viewed by others
Background
Germ cells are the raison d'être of multicellular organisms, since they are ultimately responsible for the continuation of a species from generation to generation. In many animals, germ cells arise early in development due to a specialized cytoplasmic localization called germ plasm. In other animals, germ cells arise due to interactions between cells, known as inductions. It is curious that such an important cell arises in development in two fundamentally different ways [1, 2]. Among amphibians, anurans (frogs) utilize germ plasm, while urodeles (salamanders) utilize inductions [3, 4].
Anuran germ plasm contains mitochondria, an electron-dense nuage material, and RNAs. A number of germ plasm specific RNAs have been identified in Xenopus laevis, including dazl, nanos1 (formerly Xcat2), pat, and DEADSouth[5]. During oogenesis, these RNAs are transported to the vegetal cortex of the oocyte. Following fertilization, they are grouped into islands of germ plasm, and as cleavage proceeds the germ plasm islands are segregated into a small number of cells. Cells receiving germ plasm become the primordial germ cells. They migrate from the endoderm into the genital ridges which form the gonads. Orthologues of dazl are also localized to germ plasm in the frogs Lithobates pipiens and Pelophylax lessonae, both formerly placed in the genus Rana[6, 7].
In addition to the localization of germ plasm RNAs, RNAs involved in patterning the embryo's body are localized to the vegetal cortex. These RNAs in X. laevis include vegt and vg1[5], and the vegt orthologue is also localized in the leopard frog Lithobates (Rana) pipiens[6]. The mechanism for transporting the germ plasm RNAs and the patterning RNAs is different [5, 8, 9]. Germ plasm RNAs accumulate first in the mitochondrial cloud of the small oocyte and then move vegetally via the early METRO pathway. Many patterning RNAs move vegetally via a late pathway.
In contrast, urodeles lack germ plasm [3, 10, 11] as well as localization of RNAs to the vegetal pole of the oocyte. The latter include the dazl orthologue in both Ambystoma mexicanum and Cynops pyrrhogaster[11, 12] as well as the vegt orthologue in A. mexicanum[13].
The conclusions on RNA localization in germ plasm are based on limited data from only four species, two anurans and two urodeles. Examination of other species, particularly those with diverse life histories, might reveal variations that would indicate how the anuran and urodele patterns arose from their last common ancestor 300 to 250 million years ago (MYA) [14].
In the direct developing frog Eleutherodactylus coqui, orthologues of vegt and vg1 RNAs are not localized to the vegetal cortex [15]. Direct developers like E. coqui are derived from anurans with tadpoles. Although the last common ancestor between E. coqui and X. laevis was 230 MYA, E. coqui and Bufo bufo, which has germ plasm [16], shared a common ancestor about 53 MYA [14]. E. coqui has large 3.5 mm eggs, which show significant differences in embryonic patterning compared to X. laevis[17]. These variations raise the question as to whether E. coqui has germ plasm with localized RNAs. We examine that question here.
Methods
Animals and embryos
Eleutherodactylus coqui were collected on the Big Island of Hawaii under Injurious Wildlife Export permits issued by the Department of Land and Natural Resources, Hawaii. Adults were maintained in the laboratory as breeding pairs in terraria as described previously [15, 18]. Pairs mated naturally, and clutches of embryos were collected from their guarding father. The embryos were staged according to Townsend and Stewart (TS) [19] and usually cultured in Petri dishes on filter paper, wetted with 20% Steinberg's solution. Ovarian oocytes were obtained by removal from a sacrificed female or by surgical removal from females anesthetized with 0.1% Tricaine methane sulfonate (MS222), made to pH 7.4 by addition of Na2HPO4. Procedures for Xenopus laevis were described previously [20]. Use of E. coqui and X. laevis in this research was carried out under protocols approved by the Duquesne University Institutional Animal Care and Use Committee (IACUC) and guidelines for animal experiments at the University of Hyogo.
DiOC6(3) staining
Embryos at around the two-cell stage were placed in 20% Steinberg's solution to swell the jelly. The outer jelly layers were removed using watchmaker's forceps and the inner jelly layer was removed by gentle swirling in 3% cysteine, pH8. The green fluorescent dye, DiOC6(3) (Invitrogen, Carlsbad CA, USA), was prepared according to Venkatarama et al. [21]. The stock, a saturated solution of DiOC6(3) in dimethyl sulfoxide, was diluted 2 μl/ml 20% Steinberg's and then further diluted 4 μl/ml 20% Steinberg's. Dejellied embryos were stained for 30 minutes and washed in 20% Steinberg's to remove excess stain. Embryos were prepared for viewing by incubating them in 15% Ficoll in 20% Steinberg's to remove water from the perivitelline space. To view the lower vegetal side, embryos were placed vegetal pole up in a well made of plasticene. The walls of the plasticene well were gently pushed against the inverted embryo to hold it in position. Embryos were viewed with either an Olympus SZx12 stereomicroscope or a Nikon Microphot-SA compound microscope, both equipped with a fluorescent light source. Embryos developed normally after staining and viewing.
Cloning E. coqui orthologues of genes expressed in X. laevis germ plasm
To look for RNAs that might identify germ plasm in E. coqui, we selected dazl and DEADSouth (ortholog of ddx25). Both RNAs are localized in X. laevis germ plasm [5, 22, 23], and dazl orthologue is localized in Lithobates (Rana) pipiens and Pelophylax (Rana) lessonae germ plasm [5, 7]. Our usual cloning route relies on designing degenerate PCR primers based on several orthologous sequences, so we did not try nanos1, whose close homologies were only recently recognized, or to pat, a germ plasm RNA so far unique to Xenopus[24–26]. Vasa is used as a marker for germ plasm in many species, but neither the RNA for the X. laevis vasa orthologue nor its protein is localized in X. laevis germ plasm [27–29].
RNA was extracted from pieces of ovary with Trizol (Invitrogen) with the addition of a precipitation step with 4 M (final) LiCl at -20C. The RNA was used to make cDNA for PCR amplification of dazl and DEADSouth. For dazl, we started with the degenerate primers designed by Tamori et al. [12] for C. pyrrhogaster, which yielded 202 nt. An additional round of PCR using an exact primer to the cloned fragment and a degenerate primer, based on A. mexicanum, C. pyrrhogaster, and L. pipiens, generated a 603 nt fragment. The complete cDNA was then obtained from an E. coqui ovarian library using exact primers in combination with plasmid primers for T7 or Sp6.
For DEADSouth, degenerate primers based on human, mouse, chick, and X. laevis yielded a 258 nt fragment. The complete cDNA was obtained from the E. coqui ovarian library as above.
In situ hybridization
In order to detect RNA, in situ hybridization was carried out on ovarian pieces and on whole embryos as described previously [6, 30]. In situ hybridization of cleaving embryos proved difficult due their large size and yolkiness, which prevented good fixation. The proteinase K concentration in the procedure was decreased to 5 μg/ml to preserve better the specimens.
Assaying 3'UTR activity
The 3'UTRs of E. coqui DEADSouth (EcDS3'UTR) and dazl (EcDazl3'UTR) were amplified by PCR using a PrimeSTAR enzyme (TaKaRa). They were inserted into the site following the stop codon of mCherry of pCS2-mCherry without adding any sequence using In-Fusion Advantage PCR Cloning Kit (Clontech). The mCherry coding region and Ec3'UTRs of pCS2-mCherry-EcDS3'UTR and pCS2-mCherry-EcDazl3'UTR were confirmed by sequencing. In addition to them, pCS2-Venus-DEADSouth3'UTR[20] plasmid was digested with XhoI, and all three constructs were used as templates for in vitro mRNA synthesis with mMESSAGE mMACHINE SP6 Kit (Ambion, Austin TX, USA). The mRNAs were dissolved in water, and 4.6 nL of the mRNA (0.1 microgram each mRNA/microliter) was injected into the cortical region at the vegetal pole of a X. laevis fertilized egg (460 pg each mRNA/egg). The embryos were allowed to develop at 18°C and were observed at Nieuwkoop - Faber stages 32 and 40 under a Leica MZ16F fluorescent binocular microscope.
Results
Migrating primordial germ cells
In several anurans, the primordial germ cells migrate from the endoderm along the dorsal mesentery to the genital ridges [3, 16, 31, 32]. In X. laevis, this final migration occurs at Nieuwkoop-Faber stages 41 to 45, prior to tadpole feeding at NF 46 [33, 34]. Primordial germ cells, recognizable by their high content of yolk platelets, are visible in the dorsal mesentery in E. coqui embryos at Townsend-Stewart (TS) stages 7 to 9 (Figures 1 and 2). They end up near the ventral surface of the dorsal aorta at TS9 (Figures 1 and 2) and reach the genital ridges by TS11.
PGCs in embryo sections. (A) PGCs are present in the dorsal mesentery (arrow) which attaches the yolky endoderm to the rest of the body of this TS8 embryo. The inset shows an enlargement with three PGCs in the mesentery. (B) At TS9, yolk-filled PGCs are clumped in the prospective genital ridges (arrow), ventral to the dorsal aorta. The inset shows an enlargement of this region.
The presence of E. coqui PGCs in the dorsal mesentery suggests that they arise as in other anurans and not as in urodeles. In urodeles, PGCs form by induction in the lateral plate mesoderm, so their migration to the genital ridge is not along the dorsal mesentery [3, 4, 10, 11, 35]. If E. coqui follow the anuran pattern, they should have localized germ plasm in their oocytes and early embryos. To identify E. coqui germ plasm, we used DiOC6(3) staining and in situ hybridization of orthologues of dazl and DEADSouth.
DiOC6(3) staining of E. coqui cleaving embryos
Germ plasm in X. laevis has a high concentration of mitochondria, which allows for the visualization of germ plasm islands in living zygotes using the fluorescent lipophilic dye DiOC6(3) [21, 36]. When E. coqui cleaving embryos were stained with DiOC6(3), they showed patterns similar to X. laevis germ plasm islands (Figure 3).
DiOC 6 (3) staining of cleaving embryos. (A) Only one furrow (bright green line) has reached the vegetal pole in this eight-cell embryo. A field of small DiOC6(3) stained islands is present on the vegetal surface, with most of the islands on one side of the furrow. Furrows stain brightly as they represent two closely-apposed new membranes. (B) This photograph of a different eight-cell embryo shows a cleavage furrow (bright green line) and the boundary between vegetal regions with DiOC6(3) stained islands and with no stained islands. (C, D) Vegetal (C) and animal (D) views of the same approximately 16-cell embryo. Although seven cells are present near the animal pole (D), only two furrows are present in the vegetal region (C). DiOC6(3) stained islands are present primarily in three of the four vegetal cells (C). (E, F) Vegetal (E) and equatorial (F) views of the same morula stage embryo. Although the region near the animal pole is divided into many cells, a limited number of cleavage furrows have reached the vegetal pole. DiOC6(3) islands remain on the vegetal surface, with islands lined up in the forming furrows (E). Embryos are about 3.5 mm in diameter, with (C) enlarged approximately 30% relative to (A) (D) (E) (F).
The small putative germ plasm islands covered the vegetal third of the two- to eight-cell embryo (Figure 3A, B). At fourth cleavage, there are eight smaller cells near the animal pole (Figure 3D), but cleavage is incomplete at the vegetal pole with two cleavage furrows (Figure 3C). The DiOC6(3) islands are usually located asymmetrically relative to the vegetal pole and present in two or three cells (Figure 3C). With continued cleavage, there were fewer germ plasm islands on the surface (Figure 3E, F). Islands accumulated in cleavage furrows (Figure 3E), suggesting as in X. laevis that germ plasm leaves the surface by cortical ingression.
Cloning of E. coqui orthologues of dazl and DEADSouth
To confirm the identification of the DiOC6(3) islands in E. coqui as germ plasm, we cloned E. coqui orthologues of dazl and DEADSouth, two RNAs localized to germ plasm in X. laevis. For convenience, we will refer to these as Ecdazl and EcDEADSouth. The Ecdazl cDNA has 1,698 nucleotides (nt) and contains the complete open reading frame (ORF) for a 267 amino acid protein (GenBank Accession JN130399). There is a 74 nt 5'UTR and a 820 nt 3'UTR. The Ecdazl 3'UTR is shorter than the 3'UTRs of the orthologues in the frogs X. laevis (1,352 nt) and L. pipiens (2,635 nt). Nucleotide sequence identities to dazl ORFs from the frogs X. laevis and L. pipiens, the axolotl Ambystoma mexicanum, and the mouse Mus musculus were 61 to 66%, and amino acid identities were 41 to 45%. These somewhat low identities are true for all pairwise comparisons among these animals with the exception of mouse and axolotl, an anomaly pointed out by Johnson et al. [11, 37].
Evidence supporting the identification of our clone as Ecdazl comes from domain comparisons. Dazl proteins have a conserved RNA Recognition Motif (RRM), which includes two small RNA binding domains RNP-1 and RNP-2. The Ecdazl RRM is 67% identical at the amino acid level to that of X. laevis. The seven-amino acid RNP-2 domain is identical between Ecdazl and the four above animals, as well as human, chicken, and a second urodele amphibian Cynops pyrrhogaster[12]. The only difference in the seven-amino acid RNP-1 domain among these seven animals is one amino acid difference in X. laevis dazl. A recognizable DAZ repeat is present at amino acids 152 to 177 of Ecdazl.
Our cloned EcDEADSouth cDNA has 2,024 nt with 38 nt of 5'UTR, a 1,422 nt ORF, and a 564 nt 3'UTR (GenBank Accession JN130398). The ORF is 71% identical to that of X. laevis and would code for a protein with 473 amino acids that are 70% identical to X. laevis DEADSouth protein. Evidence supporting the identification of our clone as EcDEADSouth comes from domain analysis, parallel to that by MacArthur et al. [23] for X. laevis. The 346-amino acid DEAD-box region is 78% identical and 91% similar to that of X. laevis. In one diagnostic sequence, 18/19 amino acids were identical to X. laevis. In a second diagnostic sequence of six amino acids, DEADSouth of X. laevis shared identity with three amino acids of mouse Mus DEAD5 [23], and those same three amino acids are shared with EcDEADSouth.
The 564 nt 3'UTR is shorter than the 1,597 nt 3'UTR of X. laevis DEADSouth, so we tried several approaches to confirm that we have all of the 3'UTR. Our EcDEADSouth 3'UTR ends in polyA, and numerous attempts to find a longer clone from an E. coqui oocyte cDNA library or from cDNA were unsuccessful. There is an AUUAAA sequence which is similar to the polyA signal sequence, although different by one nucleotide. An estimate of the transcript size from Northern blots gave a value slightly over 2 kb, similar to our 2,024 nt sequence. These results indicate that the EcDEADSouth 3'UTR is significantly shorter than that of X. laevis.
Ecdazl and EcDEADSouth expression during oogenesis
Pieces of ovary were subjected to whole mount in situ hybridization to see whether Ecdazl RNA was localized in oocytes, like its orthologues in X. laevis and L. pipiens. Oocytes, just starting to take up yolk, were stained completely, while small vitellogenic oocytes with yolk showed a clear localization of Ecdazl RNA on one side (Figure 4). Similar results were obtained for EcDEADSouth RNA. Although EcDazl staining asymmetry was present on 1 mm oocytes, there was no obvious staining in larger oocytes.
Ecdazl expression in oocytes. In this ovarian fragment, in situ hybridization showed Ecdazl RNA throughout small oocytes, just beginning vitellogenesis, and localized to one side of vitellogenic oocytes. The vitellogenic oocytes in this preparation are about 1 mm in diameter. Large oocytes, up to 3.5 mm, did not show clear staining and were removed prior to processing of the ovarian fragment.
This lack of staining in large oocytes could be due to the diffuse presence of Ecdazl RNA in a large volume. To test this, oocytes were cut into three pieces, animal, equatorial, and vegetal, following the protocol of Beckham et al. [15]. RNA was extracted from pooled pieces of each type, and assayed for Ecdazl RNA by RT-PCR. Although Ecdazl RNA was detected, the results were variable. In two experiments, Ecdazl RNA was only in the vegetal third. In a third experiment, the equatorial as well as the vegetal thirds were positive, and in the fourth experiment, Ecdazl RNA was found in all three pieces. These results suggest that the location of Ecdazl RNA is primarily in the vegetal third, but it may not be as tightly regulated in large oocytes.
Ecdazl and EcDEADSouth RNA in cleaving embryos
In order to confirm that the DiOC6(3) staining identified germ plasm, we carried out whole mount in situ hybridization for Ecdazl and EcDEADSouth RNAs on cleaving embryos. Good fixation of these large, yolky cleaving embryos was difficult, and the vegetal surface tended to be lost in the many changes of solutions. Nonetheless, the in situ patterns were similar to the patterns found with DiOC6(3) staining.
For both Ecdazl and EcDEADSouth, there were many small stained islands on the vegetal surface of four to eight cell embryos (Figure 5). At 16 to 32 cells, the islands were usually displaced to one side of the vegetal pole (Figure 5B, D). As with DiOC6(3), the RNAs were present within cleavage furrows (Figure 5B, D). Based on the similarity of patterns, we conclude that staining with DiOC6(3), Ecdazl, and EcDEADSouth identifies germ plasm near the vegetal surface of E. coqui cleaving embryos.
Ecdazl and EcDEADSouth RNA in cleaving embryos. (A) In this vegetal view, many islands containing Ecdazl RNA are visible near the surface of a four-cell embryo. Cleavage furrows have not reached the vegetal half. (B) In this side view of a 16-cell embryo, islands of Ecdazl RNA are present near the vegetal pole. Cleavage furrows have reached the vegetal half. (C) In this vegetal view, many EcDEADSouth islands are visible despite some surface damage. (D) In this 16-cell embryo with two furrows reaching the vegetal pole, most of the surface surrounding the vegetal pole has been lost, revealing the whiter underlying cytoplasm. The surface loss permits several observations. First, EcDEADSouth islands are visible on the intact surface. Second, the islands are located asymmetrically relative to the vegetal pole, marked by the intersecting furrows. Third, EcDEADSouth islands are present in the cleavage furrow, where internal cytoplasm is exposed.
Ecdazl RNA in primordial germ cells
We have not attempted to track the fate of cells with germ plasm in these large embryos after the cleavage stages, but we carried out in situ hybridization on sections of TS14 and TS15 embryos to see whether the primordial germ cells in the genital ridges expressed Ecdazl. TS15 is the stage at which the froglet hatches from its jelly capsule and prior to feeding. Yolk-filled cells in the genital ridges stained positively with the Ecdazl probe (Figure 6). This early gonadal expression of Ecdazl contrasts with the pattern in X. laevis or Xenopus tropicalis. In those frogs, dazl RNAs disappear before their tadpoles begin feeding, but the genes are re-expressed in the frog testis and ovary [22, 38]. Xdazl protein, however, remains in the primordial germ cells of tadpoles [20, 39]. Since the tadpole was deleted in the evolution of direct development, it is expected that the gonad of a TS15 E. coqui froglet would be more equivalent to a gonad in newly metamorphosed Xenopus than to its tadpole.
Activity of EcDEADSouth and Ecdazl 3'UTRs
The 3'UTRs of DEADSouth and dazl are sufficient to program translation of an attached ORF in the primordial germ cells of X. laevis[20, 40]. This raises the question as to whether the 3'UTRs of EcDEADSouth and Ecdazl would have activity in this assay. Both of the E. coqui 3'UTRs are much shorter than the X. laevis orthologues, and there is little overall nucleotide sequence identity.
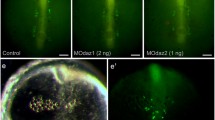
To test their activities, the 3'UTRs were placed in plasmids with mCherry, and RNAs were synthesized in vitro. Either the mCherry-EcDS3'UTR RNA or the mCherry-EcDazl3'UTR RNA was microinjected into X. laevis fertilized eggs along with Venus-XlDS3'UTR RNA. The X. laevis DEADSouth 3'UTR led to synthesis of Venus protein in PGCs, and mCherry protein co-localized with Venus using 3'UTRs of either EcDEADSouth (10/10) or Ecdazl (6/7) (Figure 7).
Activity of the EcDazl3'UTR. A X. laevis egg was injected with RNAs of both Venus-XlDS3'UTR and mCHERRY-EcDazl3'UTR. The genital ridge region of the embryo (A) was examined at stage 40 with different light sources to detect Venus (B, green) and mCHERRY (C, red) proteins. The merged image (D, yellow) shows that these two proteins co-localized. The co-localization indicates that the EcDazl 3'UTR directed PGC-specific protein expression in X. laevis.
Discussion
Large surface area covered by islands of germ plasm in E. coqui
Our present results indicate that E. coqui, with big 3.5 mm eggs, has germ plasm similar to frogs such as X. laevis, with 1.3 mm eggs, and L. pipiens, with 1.7 mm eggs. Islands of E. coqui germ plasm were identified by DiOC6(3) staining and by localization of Ecdazl and EcDEADSouth RNAs, and they are spread over a large part of the vegetal cortex of the cleaving embryo. Their distribution is similar to that found in X. laevis, but the E. coqui germ plasm covers a more extensive area both relatively and absolutely.
In relative terms, germ plasm in X. laevis becomes concentrated near the vegetal pole by the time the second cleavage furrow reaches that pole at the four- to eight-cell stage [22, 23, 25, 36, 38, 41–43]. Germ plasm is removed from the surface along the furrows by cortical ingression [36, 41], so that little remains by 32 cells. In contrast, many islands of germ plasm remain at the surface of E. coqui morulae. E. coqui germ plasm also leaves the surface along cleavage furrows, but the furrows arrive later and the vegetal cells remain larger compared to X. laevis. This less active early removal is coupled with the greater extent of surface coverage by germ plasm in E. coqui.
In absolute terms, E. coqui germ plasm extends far towards the equator from the vegetal pole. If we estimate that germ plasm is present from the pole to the 45° latitude, that surface area is 5.8 mm2, which is greater than the surface area of an entire X. laevis egg (5.3 mm2).
An important function of germ plasm in X. laevis is to repress transcription, thereby preventing primordial germ cells from responding to inductive signals from neighboring cells [21]. Transcriptional repression by germ plasm has also been documented in Drosophila and C. elegans[44], indicating that this activity may be the main role for germ plasm in the early embryo. If E. coqui has a larger area covered by germ plasm, how can it avoid the negative consequences of transcriptional repression in many vegetal cells?
One possibility is that transcriptionally repressed vegetal cells can be tolerated in E. coqui because most vegetal cells serve only a nutritional function and do not contribute to any embryonic tissues. We previously defined these cells as nutritional endoderm [45]. Direct development is a derived condition resulting from the evolutionary loss of the aquatic-feeding tadpole from the life history. To compensate for the lack of a feeding larva, the oocyte was provisioned with more yolk and increased in size. Increased egg size, however, could pose difficulties for the migration of primordial germ cells from the vegetal pole to their target, the genital ridge. One solution would be to increase the number of potential primordial germ cells, making it probable that some would be in position to migrate out of the endoderm and reach the genital ridges. While this is a speculative hypothesis to account for the large surface covered by germ plasm, a testable prediction is that there will be cells with germ plasm trapped among the cells of the nutritional endoderm at later stages. These cells will be lost along with the yolk-depleted cells of the nutritional endoderm.
Alternatively, the presence of transcriptional repressors in a greater number of vegetal cells may be part of the mechanism generating nutritional endoderm. Cells of the nutritional endoderm appear undifferentiated. While this may be due to the lack of signaling by nodal or other ligands [17], transcriptional repression could ensure that these cells remain in an embryonic state as their yolk is depleted. Venkatarama et al. [21] showed by immunocytochemistry that transcriptional elongation occurred in early somatic cells but not in primordial germ cells. A similar immunocytochemical approach would provide a test of whether repression of transcriptional elongation is widespread in the nutritional endoderm.
Evolutionary differences in RNA localization in amphibian oocytes
In X. laevis, there are three groups of RNAs, localized to the vegetal cortex of the oocyte, that are necessary for embryogenesis. RNAs for germ plasm are localized to the vegetal cortex during oogenesis via the early or METRO pathway, while RNAs involved in germ layer specification, notably vegt, are localized via the late pathway [5, 8]. The third group of localized RNAs is involved in establishing the embryo's dorsal axis, which is specified by the microtubule-dependent cortical rotation in the first cell cycle after fertilization [46, 47]. These RNAs include wnt11, fatvg (plin2), and trim36. Wnt11 plays a major role in causing movement of β-catenin into nuclei on the dorsal side [48–50]. Both fatvg and trim36 are required for the cortical rotation, and the parallel array of microtubules does not form in the absence of trim36[51, 52]. All three of these RNAs are localized to the vegetal cortex via the early pathway, although there is some movement of fatvg RNA via the late pathway [51].
The localization of Ecdazl and EcDEADSouth to the vegetal cortex of E. coqui small oocytes suggests the presence of the early localization pathway. Prior to localization, however, these RNAs were detected throughout the oocyte and not localized to an organelle, such as the mitochondrial cloud or Balbiani body as in X. laevis[53, 54]. Attempts to visualize a Balbiani body in E. coqui oocytes have not been successful. Organelle localization of a dazl orthologue was also not found in Lithobates pipiens and Pelophylax lessonae, both formerly placed in the genus Rana[6, 7].
Although the early RNA localization pathway appears to exist in E. coqui, the late localization pathway does not. Neither vegt nor vg1 RNA, are localized to the vegetal cortex, and these orthologous RNAs are present diffusely in the animal cytoplasm [15]. Their animal location correlates with the more animal origin of both mesoderm and definitive endoderm in E. coqui[17, 45].
The presence of RNAs localized to germ plasm in E. coqui raises the question as to the presence of maternal RNAs for specification of the dorsal axis. Although a cortical rotation has not been documented for E. coqui, it likely exists. There is a massive vegetal array of parallel microtubules which persists beyond first cleavage, and artificially moving cytoplasm relative to the cortex by tilting zygotes 90° with respect to gravity can specify the dorsal axis [55]. In the current work, islands of germ plasm usually extended closer to the equator on one side of the cleaving embryo, suggesting that a cortical rotation occurred and caused this asymmetry. It would be interesting to observe the behavior of wnt11 RNA in E. coqui, with the predictions that it would be localized to the vegetal cortex in oocytes and that it would be shifted to one side equatorially during cleavage.
In the urodele A. mexicanum, the orthologous vegt RNA is not localized [13], raising the possibility that as in E. coqui, the late pathway is absent. Urodeles lack germ plasm and its associated RNAs, suggesting that the early pathway also is absent. Nonetheless, specification of the dorsal axis in urodeles is likely caused by a cortical rotation. A grey crescent, which forms due to the rotation, appears after fertilization in several species [56–58], and cortical arrays of parallel microtubules have been detected in C. pyrrhogaster[59]. These observations raise the question as to whether RNA of orthologues of wnt11, fatvg, and trim 36 are localized in urodele oocytes or whether urodeles use a different mechanism to initiate development of the dorsal axis.
Conservation of PGC RNA behavior
The last common ancestor of X. laevis and E. coqui lived about 230 MYA. Although the 3'UTR nucleotide sequences of the DEADSouth and dazl orthologues are very different, the activity of the 3'UTRs in directing PGC specific translation has been conserved apparently over this long period of time. Unlike the constraints on sequence changes within an ORF, many 3'UTR functions require only short sequences. These sequences could be stems for generating secondary RNA structures, recognized by a protein, or they could be targets for microRNA binding.
Two microRNAs have been implicated in control of RNAs in X. laevis PGCs, namely miR-427 and miR-18[40, 60]. miR-427 mediates the destruction of many maternal RNAs [60] and is the orthologue of zebrafish miR-430, which is involved in regulation of RNAs in zebrafish PGCs [61–63]. miR-18 causes the destruction of the Dead end RNA in somatic cells, but Dead end RNA is protected in the PGCs [40]. Accordingly, we examined the 3'UTRs of the DEADSouth and dazl orthologues for target sites for these and other miRs [64]. The 3'UTR of X. laevis DEADSouth has four putative targets of miR427, EcDEADSouth has one, X. laevis dazl has one, and Ecdazl has two. The 3'UTR of X. laevis DEADSouth has two putative targets of miR-18, but no miR-18 targets were present in the 3'UTRs of EcDEADSouth, Ecdazl, or X. laevis dazl. These comparisons suggest that miR-427 should be the first candidate examined to account for the conservation of 3'UTR activities between X. laevis and E. coqui.
Conclusions
The great increase in yolk in the evolution of direct development in E. coqui has led to changes in the organization of its egg and early development. Compared to X. laevis and most anurans with small eggs and aquatic development, the changes include: slow and delayed cleavage of the vegetal region (Figure 3), lack of localization to the vegetal cortex of RNAs involved in germ layer patterning [15], and the origin of the novel nutritional endoderm [18, 45]. Despite these profound changes of the yolk-rich vegetal region, formation of primordial germ cells via germ plasm, localized to the vegetal cortex, has been conserved. These direct developers have not moved towards an alternative path to generate primordial germ cells, such as found in urodeles. Rather, they appear to have expanded the relative amount of germ plasm coverage of the vegetal region. Potential reasons for this expansion of germ plasm are to participate in the formation of the nutritional endoderm and to ensure that some primordial germ cells, among the huge number of vegetal cells, get to the genital ridge. These hypotheses require future experimental testing.
Abbreviations
- IACUC:
-
Institutional Animal Care and Use Committee
- nt:
-
nucleotides
- ORF:
-
open reading frame.
References
Extavour CG, Akam M: Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003, 130: 5869-5884. 10.1242/dev.00804.
Johnson AD, Richardson E, Bachvarova RF, Crother BI: Evolution of the germ line-soma relationship in vertebrate embryos. Reproduction. 2011, 141: 291-300. 10.1530/REP-10-0474.
Nieuwkoop PD, Sutasurya LA: Primordial Germ Cells in the Chordates. 1979, Cambridge: Cambridge University Press
Johnson AD, Drum M, Bachvarova RF, Masi T, White ME, Crother BI: Evolution of predetermined germ cells in vertebrate embryos: implications for macroevolution. Evol Dev. 2003, 5: 414-431. 10.1046/j.1525-142X.2003.03048.x.
King ML, Messitt TJ, Mowry KL: Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell. 2005, 97: 19-33. 10.1042/BC20040067.
Nath K, Boorech JL, Beckham YM, Burns MM, Elinson RP: Status of RNAs, localized in Xenopus laevis oocytes, in the frogs Rana pipiens and Eleutherodactylus coqui. J Exp Zool (Mol Dev Evol). 2005, 304B: 28-39. 10.1002/jez.b.21020.
Marracci S, Michelotti V, Casola C, Giacoma C, Ragghianti M: Daz- and Pumilio-like genes are asymmetrically localized in Pelophylax (Rana) oocytes and are expressed during early spermatogenesis. J Exp Zool (Mol Dev Evol). 2011, 316B: 330-338. 10.1002/jez.b.21405.
Kloc M, Etkins LD: Two distinct pathways for the localization of RNAs at the vegetal cortex in Xenopus oocytes. Development. 1995, 121: 287-297.
Kloc M, Bilinski S, Chan AP, Allen LH, Zearfoss NR, Etkin LD: RNA localization and germ cell determination in Xenopus. Int Rev Cytol. 2001, 203: 63-91.
Wakahara M: Primordial germ cell development: is the urodele pattern closer to mammals than to anurans?. Int J Dev Biol. 1996, 40: 653-659.
Johnson AD, Bachvarova RF, Drum M, Masi T: Expression of axolotl DAZL RNA, a marker of germ plasm: widespread maternal RNA and onset of expression in germ cells approaching the gonad. Dev Biol. 2001, 234: 402-415. 10.1006/dbio.2001.0264.
Tamori Y, Iwai T, Mita K, Wakahara M: Spatio-temporal expression of a DAZ-like gene in the Japaneses newt Cynops pyrrhogaster that has no germ plasm. Dev Gene Evol. 2004, 214: 615-627. 10.1007/s00427-004-0443-z.
Nath K, Elinson RP: RNA of AmVegT, the axolotl orthologue of the Xenopus meso-endodermal determinant, is not localized in the oocyte. Gene Expr Patterns. 2007, 7: 197-201. 10.1016/j.modgep.2006.07.003.
Timetree of Life. [http://timetree.org]
Beckham YM, Nath K, Elinson RP: Localization of RNAs in oocytes of Eleutherodactylus coqui, a direct developing frog, differs from Xenopus laevis. Evol Dev. 2003, 5: 562-571. 10.1046/j.1525-142X.2003.03061.x.
Blackler AW: Contribution to the study of germ-cells in the anura. J Embryol Exp Morphol. 1958, 6: 491-503.
Ninomiya H, Zhang Q, Elinson RP: Mesoderm formation in Eleutherodactylus coqui: body patterning in a frog with a large egg. Dev Biol. 2001, 236: 109-123. 10.1006/dbio.2001.0310.
Singamsetty S, Elinson RP: Novel regulation of yolk utilization by thyroid hormone in embryos of the direct developing frog Eleutherodactylus coqui. Evol Dev. 2010, 12: 437-448. 10.1111/j.1525-142X.2010.00430.x.
Townsend DS, Stewart MM: Direct development in Eleutherodactylus coqui (Anura: Leptodactylidae): a staging table. Copeia. 1985, 1985: 423-436. 10.2307/1444854.
Kataoka K, Yamaguchi T, Orii H, Tazaki A, Watanabe K, Mochii M: Visualization of the Xenopus primordial germ cells using a green fluorescent protein controlled by cis elements of the 3' untranslated region of the DEADSouth gene. Mech Dev. 2006, 123: 746-760. 10.1016/j.mod.2006.07.006.
Venkatarama T, Lai F, Luo X, Zhou Y, Newman K, King ML: Repression of zygotic gene expression in the Xenopus germline. Development. 2010, 137: 651-660. 10.1242/dev.038554.
Houston DW, Zhang J, Maines JZ, Wasserman SA, King ML: A Xenopus DAZ-like gene encodes an RNA component of germ plasm and is a functional homologue of Drosophila boule. Development. 1998, 128: 171-180.
MacArthur H, Houston DW, Bubunenko M, Mosquera , King ML: DEADSouth is a germ plasm specific DEAD-box RNA helicase in Xenopus related to eIF4A. Mech Dev. 2000, 95: 291-295. 10.1016/S0925-4773(00)00357-9.
Lai F, Zhou Y, Luo X, Fox J, King ML: Nanos1 functions as a translational repressor in the Xenopus germline. Mech Dev. 2011, 128: 153-163. 10.1016/j.mod.2010.12.001.
Hudson C, Woodland H: Xpat, a gene expressed specifically in germ plasm and primordial germ cells of Xenopus laevis. Mech Dev. 1998, 73: 159-168. 10.1016/S0925-4773(98)00047-1.
Houston DW, King ML: Germ plasm and molecular determinants of germ cell fate. Curr Topics Dev Biol. 2000, 50: 155-181.
Komiya T, Itoh K, Ikenishi K, Furusawa M: Isolation and characterization of a novel gene of the DEAD box protein family which is specifically expressed in germ cells of Xenopus laevis. Dev Biol. 1994, 162: 354-363. 10.1006/dbio.1994.1093.
Ikenishi K, Tanaka TS, Komiya T: Spatio-temporal distribution of the protein of Xenopus vasa homologue (Xenopus vasa-like gene 1, XVLG1) in embryos. Dev Growth Differ. 1996, 38: 527-535. 10.1046/j.1440-169X.1996.t01-4-00009.x.
Ikenishi K, Tanaka TS: Spatio-temporal expression of Xenopus vasa homologue, XVLG1, in oocytes and embryos: the presence of XVLG1 RNA in somatic cells as well as germline cells. Dev Growth Differ. 2000, 42: 95-103. 10.1046/j.1440-169x.2000.00493.x.
Elinson RP, Walton Z, Nath K: Raldh expression in embryos of the direct developing frog Eleutherodactylus coqui and the conserved retinoic acid requirement for forelimb initiation. J Exp Zool (Mol Dev Evol). 2008, 310B: 588-595. 10.1002/jez.b.21229.
Delbos M, Gipouloux J-D, Guennoun S: Intraendodermal and intramesenteric migration of anuran amphibian germ cells: transmission and scanning electron microscopy. J Morphol. 1982, 171: 355-360. 10.1002/jmor.1051710308.
Nishiumi F, Komiya T, Ikenishi K: The mode and molecular migration of presumptive PGC in the endoderm cell mass of Xenopus embryos. Dev Growth Differ. 2005, 47: 37-48. 10.1111/j.1440-169x.2004.00777.x.
Kamimura M, Ikenishi K, Kotani M, Matsuno T: Observations on the migration and proliferation of gonocytes of Xenopus laevis. J Embryol Exp Morphol. 1976, 36: 197-207.
Wylie CC, Heasman J: The formation of the gonadal ridge in Xenopus laevis. I. A light and transmission electron microscope study. J Embryol Exp Morphol. 1976, 35: 125-138.
Bachvarova RF, Crother BI, Johnson AD: Evolution of germ cell development in tetrapods: comparison of urodeles and amniotes. Evol Dev. 2009, 11: 603-609. 10.1111/j.1525-142X.2009.00366.x.
Savage RM, Danilchik MV: Dynamics of germ plasm localization and its inhibition by ultraviolet irradiation in early cleavage Xenopus embryos. Dev Biol. 1993, 157: 371-382. 10.1006/dbio.1993.1142.
Johnson AD, Crother B, White ME, Patient R, Bachvarova RF, Drum M, Masi T: Regulative germ cell specification in axolotl embryos: a primitive trait conserved in the mammalian lineage. Phil Trans R Soc Lond B. 2003, 358: 1371-1379. 10.1098/rstb.2003.1331.
Sekizaki H, Takahashi S, Tanegashima K, Onuma Y, Haramoto Y, Asashima M: Tracing of Xenopus tropicalis germ plasm and presumptive primordial germ cells with the Xenopus tropicalis DAZ-Like gene. Dev Dyn. 2004, 229: 367-372. 10.1002/dvdy.10448.
Mita K, Yamashita M: Expression of Xenopus Daz-like protein during gametogenesis and embryogenesis. Mech Dev. 2000, 94: 251-255. 10.1016/S0925-4773(00)00295-1.
Koebernick K, Loeber J, Arthur PK, Tarbashevich K, Pieler T: Elr-type proteins protect Xenopus Dead end mRNA from miR-18-mediated clearance in the soma. Proc Natl Acad Sci USA. 2010, 107: 16148-16153. 10.1073/pnas.1004401107.
Ressom RE, Dixon KE: Relocation and reorganization of germ plasm in Xenopus embryos after fertilization. Development. 1988, 103: 507-518.
Berekelya LA, Ponomarev MB, Luchinskaya NN, Belyavsky AV: Xenopus Germes encodes a novel germ plasm-associated transcript. Gene Expr Patterns. 2003, 3: 521-524. 10.1016/S1567-133X(03)00055-3.
Horvay K, Claussen M, Katzer M, Landgrebe J, Pieler T: Xenopus Dead end mRNA is a localized maternal determinant that serves a conserved function in germ cell development. Dev Biol. 2006, 291: 1-11. 10.1016/j.ydbio.2005.06.013.
Strome S, Lehmann R: Germ vs. soma decisions, lessons from flies and worms. Science. 2007, 316: 392-393. 10.1126/science.1140846.
Buchholz DR, Singamsetty S, Karadge U, Williamson S, Langer CE, Elinson RP: Nutritional endoderm in a direct developing frog: a potential parallel to the evolution of the amniote egg. Dev Dyn. 2007, 236: 1259-1272. 10.1002/dvdy.21153.
Vincent JP, Oster GF, Gerhart JC: Kinematics of gray crescent formation in Xenopus eggs. Dev Biol. 1986, 113: 484-500. 10.1016/0012-1606(86)90184-3.
Elinson RP, Rowning B: A transient array of parallel microtubules in frog eggs: potential tracks for a cytoplasmic rotation that specifies the dorso-ventral axis. Dev Biol. 1988, 128: 185-197. 10.1016/0012-1606(88)90281-3.
Ku M, Melton DA: Xwnt-11: a maternally expressed Xenopus wnt gene. Development. 1993, 119: 1161-1173.
Tao Q, Yokota C, Puck H, Kofron M, Birsoy B, Yan D, Asashima M, Wylie CC, Lin X, Heasman J: Maternal Wnt11 activates the canonical Wnt signaling pathway required for axis formation in Xenopus embryos. Cell. 2005, 120: 857-871. 10.1016/j.cell.2005.01.013.
Cha SW, Tadjuidje E, Tao Q, Wylie , Heasman J: Wnt5a and Wnt11 interact in a maternal Dkk-1 regulated fashion to activate both canonical and non-canonical signaling in Xenopus axis formation. Development. 2008, 135: 3719-3729. 10.1242/dev.029025.
Chan AP, Kloc M, Larabell CA, LeGros M, Etkin LD: The maternally localized RNA fatvg is required for cortical rotation and germ cell formation. Mech Dev. 2007, 124: 350-363. 10.1016/j.mod.2007.02.001.
Cuykendall TN, Houston DW: Vegetally localized Xenopus trim36 regulates cortical rotation and dorsal axis formation. Development. 2009, 136: 3057-3065. 10.1242/dev.036855.
Kloc M, Bilinski S, Etkin LD: The Balbiani body and germ cell determinants: 150 years later. Curr Top Dev Biol. 2004, 59: 1-34.
Chang P, Torres J, Lewis RA, Mowry KL, Houliston E, King ML: Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with the endoplasmic reticulum. Mol Biol Cell. 2004, 15: 4669-4681. 10.1091/mbc.E04-03-0265.
Elinson RP, Ninomiya H: Parallel microtubules and other conserved elements of dorsal axial specification in the direct developing frog, Eleutherodactylus coqui. Dev Genes Evol. 2003, 213: 28-34.
Clavert J: Symmetrization of the egg of vertebrates. Adv Morphol. 1962, 2: 27-60.
Benford HH, Namenwirth M: Precocious appearance of the gray crescent in heat-shocked axolotl eggs. Dev Biol. 1974, 39: 172-176. 10.1016/S0012-1606(74)80020-5.
Fujisue M, Sakai M, Yamana K: Subcortical rotation and specification of the dorsoventral axis in newt eggs. Dev Growth Differ. 1991, 33: 341-351. 10.1111/j.1440-169X.1991.00341.x.
Iwao Y, Yasumitsu K, Narihira M, Jiang J, Nagahama Y: Changes in microtubule structures during the first cell cycle of physiologically polyspermic newt eggs. Mol Reprod Dev. 1997, 47: 210-221. 10.1002/(SICI)1098-2795(199706)47:2<210::AID-MRD13>3.0.CO;2-3.
Lund E, Liu M, Hartley RS, Sheets MD, Dahlberg JE: Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA. 2009, 15: 2351-2363. 10.1261/rna.1882009.
Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K: Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006, 16: 2135-2142. 10.1016/j.cub.2006.08.086.
Takeda Y, Mishima Y, Fujiwara T, Sakamoto H, Inoue K: DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS ONE. 2009, 4: e7513-10.1371/journal.pone.0007513.
Mickoleit M, Banisch TU, Raz E: Regulation of hub mRNA stability and translation by miR430 and Dead end protein promotes preferential expression in zebrafish primordial germ cells. Dev Dyn. 2011, 240: 695-703. 10.1002/dvdy.22571.
Watanabe T, Takeda A, Mise K, Okuno T, Suzuki T, Minami N, Imai H: Stage-specific expression of microRNAs during Xenopus development. FEBS Lett. 2005, 579: 318-324. 10.1016/j.febslet.2004.11.067.
Acknowledgements
We thank Hiromasa Ninomiya for the sections in Figure 2 and Lance Davidson and Steve Trier for use of the stereomicroscope with fluorescent light source. This work was supported by National Science Foundation (NSF) grants 0343403 and 0841720 to RPE.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
RPE designed the study, carried out the observations with DiOC6(3), and drafted the manuscript. MCS and CF cloned the first pieces of Ecdazl and EcDEADSouth cDNA, respectively, which they began as undergraduate project students. TY and HO prepared the 3'UTR constructs and carried out the assay of their activity. KN obtained complete clones of Ecdazl and EcDEADSouth cDNA, analyzed their sequences, and carried out the in situ hybridizations. All authors read and approved the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Elinson, R.P., Sabo, M.C., Fisher, C. et al. Germ plasm in Eleutherodactylus coqui, a direct developing frog with large eggs. EvoDevo 2, 20 (2011). https://doi.org/10.1186/2041-9139-2-20
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/2041-9139-2-20