Abstract
Background
The Chilean Altiplano and the Atacama Desert in northern Chile harbor isolated hydrological systems or oases where it is possible to find minute aquatic snails of the genus Heleobia whose taxonomy is uncertain and where many populations remain unknown. Here, we obtained samples from 30 localities distributed in the region and used molecular (12S and 16S mitochondrial genes) and morphological (penis) characters to investigate diversity of this poorly known fauna.
Results
Molecular phylogenetic analysis consistently recovered five clades, one of which constitutes a cryptic species previously assigned to a species recognized in the area. Four other clades contained sequences of one nominal species consistent with its type locality and at least two additional candidate species, which were corroborated by a particular penis morphology. Furthermore, some morphological differences in penis morphology were observed in two Altiplano populations not resolved by the DNA sequences, providing support for two additional candidate species in the genus. A molecular clock analysis allowed tracing the origin of lineages back to the Early Pleistocene.
Conclusions
We found support for recognizing four nominal species, one undescribed species and at least other four candidate species of the genus Heleobia in northern Chile. We also suggest that the current level of species diversity of Heleobia in the region is underestimated by the use of conchological criteria to recognize species and by the limited sampling conducted to date.
Similar content being viewed by others
Background
The practical matter of species delimitation is receiving increased attention considering the high rate of extinction of species along with the huge undiscovered biodiversity in a number of taxa (Sites and Marshall 2003, 2004; Bickford et al. 2007; Wiens 2007; Puillandre et al. 2011). In the Andean Altiplano, the second highest plateau on Earth (Babeyko and Sobolev 2005), the dynamic and complex geological history of the region has produced ecosystems with a high degree of endemism (Veloso and Bustos-Obregón 1982; Dyer 2000; Vargas et al. 2004), probably related to the extensive terraces and restricted mountain ranges that have originated since the Miocene (Wörner et al. 2000; Risacher et al. 2003; Strecker et al. 2007). For example, the diversity of the killifish of the genus Orestias Valenciennes, 1839 and the lunged aquatic snails of the genus Biomphalaria Preston, 1910 is hypothesized to be principally a consequence of the fragmentation of populations during the Middle and Late Pleistocene after the regression of several paleolakes that existed in the area (Lüssen et al. 2003; Vila 2006; Vila et al. 2011, 2013; Collado et al. 2011a). West of this area, the Atacama Desert, the driest place in the world (McKay 2002; Vesilind 2003) with thousands of square miles of arid desert interrupted by occasional oases, is a hyper-arid system thought to be relatively simple ecologically and with low productivity (Noy-Meier 1973), but containing endemic species, for example, within the genus Basilichthys Girard, 1855 (Atheriniformes), that has allopatric distribution (Dyer 2000). Both regions harbor restricted and isolated hydrological systems ranging from springs on the banks of the Pacific coast along the Atacama Desert to watersheds situated at more than 4,000 m altitude in the Andes. In some of these watersheds, it is possible to find gilded snails of the genus Heleobia Stimpson, 1865 (Courty 1907; Biese 1944, 1947; Hershler and Thompson 1992; Collado et al. 2011b), subfamily Semisalsinae Giusti and Pezzoli, 1980 (Bouchet and Rocroi 2005), sometimes living in amazing conditions of water stress and habitat degradation (Collado 2012). The group is characterized by small species that live in marine, brackish, and freshwater environments, and whose shells generally are less than 10 mm in length (Biese 1944, 1947; Hershler and Thompson 1992). The species have a wide range of ecological tolerances and reproductive strategies (Marcus and Marcus 1963, 1965; Cazzaniga 1982a; Martín 2002; Neves et al. 2010; Collado and Méndez 2011), and some of them serve as intermediate hosts in the life cycles of digenean trematodes (Etchegoin and Martorelli 1997; Simões et al. 2008, 2009, 2010; Alda et al. 2010; Merlo and Etchegoin 2011). Kroll et al. (2012) studied the phylogenetic relationships of species mainly distributed in the Altiplano region and its major internal Lake Titicaca, and this is the only phylogenetic study performed in the genus Heleobia.
The systematics of the Chilean Semisalsinae fauna has been contentious at different taxonomic levels (De Francesco and Isla 2004;Cazzaniga 2011; Collado et al. 2011b). While some authors have included the species in the genus Heleobia Stimpson, 1865 (Davis et al. 1982; Hershler and Thompson 1992; Kabat and Hershler 1993), others have preferentially assigned them to the genus Littoridina Souleyet, 1852 (e.g., Pilsbry 1911; Preston 1915; Biese 1944, 1947; Haas 1955; Hubendick 1955; Stuardo 1961; Weyrauch 1963; Figueroa et al. 2003; Sielfeld 2001; Valdovinos 1999, 2006, 2008). At the species level, the knowledge of the group has not been clarified, so alpha taxonomic work is still needed (see Collado et al. 2011b).
In the Chilean Altiplano and the Atacama Desert, eight species and seven subspecies of Heleobia have been described based on conchological characters: Heleobia atacamensis (Philippi 1860, Heleobia loaensis (Biese 1947), Heleobia opachensis (Biese 1947), Heleobia striata (Biese 1944), Heleobia transitoria (Biese 1947), Heleobia ascotanensis (Courty 1907) with five subspecies, and Heleobia chimbaensis (Biese 1944) and Heleobia copiapoensis (Biese 1944) with one subspecies (Hershler and Thompson 1992; Collado et al. 2011b). The taxonomic status of these species and several previously unexamined populations distributed in the region has never been investigated. Additionally, the distribution ranges and relationships among the species have been addressed by few studies (Biese 1944, 1947; Kroll et al. 2012). Considering that these two geographical areas are difficult to access and concomitantly poorly explored, it is highly possible that the biodiversity of Heleobia is underestimated. In the present study, we use DNA sequence data of the large (16S) subunit and small subunit (12S) of ribosomal RNA mitochondrial genes and penis morphology to investigate diversity of Heleobia, using an extensive sampling covering the majority of the type localities and distribution range of the genus in the region and to test the hypotheses of nominal species proposed until now. We investigate penis morphology considering that the organ is discriminatory in Heleobia species (Gaillard and de Castellanos 1976; Cazzaniga 1980, 1982a, b; Hershler and Thompson 1992; Pons da Silva 1993; Collado et al. 2011b; Ovando and De Francesco 2011). Hubendick (1955) described the penis of H. chimbaensis from northern Chile, and Collado et al. (2011b) studied this organ in this and other species of the genus.
Methods
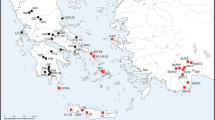
The snails were collected from macrophyta or sediment using a sieve, from 2010 to 2012 from 30 localities situated in the Chilean Altiplano and the Atacama Desert (Figure 1, Table 1). The snails were preserved in 70% to 100% ethanol. The material included specimens collected from Quebrada La Chimba, the type locality of H. chimbaensis, a ravine near Antofagasta city, Vertiente Opache, the type locality of H. opachensis in San Salvador River, and Las Cascadas, the type locality of H. loaensis in Loa River. The last two localities are situated near the city of Calama in Loa Basin. The snails were also obtained from Tilopozo in the Salar de Atacama, Chilean Altiplano, the type locality of H. atacamensis, and from Quebrada Cachina, the type locality of H. transitoria, a ravine located in the Pacific coast in the Atacama Desert. Heleobia copiapoensis, H. copiapoensis costata, and H. striata were described from the Copiapó River and from its tributary stream Ojancos, the type locality of the species (Biese 1944, 1947; we sampled the Copiapó River at four sites located over the majority of its length. We also obtained snails from two isolated springs from the Salar de Ascotán, the type locality of H. ascotanensis (see Courty 1907); Collado and Méndez 2012a). Because we assigned snails to H. ascotanensis in a previous study with samples of Spring 2 from this salt pan, we have kept this name for a set of snails obtained from this site which formed a monophyletic group. Additional internal subclades formed with samples of this site and from Spring 11 from the Salar de Ascotán were treated as Heleobia sp. Voucher specimens were deposited in the Laboratorio de Genética y Evolución (GEVOL), Facultad de Ciencias, Universidad de Chile and the malacology section of the Museo Nacional de Historia Natural, Santiago, Chile (MNHNCL).
Collection sites for the genus Heleobia in northern Chile. As S2: Salar de Ascotán (Spring 2), As S11: Salar de Ascotán (Spring 11), Br: Las Breas (Quebrada de Taltal), Ca: Las Cascadas, Car: Salar de Carcote (Spring 1), Cb: Quebrada Cascabeles, Cc: Quebrada Cachina, Ch: Chiu-Chiu, Cho: Aguada de Chorrillos, Col: Colpa, Cop: Copiapó, Copiapó River, CP: Carrera Pinto, Cz: Quebrada Carrizo (= Quebrada La Negra), Fi: La Finca, Ho: Hornitos, Copiapó River, Hum: Humedal, Copiapó River, Is: Isluga, LC: Quebrada La Chimba, Le: Quebrada El León, LP: Los Perales (Quebrada Paposo), Op: Laguna Opache, Pab: Pabellón, Copiapó River, Par: Parinacota, Pe: Peine, Tat: El Tatio, Tm: Tilomonte, Tot: El Totoral, Tp: Tilopozo, Ve: Las Vertientes, Vo: Vertiente Opache.
For morphological data, the shell of adult specimens was cleaned in 1% sodium hydroxide for 10 h to remove adhered sediment, washed in distilled water, and finally preserved in 70% ethanol. The snails were photographed at the same magnification with a Motic SMZ-168 Stereo Microscope with a Moticam 2000 (British Columbia, Canada) integrated digital camera. For the study of penis morphology, we followed the method implemented by Collado et al. (2011b), dissecting three male specimens for each population surveyed when possible. The penis was photographed using the same equipment. Considering the similarity of the characters of the shell and penis between some populations of the same species (see for instance Collado 2012), we do not show these data for all locations, but only those representatives of geographic areas or clades, or when it is required to emphasize the possibility of finding candidate species. For the molecular analyses, genomic DNA was isolated from the gill and mantle tissue of the snails fixed in 100% ethanol using the cetyltrimethyl ammonium bromide method (CTAB) (Winnepennickx et al. 1993). The mitochondrial 16S rRNA gene was amplified by polymerase chain reaction (PCR) using the primers 16Sar-L (5′-CGCCTGTTTATCAAAAACAT-3′) and 16Sbr-H (5′-CCGGTCTGAACTCAGATCACGT-3′) (see Palumbi 1996). PCR conditions were described in Collado and Méndez (2012b) for this molecular marker. The mitochondrial 12S rRNA gene was amplified by PCR using the primers L1091 (5′-AAAAAGCTTCAAACTGGGATTAGATACCCCACTAT-3′) and H1478 (5′-TGACTGCAGAGGGTGACGGGCGGTGTGT-3′) (Kocher et al. 1989). The PCR reaction cycle was 94°C for 3 min followed by 40 cycles of 94°C for 30 s, 45°C for 45 s, and 72°C for 60 s. Nucleotide sequences were obtained from the Macrogen Company (South Korea), edited in BioEdit (Hall 2001), and aligned in Clustal X (Thompson et al. 1997), with final visual inspection. Phylogenetic analyses were performed using maximum parsimony (MP) and Bayesian inference (BI) for the separate and combined analyses. Alternative analyses were performed using the neighbor-joining (NJ) method in Mega 5 (Tempe, AZ, USA) (Tamura et al. 2011). The MP analysis was performed with PAUP* 4.0 (MA, USA) (Swofford 2003) using a heuristic search with the TBR algorithm and the addition of random sequences. Characters not informative were excluded from the analyses. The statistical confidence of the nodes was evaluated using 100 bootstrap pseudoreplicates (Felsenstein 1985). The BI was performed in the MrBayes v. 3.1.2 program (Ronquist and Huelsenbeck 2003), selecting the best evolutionary model with jModelTest (Posada 2008) for each data partition based on the Bayesian information criterion. This analysis was run three times for three million generations for the combined analyses. Trees were sampled every 1,000 generations, and posterior probabilities were obtained after a burn-in period of 10%.
Divergence times were estimated using the BEAST program (version 1.5.3, GNU Lesser General Public License, Boston, MA, USA) (Drummond and Rambaut 2007). We used a nucleotide substitution rate of 1.9% per million years for the 16S gene, the average divergence rate of this gene estimated for invertebrates (Cunningham et al. 1992; Patarnello et al. 1996) and previously used in gastropods (DeJong et al. 2001; Collado and Méndez 2012b). The analysis was performed using a lognormal molecular clock, the general time reversible (GTR) substitution model plus the gamma distribution and the Yule process of speciation. Convergence of posterior distributions was analyzed in Tracer (version 1.5, GNU Lesser General Public License) (Rambaut and Drummond 2007), and the Bayesian tree was obtained after removing burn-in (10%) with TreeAnnotator (version 1.5.3, GNU Lesser General Public License).
Original 16S rRNA and 12S rRNA sequences obtained in this study were deposited in the National Center for Biotechnology Information database [GenBank: KF285827 to KF285867 for 16S; KF285868 to KF285926 for 12S] (Table 1). The 12S rRNA and 16S rRNA sequences of the Lithoglyphidae Lithoglyphus naticoides (Pfeiffer, 1828) [GenBank: AF445341 and AF445351, respectively] and the Pyrgulidae Pyrgula annulata (Linnaeus, 1758) [GenBank: AF445340 and AF445350, respectively] were used as outgroup in the molecular analyses (Hausdorf et al. 2003). The matrix used in the combined analysis included taxa for which we did not have sequences for some of the two loci; in this case, the taxa were coded as “missing data”, which has shown not to unduly influence the phylogenetic resolution (Wiens and Reeder 1995; Wiens 1998; Collado and Méndez 2012b).
Results
Morphology
With few exceptions, the morphology of the penis showed differences between populations and species of Heleobia from northern Chile (Figure 2). Conversely, the shell of the snails presented little differentiation (Figure 3), with a size always greater than 3.5 mm (and less than 8 mm), so they belong to the large-sized species group proposed in the genus (see Biese 1944, 1947).
Penis morphology of species and populations of Heleobia from northern Chile. (A) Los Perales, (B) Salar de Carcote, (C) Isluga, (D) Aguada de Chorrillos, (E) Heleobia transitoria, (F) Heleobia ascotanensis, (G) El Tatio, (H) Tilomonte, (I) Heleobia atacamensis. Left, dorsal view; right, ventral view. Scale bar = 0.5 mm.
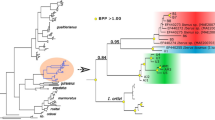
One of 100 equally most parsimonious trees obtained by the combined MP analysis. The tree was constructed using concatenated 12S and 16S mitochondrial sequences. Numbers above the nodes indicate bootstrap values obtained under the MP analysis (only values above 50% are shown), followed by the posterior probability values obtained in the BI (only values equal to or above 0.95 are shown). To the right of the figure are shown some shells of nominal species and previously unknown populations of Heleobia from northern Chile.
12S rRNA gene
We amplified 369 bases of the 12S rRNA gene from 59 Heleobia specimens, the final length of the alignment including the two outgroup sequences. The base composition was A = 0.41, C = 0.14, G = 0.16, and T = 0.29. For MP, 48 characters were informative, and 321 were excluded from the analysis. This analysis recovered 100 trees (not shown) with a length of 65 steps, consistency index = 0.83, retention index = 0.93, and rescaled consistency index = 0.78. For the BI (tree not shown), the best model of evolution for this data set was the general time reversible model (Rodríguez et al. 1990) and gamma distribution rate heterogeneity (GTR + G). The mean genetic divergence among all the populations and species of Heleobia from northern Chile was 1.20% using the two-parameter model of Kimura (1980) (K2P). The greatest distance among the nominal species occurred between H. ascotanensis and H. atacamensis (1.95%), and between the latter species and H. chimbaensis (1.95%, data not shown).
16S rRNA gene
We amplified 497 to 499 bases of the 16S rRNA gene from 41 Heleobia specimens. The alignment was 499 nucleotide sites in length considering the two outgroup sequences. The base composition was A = 0.34, C = 0.14, G = 0.19, and T = 0.33. For MP, 54 characters were informative, and 445 were excluded from the analysis. This analysis recovered two trees with the same topology (not shown) and a length of 88 steps, consistency index = 0.76, retention index = 0.89, and rescaled consistency index = 0.68. For the BI, the best evolutionary model was GTR + G. The mean genetic divergence among all populations and species of Heleobia was 1.30% using the K2P model. Genetic distance analysis was performed between Heleobia species included in the present study plus 16S sequences of the Semisalsinae taxa, Semisalsa dalmatica Radoman, 1974 [GenBank: AY676119] (Wilke 2005) and Semisalsa stagnorum Gmelin, 1791 [GenBank: JX970535] (Wilke et al. 2013) for comparative purposes (Table 2). The greatest distance occurred between H. atacamensis and H. transitoria (2.63%), followed by H. atacamensis and H. chimbaensis (2.40%). The percentage of genetic divergence between Semisalsa dalmatica and Semisalsa stagnorum was 0.43%, much lower than several pair-wise comparison values obtained in the present study (Table 2). The BEAST analyses showed that the origin of the lineages occurred in the Early Pleistocene, with an average of 2.1 million years to the most ancestral node of Heleobia.
Combined analysis
For the combined phylogenetic analyses, 5 sequences were left as missing data within the 12S rRNA and 16 within the 16S rRNA data partition for which these sequences were unavailable (the alignment data are available from the corresponding author). The data matrix included 55 Heleobia sequences and 868 characters, of which 102 were parsimony informative. This analysis recovered 100 trees (one of them shown in Figure 3) with a length of 148 steps, consistency index = 0.82, retention index = 0.92, and rescaled consistency index = 0.75. These indexes were slightly lower than those obtained with the 12S gene but considerably higher than those obtained with the 16S gene. Base frequencies were A = 0.38, C = 0.13, G = 0.17, and T = 0.31. Most of the major clades retained in the separate analyses were recovered in the combined MP analysis, with similar support values for most nodes. We used the GTR + G model to perform the combined BI analyses (tree not shown).
MP analysis of the combined data set and the BI recovered five major clades, four of which were composed exclusively of one each of the nominal species recognized in the region: H. ascotanensis, H. chimbaensis, H. transitoria, and H. atacamensis. The first clade (clade A, Figure 3) was integrated by H. atacamensis from Tilopozo and the allopatric snails from Peine and Tilomonte. This clade was inferred as the sister group of all the rest of the species and populations of Heleobia sequenced (100% bootstrap, 1.00 posterior probability). However, the sister group position of all Heleobia snails from northern Chile was occupied by a different taxon or lineage in the 12S and 16S analyses performed, suggesting that further work is needed to resolve this issue. Likewise, relationships among the remaining species were not resolved. The monophyly of the sequences of H. atacamensis and those from Peine was supported by 91% bootstrap and 0.96 posterior probability. Within this clade, the snails from Tilomonte were recovered as the sister group of these sequences with high support values (99% bootstrap, 1.00 posterior probability). The sequences of the snails from Salar de Ascotán formed a monophyletic group (clade B in Figure 3) with the sequences from the allopatric locality of El Tatio with moderate bootstrap values (67%). Within this clade, two subclades were inferred, one containing snails from Spring 2 from this salt pan (assigned to H. ascotanensis) plus a group of snails from this same spring, which was supported by 54% bootstrap, and another composed of snails from Spring 11 from Salar de Ascotán and those from El Tatio, with moderate bootstrap values (60%).
The monophyly of the sequences of H. transitoria (clade C in Figure 3) from its type locality Quebrada Cachina plus snails from Quebrada El León was supported by high bootstrap values (85%), but low posterior probability. The snails from Carrera Pinto and Aguada de Chorrillos were recovered as the sister group to this subclade but without node support.
The sequences of H. chimbaensis from the type locality Quebrada La Chimba nested in a clade together with snails from the Atacama Desert (74% bootstrap, 0.98 posterior probability) (clade D in Figure 3). Within this clade, the sequences from El Totoral formed a monophyletic group with a 78% bootstrap and high posterior probability values (0.99). The sequences of snails from Los Perales (clade red in Figure 3) formed a monophyletic group supported by 100% bootstrap and 1.00 posterior probability.
The sequences of snails from the Salar de Carcote in the Chilean Altiplano nested in a clade together with sequences from the Copiapó River but without support. The penis morphology of these snails, however, showed similarities with those of the species H. loaensis, H. opachensis, and H. atacamensis, in the latter case suggesting convergence of characters (Figure 2).
The sequences of snails from Las Cascadas, the type locality of H. loaensis and Vertiente Opache, the type locality of H. opachensis, were not resolved by the combined analysis, as well as the sequences of the snails from Chiu-Chiu, La Finca, Laguna Opache, and Las Vertientes in the Loa basin, and those from Colpa, Parinacota and Isluga in the Chilean Altiplano. However, we found the penis morphology of snails from Isluga to be different from those of other localities (Figure 2).
Discussion
The combined and separate phylogenetic analyses recovered the populations of Heleobia from Los Perales as a monophyletic group with high support values. These snails were originally assigned to H. chimbaensis by Biese (1947) based on external shell morphology, but our results show that they constitute an independent lineage phylogenetically separated from this species, and the male copulatory organ (Figure 2) is markedly different (see Hubendick 1955; Collado et al. 2011a; Collado 2012); it is apparent that a description and a new scientific name is required for this cryptic species. Apart from this clade, our molecular analyses show that the populations of Heleobia from northern Chile form four other generally well supported clades. The division of the clades suggests that a relatively long amount of time elapsed to produce genetic lineage diversification in the region, which is not reflected in the morphology of the shell (see Figure 3). This is not infrequent in rissooidean snails, a taxon with a large number of genera, many of which include species morphologically similar but genetically divergent (e.g., Liu et al. 2003; Hershler et al. 1999; Hershler et al. 2003; Bichain et al. 2007; Hershler et al. 2007; Falniowski et al. 2012). The percentage sequence divergence between the Heleobia species observed in the present study was generally greater than those found in other rissooidean taxa. For example, there is no variation between 12S sequences of the species of the family Tateidae Tatea huonensis (Tenison-Woods, 1876) and Tatea rufilabris (A. Adams, 1862) [GenBank: FJ619852 and FJ619856, respectively] (see Colgan and da Costa 2009), while divergence between the Lithoglyphidae Benedictia baicalensis (Gerstfeld) and Benedictia maxima (W. Dybowski) [GenBank: AF445349 and AF445348, respectively] (Hausdorf et al. 2003) is 0.83% (K2P), and the divergence between Potamopyrgus antipodarum (Gray, 1843) and Potamopyrgus estuarinus (Winterbourn, 1971) [GenBank: HQ875146 and GQ996415, respectively] (Neiman et al. 2010) is 2.25%. In the case of 16S sequences, the divergence between the Hydrobiidae Sulawesidrobia bonnei (Abbott, 1945) and Sulawesidrobia botak Haase and Bouchet, 2006 [GenBank: HM587413 and HM587411, respectively] (Zielske et al. 2011) is 0.19%, while the divergence between the Tateidae Potamopyrgus antipodarum and Potamopyrgus oppidanus Haase, 2008 [GenBank: AY634104 and AY634090, respectively] (Haase 2005) is 0.21%.
In the genus Heleobia, several studies, including original descriptions of species, show overlapping characters in the external morphology of the shell (e.g., Bavay 1904; Courty 1907; Pilsbry 1911, 1924; Biese 1944 1947; Haas 1955; Preston 1915; Weyrauch 1963). In fact, the Heleobia fauna from Lake Titicaca and closely related taxa from the surrounding areas form a species flock, in some cases including morphologically parallel species (Kroll et al. 2012). The clades inferred by our molecular analyses are concordant with the distribution patterns, and some of them include candidate species or new lineages within the genus not detected previously and whose copulatory organs show a particular morphology.
The Salar de Atacama contains snails from Tilopozo, Peine, and Tilomonte (clade A, Figure 3). Philippi (1860) described H. atacamensis from Tilopozo, and since then, it has not been addressed in the literature, with the exception of checklists of the species (Stuardo 1961; Sielfeld 2001; Valdovinos 1999, 2006, 2008) or the description of its penis (Collado et al. 2011a). The monophyly of the sequences of H. atacamensis from Tilopozo and Peine was well supported in the MP and BI analyses, while the sequence from Tilomonte appears as the sister to this group in all analyses performed. Likewise, the penis morphology shows differences between H. atacamensis and snails from Tilomonte (Figure 2), suggesting that this population represents a candidate species of the genus. These three localities are located on the east bank of the Salar de Atacama in the homonymous basin. Tilopozo is separated from Peine by about 20 km, while Tilomonte is located in the middle, at a distance of about 13 km from each of these locations. The Tilomonte oasis includes a small stream that dries at a distance of approximately 6.5 km of the shore of the Salar de Atacama, while the oasis of Peine is formed by a stream with a greater flow of water that flows at irregular intervals into the salt pan. It is apparent that seasonal flooding could connect Tilopozo and Peine, producing dispersal of the snails between these locations, while the partial isolation of Tilomonte could explain the divergence of these populations as a product of vicariance.
Clade B (Figure 3) clusters populations from Springs 2 and 11 in Salar de Ascotán and the allopatric snails from the locality of El Tatio. Heleobia ascotanensis was described by Courty (1907) (under the genus Paludestrina d’Orbigny, 1840) from the Salar de Ascotán together with five subspecies and two species of the genus Bythinella Moquin-Tandon, 1856 based on shell morphology (see Collado and Méndez 2012a). The monophyly of the sequences and support values suggest that two distinct lineages co-occur in Spring 2 from this salt pan, but we have no evidence of reproductive isolation between them. Additionally, the snail sequences from Spring 11 form a subclade together with the snails from El Tatio. In the 12S analysis, the separation of this subclade from the sympatric snails from Spring 2 was supported by a moderate bootstrap value in the MP analysis (66%) but with a high posterior probability value in the BI (0.97). Furthermore, the penis morphology of H. ascotanensis (Collado et al. 2011b; present study) is different from those of the snails of El Tatio (Figure 2), suggesting the occurrence of an additional candidate species in our data set. At present, we cannot establish if the snails from Spring 11 from Salar de Ascotán constitute a different species from those of El Tatio, although the support values for their monophyly recovered in the 12S separate analysis seem to indicate this. There is no evidence of subterranean or surface water connection between Spring 2 in the north of the Salar de Ascotán and Spring 11 in the south; these two springs are separated by a distance of approximately 21.5 km. Structuring of populations of the killifish Orestias ascotanensis Parenti, 1984 in Salar de Ascotán was recently reported, including those fish located in Spring 11, a process probably stimulated by habitat fragmentation occurring from the Late Pleistocene to the beginning of the Holocene (Morales et al. 2011). Keller and Soto (1998) suggested that in this salt pan, there is no evidence of major changes in the water level in the last 12,000 years. Recently, we also detected microvicariance processes structuring the populations of the aquatic gastropod Biomphalaria crequii (Courty 1907) inhabiting the southernmost springs of the system (Collado and Méndez 2013). There is no evidence either of subterranean or surface water connection between Spring 11 and El Tatio, a geyser field located 75 km to the south in a different basin.
Heleobia transitoria integrates a clade (clade C, Figure 3) with allopatric Atacama populations that are separated by arid desert. After its original description, this species has not been addressed in the literature, with the exception of local checklists. The type locality of the species, a small spring in Quebrada Cachina, is separated by approximately 151 km from the Carrera Pinto oasis, another small southwestern spring, 117 km from a small southern spring in Quebrada El León and 147 km from the southernmost spring Aguada de Chorrillos. Within this clade, the monophyly of H. transitoria and the snails from Quebrada El León was well supported in the MP 16S and the combined analyses; they are probably conspecific. The snails from Aguada de Chorrillos (and Carrera Pinto) never formed a monophyletic group with H. transitoria in the analyses performed, and the penis morphology shows conspicuous differences between them (Figure 2).
Clade D (Figure 3) clusters the sequences of H. chimbaensis from Quebrada La Chimba and snails from other Atacama localities, almost all of them with allopatric distribution. The monophyly of snails from Las Breas in Quebrada de Taltal and Quebrada Cascabeles was weakly supported in the combined analysis, but was recovered with relatively high support values in the 12S analysis (65% bootstrap, 0.97 posterior probability), suggesting the presence of a single species within this assemblage. The monophyly of the sequences from El Totoral and the Atacama populations from Copiapó River was supported by 70% bootstrap, suggesting some phylogenetic structure. This is consistent with the location of El Totoral, an intermittent stream in the Atacama Desert 63 km south of the Copiapó River. The snails disseminated within the Copiapó River (Humedal, Copiapó, Hornitos, and Pabellón) never clustered together within the subclade integrated by H. chimbaensis or another described species of the genus. The lack of monophyly suggests the presence of more than one taxon in this hydrological system, for which the names H. copiapoensis, H. copiapoensis costata, and H. striata are available (Biese 1944, 1947).
From the taxonomic point of view, Biese (1947) considered H. loaensis and H. opachensis as different species based on external shell morphology. The sequences of these species were not resolved by any analyses performed in the present study, with the exception of the NJ analysis (tree not shown), which recovered these taxa in a moderately well-supported clade (65% bootstrap). Considering that these taxa have similar shell and penis morphology (Biese 1944, 1947; Collado et al. 2011a), their validity needs to be tested using additional morphological characters or a distinct, faster-evolving DNA locus. Apart from snails from Salar de Carcote and Isluga, which probably represent other candidate species judging from penis morphology, the identity and systematic position of snails from Colpa and Parinacota in the Chilean Altiplano and La Finca, Laguna Opache, Chiu-Chiu, and Las Vertientes from the Loa basin in the Atacama Desert need to be clarified.
Heleobia carinata (Biese), a name that does not meet the criteria of availability according to the Code (International Commission on Zoological Nomenclature ICZN 1999) (see Collado et al. 2011b), was found in Pleistocene lacustrine facies in northern Chile (Biese 1961; Ochsenius 1974). Regardless of the availability of the name, these Heleobia record fossil data agree with our molecular calibration that places the origin of the Heleobia in the region in the Early Pleistocene, implying a recent origin of the group. Kroll et al. (2012) also found Pleistocene divergence using the mitochondrial cytochrome c oxidase subunit 1 (COI) gene among the Heleobia populations distributed in northern Chile and the Altiplano region.
According to our results, the speciation of Heleobia fauna in the Chilean Altiplano and the Atacama Desert is suggested as being allopatric from the Early Pleistocene. We consider the diversity of Heleobia at the species level in the region to be underestimated. We suggest that this is a consequence of the use of conchological criteria to delineate species boundaries and the limited sampling conducted to date. The discovery of a cryptic lineage in Los Perales and other candidate species in different localities in the region supports this statement.
Conclusions
The present results support the conclusion that the snails from Tilomonte, El Tatio, Aguada de Chorrillos, Isluga, and Salar de Carcote (Spring 1) represent candidate species of the genus Heleobia. The molecular analysis provides support for recognizing the nominal species H. chimbaensis, H. atacamensis, H. transitoria, and H. ascotanensis as valid taxa, while the specific status of H. opachensis and H. loaensis could not be resolved. The phylogenetic analysis and penis morphology revealed that the snails from Los Perales constitute a cryptic species of the genus. We also suggest that vicariance may largely explain the distribution patterns of Heleobia populations disseminated in the Chilean Altiplano and the Atacama Desert. A more intensive effort will be necessary to test the cryptic species boundaries thoroughly in Heleobia.
References
Alda P, Bonel N, Cazzaniga NJ, Martorelli SR: Effects of parasitism and environment on shell size of the South American intertidal mud snail Heleobia australis (Gastropoda). Estuar Coast Shelf Sci 2010, 87: 305–310. 10.1016/j.ecss.2010.01.012
Babeyko AY, Sobolev SV: Quantifying different modes of the Late Cenozoic shortening in the central Andes. Geology 2005,33(8):621–624. 10.1130/G21126.1
Bavay A: Mission de Crequi-Montfort et Senechal de la Grange en Amerique du Sud. Mollusques terrestres et fluviatilis recoltes par le Dr Neveu-Lemaire. Bull Soc Zool France 1904, 29: 152–156.
Bichain J-M, Gaubert P, Samadi S, Boisselier-Dubayle M-C: A gleam in the dark: phylogenetic species delimitation in the confusing spring-snail genus Bythinella Moquin-Tandon, 1856 (Gastropoda: Rissooidea: Amnicolidae). Mol Phyl Evol 2007, 45: 927–941. 10.1016/j.ympev.2007.07.018
Bickford D, Lohman DJ, Sohdi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I: Cryptic species as a window on diversity and conservation. Trends Ecol Evol 2007,22(3):148–155. 10.1016/j.tree.2006.11.004
Biese WA: Revisión de los moluscos terrestres y de agua dulce provistos de concha de Chile. Parte I, Familia Amnicolidae. Boletín del Museo Nacional de Historia Natural 1944, 22: 169–190.
Biese WA: Revisión de los moluscos terrestres y de agua dulce provistos de concha de Chile. Parte II, Familia Amnicolidae (continuación). Boletín del Museo Nacional de Historia Natural 1947, 23: 63–77.
Biese WA: El Jurásico de Cerritos Bayos. Instituto Geológico de la Universidad de Chile 1961, 19: 1–61.
Bouchet P, Rocroi J-P: Classification and nomenclator of gastropod families. Malacologia 2005,47(1–2):1–397.
Cazzaniga N: Nota sobre los hidróbidos Argentinos. I (Gastropoda: Rissoidea), Acerca de Littoridina occidentalis (Doering, 1884). Neotropica 1980,26(76):187–191.
Cazzaniga N: Nota sobre los hidróbidos Argentinos. 5. Conquiliometría de Littoridina parchappii (d’Orbigny, 1835) (Gastropoda Rissoidea) referida a su ciclo de vida en poblaciones Australes. Iheringia. Série Zoologia 1982a, 61: 97–118.
Cazzaniga N: Nota sobre los hidróbidos Argentinos. II (Gastropoda: Rissoidea), Una Littoridina del “grupo parchappii ” en Península Valdés (Chubut). Revista del Museo de la Plata (Nueva Serie) 13. Sección Zoología 1982b, 129: 11–16.
Cazzaniga NJ: Heleobia Stimpson, 1865: taxonomía. In El género Heleobia (Caenogastropoda: Cochliopidae). Edited by: Cazzaniga NJ. Am Moll (número especial); 2011:12–17.
Colgan DJ, da Costa P: DNA haplotypes cross species and biogeographic boundaries in estuarine hydrobiid snails of the genus Tatea . Mar Freshw Res 2009, 60: 861–872. 10.1071/MF08200
Collado GA: Nuevo registro de distribución geográfica y antecedentes de historia natural de Heleobia chimbaensis (Biese, 1944) (Caenogastropoda: Cochliopidae) en la costa del desierto de Atacama: implicancias para su conservación. Am Moll 2012,20(2):13–18.
Collado GA, Méndez MA: Estrategias reproductivas y tipos de desarrollo en especies endémicas del género Heleobia Stimpson, 1865 (Caenogastropoda: Cochliopidae) de Chile. Am Moll (número especial); 2011:67–71.
Collado GA, Méndez MA: Los taxa nominales descritos por Courty del Salar de Ascotán, Altiplano Chileno. Rev Chil Hist Nat 2012a, 85: 233–235. 10.4067/S0716-078X2012000200010
Collado GA, Méndez MA: Phylogenetic relationships and taxonomy of Altiplano populations of Biomphalaria (Gastropoda: Planorbidae): inference from a multilocus approach. Zool J Linn Soc 2012b, 165: 795–808. 10.1111/j.1096-3642.2012.00829.x
Collado GA, Méndez MA: Microgeographic differentiation among closely related species of Biomphalaria (Gastropoda: Planorbidae) from the Andean Altiplano. Zool J Linn Soc 2013, 169: 640–652. 10.1111/zoj.12073
Collado GA, Vila I, Méndez MA: Monophyly, candidate species and vicariance in Biomphalaria snails (Mollusca: Planorbidae) from the Southern Andean Altiplano. Zool Scr 2011a,40(6):613–622. 10.1111/j.1463-6409.2011.00491.x
Collado GA, Méndez MA, Letelier S, Veliz D, Sabando MC: Morfología peniana y taxonomía del género Heleobia Stimpson, 1865 en Chile junto a una revisión de los ejemplares tipo del Museo Nacional de Historia Natural de Chile. Am Moll (número especial); 2011b:49–58.
Courty G: Explorations géologiques dans l’Amerique du Sud. Mission Scientifique de G. de Crèqui Montefort et E. Sénéchal de la Grange 14:1–208. Paris: Impremiere Nationale; 1907.
Cunningham CW, Blackstone NW, Buss LW: Evolution of king crabs from hermit crab ancestors. Nature 1992, 355: 539–542. 10.1038/355539a0
Davis GM, Mazurkiewicz M, Mandracchia M: Spurwinkia : morphology, systematics, and ecology of a new genus of North American marshland Hydrobiidae (Mollusca: Gastropoda). Proc Acad Nat Sci Philad 1982, 134: 143–177.
De Francesco CG, Isla FI: The life cycle and growth of Heleobia australis (d’Orbigny, 1835) and H. conexa (Gaillard, 1974) (Gastropoda: Rissooidea) in Mar Chiquita coastal lagoon (Argentina). J Moll Stud 2004, 70: 173–178. 10.1093/mollus/70.2.173
DeJong RJ, Morgan JAT, Paraense L, Pointier J-P, Amarista M, Ayeh-Kumi PFK, Babiker A, Barbosa CS, Brémond P, Canese AP, de Souza CP, Dominguez C, File S, Gutierrez A, Incani RN, Kawano T, Kazibwe F, Kpikpi J, Lwambo NJS, Mimpfoundi R, Njiokou F, Poda JN, Sene M, Velásquez LE, Yong M, Adema CM, Hofkin BV, Mkoji GM, Loker ES: Evolutionary relationships and biogeography of Biomphalaria (Gastropoda: Planorbidae) with implications regarding its role as host of the human bloodfluke Schistosoma mansoni . Mol Biol Evol 2001, 18: 2225–2239. 10.1093/oxfordjournals.molbev.a003769
Drummond AJ, Rambaut A: BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 2007, 7: 214. 10.1186/1471-2148-7-214
Dyer BS: Revisión sistemática de los pejerreyes de Chile (Teleostei, Atheriniformes). Estud Oceanol 2000, 19: 99–127.
Etchegoin JA, Martorelli SR: Description of a new species of Maritrema (Digenea: Microphallidae) from Mar Chiquita coastal lagoon (Buenos Aires, Argentina) with notes on its life cycle. J Parasit 1997, 83: 709–713. 10.2307/3284251
Falniowski A, Szarowska M, Glöer P, Pešić V, Georgiev D, Horsák M, Sirbu I: Radiation in Bythinella Moquin-Tandon, 1856 (Mollusca: Gastropoda: Rissooidea) in the Balkans. Fol Malacol 2012,20(1):1–10. 10.2478/v10125-012-0006-2
Felsenstein J: Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985, 39: 783–791. 10.2307/2408678
Figueroa R, Valdovinos C, Araya E, Parra O: Macroinvertebrados bentónicos como indicadores de calidad de agua de ríos del sur de Chile. Rev Chil Hist Nat 2003,76(2):275–285.
Gaillard MC, de Castellanos ZA: Mollusca, Gasteropoda, Hydrobiidae. In: Ringuelet RA (ed) Fauna de agua dulce de la República Argentina. 15(2):1–39. Buenos Aires: Fundación para la Educación, la Ciencia y la Cultura (FECIC); 1976.
Haas F: XVII. Mollusca: Gastropoda. In: The Percy Sladen Trust Expedition to Lake Titicaca in 1937. Trans Linn Soc Lon 1955,1(3):275–308.
Haase M: Rapid and convergent evolution of parental care in hydrobiid gastropods from New Zealand. J Evol Biol 2005, 18: 1076–1086. 10.1111/j.1420-9101.2005.00894.x
Hall TA: BioEdit: a user-friendly biological sequence alignment editor and analysis, vers. 5.09. Raleigh, NC: Department of Microbiology, North Carolina State University; 2001.
Hausdorf B, Röpstorf P, Riedel F: Relationships and origin of endemic Lake Baikal gastropods (Caenogastropoda: Rissooidea) based on mitochondrial DNA sequences. Mol Phyl Evol 2003,26(3):435–443. 10.1016/S1055-7903(02)00365-2
Hershler R, Thompson FG: A review of the aquatic gastropod subfamily Cochliopinae (Prosobranchia: Hydrobiidae). Malacol Rev 1992, 5: 1–140.
Hershler R, Liu H-P, Mulvey M: Phylogenetic relationships within the aquatic snail genus Tryonia : implications for biogeography of the North American Southwest. Mol Phyl Evol 1999, 13: 377–391. 10.1006/mpev.1999.0659
Hershler R, Liu H-P, Thompson FG: Phylogenetic relationships of North American nymphophiline gastropods based on mitochondrial DNA sequences. Zool Scr 2003, 32: 357–366. 10.1046/j.1463-6409.2003.00115.x
Hershler R, Liu H-P, Frest TJ, Johannes EJ: Extensive diversification of pebblesnails (Lithoglyphidae: Fluminicola ) in the upper Sacramento River basin, northwestern USA. Zool J Linn Soc 2007, 149: 371–422.
Hubendick B: XVIII. The anatomy of the gastropoda. In: The Percy Sladen Trust Expedition to Lake Titicaca in 1937. Trans Linn Soc Lon 1955,1(3):309–327.
International Commission on Zoological Nomenclature (ICZN): International Code of Zoological Nomenclature. 4th edition. London: International Trust for Zoological Nomenclature; 1999:306.
Kabat AR, Hershler R: The prosobranch family Hydrobiidae (Gastropoda: Rissooidea): review of classification and supra-specific taxa. Smith Cont Zool 1993, 547: 1–94.
Keller B, Soto D: Hydrogeologic influences on the preservation of Orestias ascotanensis (Teleostei: Cyprinodontidae), in Salar de Ascotán, northern Chile. Rev Chil Hist Nat 1998, 71: 147–156.
Kimura M: A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980, 16: 111–120. 10.1007/BF01731581
Kocher TD, Thomas WK, Meyer A, Pääbo S, Villablanca FX, Wilson AC: Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci U S A 1989, 86: 6196–6200. 10.1073/pnas.86.16.6196
Kroll O, Hershler R, Albrecht C, Terrazas EM, Apaza R, Fuentealba C, Wolff C, Wilke T: The endemic gastropod fauna of Lake Titicaca: correlation between molecular evolution and hydrographic history. Ecology and Evolution 2012,2(7):1517–1530. 10.1002/ece3.280
Liu H-P, Hershler R, Clift K: Mitochondrial DNA sequences reveal extensive cryptic diversity within a western American springsnail. Mol Ecol 2003, 12: 2771–2782. 10.1046/j.1365-294X.2003.01949.x
Lüssen A, Falk TM, Villwock W: Phylogenetic patterns in populations of Chilean species of the genus Orestias (Teleostei: Cyprinodontidae): results of mitochondrial DNA analyses. Mol Phyl Evol 2003, 29: 151–160. 10.1016/S1055-7903(03)00077-0
Marcus E, Marcus E: On Brazilian supralittoral and brackish water snails. Bol Inst Ocean (São Paulo) 1963, 13: 41–52.
Marcus E, Marcus E: On Brazilian supralittoral and estuarine snails. Boletim da Faculdade de Filosofia Ciências e Letras da Universidade de São Paulo, Zoologia 1965, 25: 19–82.
Martín PR: Evidence for parthenogenesis and natural imposex in the Patagonian freshwater snail Heleobia hatcheri (Gastropoda: Hydrobiidae). J Moll Stud 2002, 68: 291–295. 10.1093/mollus/68.3.291
McKay CP: Two dry for life: the Atacama Desert and Mars. Ad Astra 2002,14(3):30–33.
Merlo MJ, Etchegoin JA: Testing temporal stability of the larval digenean community in Heleobia conexa (Mollusca: Cochliopidae) and its possible use as an indicator of environmental fluctuations. Parasitology 2011, 138: 249–256. 10.1017/S0031182010001150
Morales P, Vila I, Poulin E: Genetic structure in remnant populations of an endangered cyprinodontid fish, Orestias ascotanensis , endemic to the Ascotán salt pan of the Altiplano. Conserv Gen 2011, 12: 1639–1643. 10.1007/s10592-011-0245-6
Neiman M, Hehman G, Miller JT, Logsdon JM Jr, Taylor DR: Accelerated mutation accumulation in asexual lineages of a freshwater snail. Mol Biol Evol 2010,27(4):954–963. 10.1093/molbev/msp300
Neves RAF, Valentin JL, Figueiredo GM: Morphological description of the gastropod Heleobia australis (Hydrobiidae) from egg to hatching. Braz J Ocean 2010,58(3):247–250.
Noy-Meier I: Desert ecosystems: environment and producers. Annu Rev Ecol Syst 1973, 4: 25–51. 10.1146/annurev.es.04.110173.000325
Ochsenius C: Relaciones paleogeográficas y paleoecológicas entre los ambientes lénticos de la Puna de Atacama y el Altiplano boliviano, Trópico de Capriconio. Bol Preh Chil 1974,7(8):99–138.
Ovando XMC, De Francesco CG: El género Heleobia en el noroeste Argentino. In El género Heleobia (Caenogastropoda: Cochliopidae) en América del Sur. Edited by: Cazzaniga NJ. Am Moll (Número Especial); 2011:22–25.
Palumbi SR: Nucleic acids II: the polymerase chain reaction. In Molecular systematics. Edited by: Hillis DM, Moritz C, Mable BK. Sunderland: Sinauer Associates, Inc; 1996:205–247.
Patarnello T, Bargelloni L, Varotto V, Battaglia B: Krill evolution and the Antarctic ocean currents: evidence of vicariant speciation as inferred by molecular data. Mar Biol 1996, 126: 603–608. 10.1007/BF00351327
Philippi RA: Reise durch die Wueste Atacama auf Befehl der chilenischen Regierung im Sommer 1853–54. Halle: E Anton; 1860:62.
Pilsbry HA: Non-marine Mollusca of Patagonia. In Reports of the Princeton University Expedition to Patagonia 1896–1899. Part 5, Zoology. Edited by: Scott WB. Princeton, NJ, USA: The University of Princeton; 1911:513–633.
Pilsbry HA: South American land and freshwater mollusks. Notes and descriptions. Proc Acad Nat Sci Phila 1924, 76: 49–66.
Pons da Silva MCP: Dados morfológicos de Heleobia parchappei (Orbigny, 1835) (Prosobranchia, Hydrobiidae, Littoridininae). Iheringia, Série Zoologia 1993, 75: 81–87.
Posada D: jModelTest: phylogenetic model averaging. Mol Biol Evol 2008, 25: 1253–1256. 10.1093/molbev/msn083
Preston HB: List of the Mollusca, with descriptions of six new species. J Conchol 1915,14(10):347–352.
Puillandre N, Modica MV, Zhang Y, Sirovich L, Boisselier M-C, Cruaud C, Holford M, Samadi S: Large-scale species delimitation method for hyperdiverse groups. Mol Ecol 2011, 21: 2671–2691.
Rambaut A, Drummond AJ: Tracer v1.5. 2007. . Accessed 20 June 2013 http://beast.bio.ed.ac.uk/Tracer
Risacher F, Alonso H, Salazar C: The origin of brines and salts in Chilean salars: a hydrochemical review. Earth-Sci Rev 2003, 63: 249–293. 10.1016/S0012-8252(03)00037-0
Rodríguez F, Oliver JL, Marín A, Medina JR: The general stochastic model of nucleotide substitution. J Theor Biol 1990,142(4):485–501. 10.1016/S0022-5193(05)80104-3
Ronquist F, Huelsenbeck JP: MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19: 1572–1574. 10.1093/bioinformatics/btg180
Sielfeld W: Phylum Mollusca. Guías de identificación y biodiversidad fauna Chilena. Iquique: Apuntes de Zoología, Universidad Arturo Prat; 2001:15.
Simões SBE, Neves RFC, Santos CP: Life history of Acanthocollaritrema umbilicatum Travassos, Freitas and Bührnheim, 1965 (Digenea: Cryptogonimidae). Parasitol Res 2008, 103: 523–528. 10.1007/s00436-008-1000-x
Simões SBE, Barbosa HS, Santos CP: The life history of Pygidiopsis macrostomum Travassos, 1928 (Digenea: Heterophyidae). Mem Inst Oswaldo Cruz 2009, 104: 106–111. 10.1590/S0074-02762009000100016
Simões SB, Barbosa HS, Santos CP: The life cycle of Ascocotyle ( Phagicola ) longa (Digenea: Heterophyidae), a causative agent of fish-borne trematodosis. Acta Trop 2010, 113: 226–233. 10.1016/j.actatropica.2009.10.020
Sites JWJ, Marshall JC: Delimiting species: a Renaissance issue in systematic biology. Trends Ecol Evol 2003, 18: 462–470. 10.1016/S0169-5347(03)00184-8
Sites JW Jr, Marshall JC: Operational criteria for delimiting species. Annu Rev Ecol Evol Syst 2004, 35: 199–227. 10.1146/annurev.ecolsys.35.112202.130128
Strecker MR, Alonso RN, Bookhagen B, Carrapa B, Hilley GE, Sobel ER, Trauth MH: Tectonics and climate of the southern central Andes. Annu Rev Earth Planet Sci 2007, 35: 747–787. 10.1146/annurev.earth.35.031306.140158
Stuardo J: Contribución a un catálogo de los moluscos chilenos de agua dulce. Gayana 1961, 1: 7–32.
Swofford DL: PAUP*: phylogenetic analysis using parsimony (*and other methods). ver. 4. Sunderland: Sinauer Associates; 2003.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S: MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011,28(10):2731–2739. 10.1093/molbev/msr121
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG: The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acid Res 1997, 25: 4876–4882. 10.1093/nar/25.24.4876
Valdovinos C: Biodiversidad de moluscos Chilenos: base de datos taxonómica y distribucional. Gayana 1999,63(2):111–164.
Valdovinos C: Estado de conocimiento de los gastrópodos dulceacuícolas de Chile. Gayana 2006,70(1):88–95.
Valdovinos C: Invertebrados dulceacuícolas, CONAMA. Biodiversidad de Chile, patrimonio y desafíos: 201–225. Santiago (Chile): Ocho Libros Editores; 2008:640.
Vargas C, Acuña P, Vila I: Relación entre la calidad del agua y la biota en la cuenca Salar de Huasco. In El agua en Iberoamérica: experiencias en gestión y valoración del agua. Edited by: Fernández A, Sánchez V. Buenos Aires: CYTED-XVII, Programa Iberoamericano de Ciencia y Tecnología para el Desarrollo; 2004:145–152.
Veloso A, Bustos-Obregón E: El ambiente natural y las poblaciones humanas de los Andes del norte de Chile (Arica, Lat. 18° 28′ S), vol 1: la vegetación y los vertebrados inferiores de los pisos altitudinales entre Arica y el Lago Chungará. Montevideo: Proyecto MAB-6, UNEP-UNESCO 1105–77–01, ROSTLAC; 1982.
Vesilind PJ: The driest place on Earth. National Geographic Magazine. 2003. . Accessed 18 May 2013 http://ngm.nationalgeographic.com/ngm/0308/feature3/
Vila I: A new species of killifish in the genus Orestias (Teleostei: Cyprinodontidae) from the southern high Andes, Chile. Copeia 2006, 3: 472–477.
Vila I, Scott S, Mendez MA, Valenzuela F, Iturra P, Poulin E: Orestias gloriae , a new species of cyprinodontid fish from saltpan spring of the southern high Andes (Teleostei: Cyprinodontidae). Ichthyol Explor Freshwaters 2011,22(4):345–353.
Vila I, Morales P, Scott S, Poulin E, Véliz D, Harrod C, Méndez MA: Phylogenetic and phylogeographic analysis of the genus Orestias (Teleostei; Cyprinodontidae) in the Southern Chilean Altiplano: the relevance of ancient and recent divergence processes in speciation. J Fish Biol 2013, 82: 927–943. 10.1111/jfb.12031
Weyrauch W: Cuatro nuevas especies de Hydrobiidae de Argentina y Perú. Acta Zool Lilloana 1963, 19: 243–259.
Wiens JJ: Does adding characters with missing data increase or decrease phylogenetic accuracy? Syst Biol 1998, 47: 625–640. 10.1080/106351598260635
Wiens JJ: Species delimitation: new approaches for discovering diversity. Syst Biol 2007, 56: 875–878. 10.1080/10635150701748506
Wiens JJ, Reeder TW: Combining data sets with different numbers of taxa for phylogenetic analysis. Syst Biol 1995, 44: 548–558.
Wilke T, Haase M, Hershler R, Liu H-P, Misof B, Ponder W: Pushing short DNA fragments to the limit: phylogenetic relationships of ‘hydrobioid’ gastropods (Caenogastropoda: Rissooidea). Mol Phyl Evol 2013,66(3):715–736. 10.1016/j.ympev.2012.10.025
Winnepennickx B, Backeljau T, De Wachter R: Extraction of high molecular weight DNA from molluscs. Trends Genet 1993, 9: 407.
Wörner G, Hammerschmidt K, Henjes-Kunst F, Lezaun J, Wilke H: Geochronology (40Ar/39Ar, K-Ar and He-exposure ages) of Cenozoic magmatic rocks from Northern Chile (18–22°S): implications for magmatism and tectonic evolution of the central Andes. Rev Geol Chile 2000, 27: 205–240.
Zielske S, Glaubrecht M, Haase M: Origin and radiation of rissooidean gastropods (Caenogastropoda) in ancient lakes of Sulawesi. Zool Scr 2011,40(3):221–237. 10.1111/j.1463-6409.2010.00469.x
Acknowledgments
This study was supported by a grant from Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT, 3110072) to G. Collado. We also thank Claudio Correa, Francis Miño, and Cristian Araya for specimen collection, and the anonymous reviewers who improved the original manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GAC participated in the field trips for sampling, obtained the sequences, performed the analyses, and wrote the manuscript. MAV performed the morphological study, made the figures and reviewed the manuscript. MAM made intellectual contributions, reviewed the manuscript and provided technical and financial support. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Collado, G.A., Valladares, M.A. & Méndez, M.A. Hidden diversity in spring snails from the Andean Altiplano, the second highest plateau on Earth, and the Atacama Desert, the driest place in the world. Zool. Stud. 52, 50 (2013). https://doi.org/10.1186/1810-522X-52-50
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1810-522X-52-50