Abstract
Background
Dengue fever is the most important arthropod born viral disease of public health significance. Although most patients suffer only from flu-like symptoms, a small group of patient experiences more severe forms of the disease. To contribute to a better understanding of its pathogenesis this study aims to identify proteins differentially expressed in a pool of five viremic plasma from severe dengue patients relative to a pool of five non-severe dengue patients.
Results
The use of Isotope Coded Protein Labeling (ICPLTM) to analyze plasma depleted of twenty high-abundance proteins allowed for the identification of 51 differentially expressed proteins, which were characterized by mass spectrometry. Using quantitative ELISA, three of these proteins (Leucine-rich glycoprotein 1, Vitamin D binding-protein and Ferritin) were confirmed as having an increased expression in a panel of severe dengue plasma. The proteins identified as overexpressed by ICPLTM in severe dengue plasma involve in clear up action after cell injury, tissue coherence and immune defense.
Conclusion
This ICPLTM study evaluating differences between acute severe dengue plasmas and acute non-severe dengue plasmas suggests that the three proteins identified are overexpressed early in the course of the disease. Their possible use as biomarkers for the prognostic of disease severity is discussed.
Similar content being viewed by others
Background
Dengue virus (DV) is a tropical mosquito-born flavivirus (family Flaviviridae) which infects an estimated 50 to 100 million people per year in tropical and subtropical areas. After an incubation period of 3–7 days, symptoms start suddenly and follow three phases: the febrile/acute phase, the critical phase and the recovery phase. Clinical manifestations of the febrile phase include biphasic fever, body pain, maculopapular rash and minor hemorrhages [1, 2]. For a small proportion of patients, essentially children and young adults, the critical phase corresponds to the appearance of more severe symptoms, the dengue hemorrhagic fever (DHF), with or without Dengue shock syndrome (DSS). The pathogenesis of DHF/DSS has not been fully elucidated although several hypotheses have been proposed [3].
Immune components, and in particular antibodies, have been associated with pathogenicity through several mechanisms. Antibody-dependent enhancement of a secondary infection, immune complex formation and complement activation, cross-reactivity with proteins such as endothelial cells or coagulation proteins, inflammatory activation or apoptosis are the mechanisms proposed for antibody-mediated immunopathogenesis [4–7]. Moreover, several authors have reported that antibodies also play a central role in allergic processes leading to hemorrhagic syndrome [8, 9].
Other immune components including memory T-cells, innate immunity effectors and complement factors have been shown to modulate the outcome as well [10, 11]. The dengue non structural protein 1 (NS1) can also participate to the pathogenicity in cooperation with the complement proteins [10].
No specific antiviral treatment for DF and DHF exists at the moment. Available therapies are symptomatic and are administered to correct and control the clinical manifestations of hemorrhages and shock. Early diagnosis of virus infection and successful prognosis of DF complications are essential to adjust patient management. Commonly used laboratory diagnosis methods consist of virus isolation, viral nucleic acid amplification, detection of viral antigen (NS1) or detection of anti-dengue antibodies (IgG and/or IgM). No test to monitor and predict disease severity and outcome is currently available.
The identification of proteins specifically present in plasma before DHF/DSS symptoms appearance may lead to the development of new biomarkers for prognosis. Quantitative or semi-quantitative proteomic approaches such as SILAC (Stable-Isotope Labeling by Amino acids in Cell culture) or ICPLTM (Isotope Coded Protein Labeling TM) are potentially useful to find pattern of protein markers differentially expressed for a given disease [12, 13].
In the present study, ICPLTM has been applied to profile the protein expression differences existing between non-severe (DF) and severe dengue (SD) plasma samples taken early in the course of the disease. Quantitative ELISA were performed to confirm the proteomic data. The identification of these markers might contribute to improve prognosis of severe dengue and also patient care and management.
Results
Preparation and controls of sample pools for ICPLTM
Two groups of five plasma specimens (28 μL each; total volume of 140 μL), representative either of DF or SD, were constituted. The composition of each group is summarized in Table 1. All plasma came from subjects identified as having a secondary infection to dengue virus serotype 2 (DV2) and were taken between the onset and the defervescence, which corresponds to the beginning of the critical phase. The criteria used to determine that the infections were secondary were both based on the patient medical record and, when possible, on the immunoglobulin M (IgM)/IgG specific level. All samples were both positive for the presence of virus and for the presence of NS1 protein, indicating they were taken during the acute phase of the disease. For some of the plasma samples, only a low level of IgM was found. The sex ratio (M/F=3/2), the age (9–14 years old), and the collection day after onset (2–5 days) were similar between the two groups. All the SD patients were hospitalized and presented signs of plasma leakage and hemorrhages [3]. No comorbidity was reported.
After depletion of the twenty most abundant proteins using immunodepletion columns (Proteoprep20, Sigma), each pool was controlled on polyacrylamide gel after an EZBlue dying. As an example, the depletion of the serum albumin was estimated around 98% (data not shown).
Identification of differentially expressed proteins by ICPLTM and MS/MS
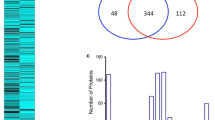
In ICPL analysis, the relative expression is measured using the H/L ratio which corresponds to the global average ratio of peptides labelled with heavy (H) or light (L) reagent, as explained in the Material and Methods section. Among the 124 proteins identified by mass spectrometry after ICPLTM experiment, only 51 had a H/L ratio calculated with a minimum of two peptides. For another group of 18 proteins, the H/L ratio was calculated with an unique peptide. For these proteins the results were considered as not significant. For a third group of 55 proteins, no information on the relative quantity was available. These two last groups were consequently excluded from subsequent analysis.
The results obtained for the 51 proteins with reliable data are summarized on the Table 2. An average H/L ratio (cf. Average column) higher than 1 meant that the signal obtained on SD pool was higher than the signal monitored for the DF pool. A ratio lower than 1 meant that the signal was higher for DF samples. The mean (m) of all these ratios and the corresponding standard deviation (s) were 1.4 and 0.83, respectively. Only four proteins (Leucine-rich alpha-2 glycoprotein 1, Galectin 3 Binding-Protein, C-Reactive Protein and the Ferritin Light-chain) had an average H/L ratio higher than 2.23, corresponding to (m+1 s). However, in order to increase the probability to find potential biomarkers for severe dengue, we choose to include three additional proteins (Vitamin D Binding-Protein, Afamin and Fibronectin). For these proteins, the average ratio was higher than 1.6. The Ferritin Light chain with an average ratio of 5.75 had the highest value. Quite the opposite, three proteins, Peroxiredoxin-2, Haptoglobin and Complement component C7 had an average ratio lower than 0.57, corresponding to (m-1 s). Haptoglobin and Complement component C7 were two contaminant proteins, imperfectly removed by the kit of depletion. Thus, the Peroxiredoxin-2 was the unique protein which had a ratio lower than 0.57. Finally, the selected proteins were Peroxyredoxin-2, Vitamin D Binding-Protein (VitDBP), Afamin, Fibronectin, Leucine-rich alpha-2 glycoprotein 1 (LRG1), Galectin 3 Binding-Protein, C-Reactive Protein and Ferritin Light-chain. These proteins, with an average ratio higher than 1.6 or lower than 0.57, represented 20% of the 51 selected proteins. The Table 2 also gave the standard deviation (cf. Max. average column). This standard deviation corresponded to a percentage generally included between 10 and 30% of the average ratio, even though sometimes it was higher: 35% for Haptoglobin, 40-45% for Apolipoprotein A, VitDBP and LRG1, or more for Complement factor H-related protein or Histidine-rich glycoprotein. The seven selected proteins were classified into several categories based both on their differential expression and on their functional significance (Table 3). They belong to a variety of functional categories (transport protein, inflammatory protein, adhesion, cell organisation and interaction). Interactions have been identified between G3BP and Peroxiredoxin 2 or LRG1 and Fibronectin using the STRING protein-protein interactions database ( http://string-db.org). All are soluble proteins either located in the cytosol or secreted.
Despite depletion of the twenty most abundant proteins, Fibrinogen, Serum Albumin, Immunoglobulins and Complement proteins were also identified. However, for these proteins the H/L average ratio was not considered as significant.
Validation of differentially expressed proteins by quantitative ELISA
To confirm the ICPLTM data, specific ELISA were performed using individual plasma samples from SD or DF patients. For each pathology, between 12 and 20 specimens were tested. Testing was also performed on three healthy plasmas (not dengue virus-infected). All dengue samples corresponded to secondary dengue infections and had been taken between onset and defervescence (between days 2 and 5) from patients infected with various DV serotypes (DV1, DV2 and DV3). The patients were between 4 and 16 years old. The M/F sex ratios for DF and SD samples were 5/12 and 7/16 respectively. These samples were both controlled positive for the presence of virus by RT-PCR and for the NS1 protein by a commercial ELISA (NS1 PlateliaTM). The SD patients were hospitalized and presented signs of plasma leakage and hemorrhages. No comorbidity was reported.
For Peroxiredoxin-2 detection, a specific ELISA was set-up in-house. Six commercial quantitative ELISA were used for the other proteins. For Peroxiredoxin, Afamin, Galectin-3 Binding Protein and C-Reactive Protein, no significant difference was found between DF and SD samples (data not shown). For VitDBP, Ferritin, LRG1, the results were illustrated in the Figure 1. These results showed that for these proteins a difference in the expression level existed and was significantly higher in SD patients.
Comparison of protein concentration detected by quantitative ELISA. Acute plasma from classical dengue patient (DF), acute plasma from severe dengue patients (DHF) or healthy plasma from non-dengue patients (HLTY) were tested by ELISA for (a) VitDBP, (b) LRG1 and (c) Ferritin. Each value corresponds to the mean of two experiments in duplicate.
Discussion
Dengue is an important health problem in tropical and sub-tropical areas. Although the pathogenesis of the disease has been studied intensively, some aspects remain not well understood. Genome-wide expression profile analysis on microarrays or 2D-electrophoresis protein difference analysis in combination with MALDI-TOF/TOF Mass Spectrometry have been used to search for biomarkers that could serve as prognostic tools or therapeutic targets for severe dengue. Proteins belonging to the complement pathway or involving in processes such as inflammation, signal transduction or translation/transcription have been identified but there is no clear consensus on their potential use in the severity prognostic [14–17]. More recently, Brasier et al. [18] described a new pre-fractionation method on acute dengue Venezuelan plasmas followed by a 2D-gel electrophoresis in tandem with LC-MS/MS and a non-parametric analysis. Previously unidentified biomarkers having biological significance in process underlying severe dengue such as capillary leakage or hepatic injury, have been found.
The plasma, which corresponds to the unclotted blood after removal of cellular components, has constant intimacy with different body parts and contains proteins released by diseased tissues [19]. The comparison of circulating proteins in plasma from patients developing different aspects of a same disease may lead to the discovery of biomarkers for diagnosis, prognosis and disease monitoring [20]. The present study aims to identify differences in circulating proteins during different degrees of severity of dengue virus acute infections.
Stable isotope protein labeling combined with Mass Spectrometry is a powerful tool to identify and relatively quantify proteins within complex protein mixtures such as tissue extracts or body fluids. The ICPLTM technique allows high-throughput quantitative proteome profiling on a global scale. It is used for protein profile comparison [21, 22].
Here, ICPLTM was used to compare mild and severe dengue plasmas for the identification of differentially expressed proteins. To increase the chance of finding low-abundance proteins the study was conducted on plasma depleted of the twenty most abundant proteins. For this purpose, it has previously been proven that immune-capture columns are useful [23]. However, the higher the number of depleted proteins is, the more important the drawback in removing associated proteins and peptides is. This drawback could have an impact on the list of identified markers of severity [23].
For each pathology, the use of pool of five plasma specimen homogeneous in age, sex ratio, days of collect after onset of symptoms and serotype would limit the ICPL identification of idiotypic markers specific of each individual plasma sample. It would also promote the discovery of markers that should be common to all samples, such as severe dengue markers. Although sometimes criticized [24], the use of a pool instead of several separate samples for proteomic analysis has been reported in recent studies [25, 26].
Three proteins, identified by mass spectrometry, were confirmed by ELISA to have significant higher expression levels in SD plasmas. These proteins were Leucine-Rich alpha-2 Glycoprotein 1, Ferritin and Vitamin D Binding-Protein.
The physiological function of LRG1 is still unknown. In 2002, O'Donnell et al. [27] have suggested a physiological role in neutrophilic granulocytes. In 2006, Cummings et al. [28] identified LRG1 as cytochrome c inhibitor in sera. More recently, Codina et al. [29] have shown that LRG1 protects against Cytc-induced lymphotoxicity. LRG1 has also been shown to be involved in protein-protein interaction, signal transduction, cell adhesion and development. LRG1 was identified as a serum biomarker that identifies patients with heart failure [30]. In the literature, LRG1 has never been associated to the dengue severity. Interestingly, LRG1 has been shown to interact with the Fibronectin also identified in the present study as being over-expressed in SD plasmas [31].
VitDBP is a 52 kDa protein that binds to vitamin D as well as its plasma metabolites and transports them to target tissues. VitDBP binds monomeric actin in addition to Vitamin D. The protein forms three domains. The structure of VitDBP is similar to the Human Serum Albumin [32, 33]. It is presumed that the function of VitDBP binding actin is to clear up any actin that enters the blood stream as a result of cell injury. Its affinity for actin monomers is high [34] and binding involves residues from all three domains [32, 33]. VitDBP also participates to macrophage activation and chemotaxis. The protein has been already associated to the pathogenesis of dengue by a Two-dimensional difference gel electrophoresis (DiGE) analysis on Brazilian plasma specimens [14].
Ferritin is a globular protein which serves to store iron in a non-toxic form, and to transport it to areas where it is required [35]. Free iron is toxic to cells because it catalyzes the formation of free radicals from reactive oxygen species [36]. Within cells, iron is stored in a protein complex as ferritin or hemosiderin. Ferritin concentrations increase drastically in the presence of an infection or with cancer [37]. The inflammatory response may cause ferritin to migrate from the plasma to the cells in order to deny iron to the infectious agent [37]. The concentration of ferritin has also been shown to increase in response to stresses such as anoxia [38]. In a previous study, high serum ferritin levels have been demonstrated by ELISA to be a biomarker of dengue hemorrhagic fever in Thai children [39]. Interestingly, proteins involved in inflammatory process have often been identified as potential markers of dengue severity [14–17].
All these proteins are related to several functions: transportation, adhesion and iron storage and seem to participate in the preservation of the body homeostasis during dengue infection.
Conclusions
This is the first report of a study using ICPLTM to evaluate differences between acute severe dengue plasma and acute non-severe dengue plasma. Among 8 proteins found having significant differential expression, we confirmed by ELISA that 3 proteins (Leucine-Rich alpha-2 Glycoprotein 1, Ferritin and Vitamin D Binding-Protein) were present at higher concentrations in SD samples early in the course of the disease. The contribution of these proteins to disease evolution and final outcome needs to be further investigated. Interestingly and more importantly the study suggests that these proteins could be used as potential markers for dengue severity and could be part of a serological assay as an aid for better patient stratification and disease management. Moreover, it will be necessary to validate these biomarkers with samples from other cohorts coming from different parts of the globe where dengue is endemic.
Methods
Plasma specimens
Plasma samples from secondary dengue-virus serotype 2 (DV2) infection were obtained from the Institut Louis Malardé, Papeete, Tahiti (French Polynesia). These samples were collected from patients being part of retrospective studies reviewed and approved by the local Medical Ethic Committee in compliance with the ethical standards of the Declaration of Helsinki. Alternatively, other plasma samples of mixed serotypes (DV1, DV2 and DV3) coming from the same retrospective studies, were used in ELISA. All the samples were taken between the onset of symptoms and the defervescence. Dengue-negative plasma specimens were obtained from healthy donors through the National French Bloodbank (Etablissement Français du Sang, Lyon, France). Dengue samples were tested for the presence of the viral NS1 protein with the PlateliaTM ELISA following the manufacturer’s instructions (BioRad, Marnes-la-Coquette, France).
Depletion of high-abundance plasma proteins
The twenty most abundant plasma protein were removed from 140 μL (5 plasmas × 28 μL) of the plasma pools using ProteoPrep20 Plasma immunodepletion Kit (Sigma Aldrich, Saint-Louis, USA) as recommended by the manufacturer. After filtration on a 0,2 μM device, the total proteins were quantified using the protein assay (Bio-Rad, Marnes-la-Coquette, France).
ICPLTM and protein identification by mass spectrometry
The ICPLTM technique allows the comparative analysis of complex samples by an isotopic labeling of the intact proteins. ICPLTM and protein identification have been conducted by InnovaProteomic (Rennes, France). Before ICPLTM-labeling, acetone precipitation of proteins from either DF or DHF depleted pool of plasma was carried out. To one volume of sample 5 volumes of acetone cooled to −20°C were added and incubated overnight at −20°C. Precipitated proteins were pelleted by centrifugation 15 min at 15,000 g at 4°C. Pellets were washed with cold 80% (v/v) acetone and dried. The protein concentration, previously determined by a Bradford assay, was used to adjust sample concentration at 3 mg/ml in ICPLTM lysis buffer containing 6 M Guanidine-HCl pH8.5 (SERVA, ICPLTM Kit). For equal amounts of proteins (60 μg for each sample), cysteines were alkylated by addition of 0.4 M of iodoacetamide and differentially labeled using the ICPLTM kit containing 12C- (light labeling) and 13C6–Nicotinoyloxysuccinimide (heavy labeling) from Serva Electrophoresis GmbH (Heidelberg, Germany) as described by the manufacturer [40]. Thereafter the two ICPLTM labeled samples were combined and proteins were precipitated by acetone to remove excess labeling reagent. Proteins were solubilized in Laemmli sample buffer, separated on 12% SDS-PAGE (GeBa, Interchim) in duplicate and stained with EZblue (Sigma Aldrich, Saint Louis, USA). After staining, the gel lanes were cut into 20 pieces and in-gel digestion by Trypsin was performed overnight at 37°C, according to the conventional method [41]. Finally, the peptides were extracted with 50% (v/v) acetonitrile containing 0.1% of formic acid [42]. The extracts were evaporated using a vacuum concentrator. The dried peptide samples were stored at −80°C until analysis.
Peptide separation was done by liquid chromatography using a nanoLC Ultimate system (Dionex GmbH, Idstein, Germany) coupled to an ion trap Esquire HCT ultra (Bruker Daltonics GmbH, Bremen, Germany). Mass spectra were recorded and the quantification was operated by the WARP-LC 1.1 software. To identify proteins, searches against the MSDB database (version 2007, 148 210 sequences for human) using Mascot 2.3 software (Matrix Science, in-house server) were carried out via Biotools software (Bruckner Daltonic). Proteins were identified with a score higher than 36 (p<0.05).
In order to eliminate false matches and incorrect protein identification, search on the SwissProt-Trembl_Decoy database was performed using the Mascot 2.3 software. The IRMa software [43] was used to filter the results so that the rate of false positives was lower than 1%.
The change in protein abundance was expressed by the calculation of a ratio between the two type of labeled peptides (heavy and light). To calculate the heavy/light (H/L) ratios of a protein at least two unique peptides labeled with heavy and light reagents were chosen respectively and a global average ratio was determined. For each result, standard deviation was calculated.
Three independent analytical replicates were performed for each 1D-gel band.
Quantitative ELISA
Protein quantities were measured twice in duplicate on each DF or SD plasma specimens using commercial ELISA. Human Ferritin and LRG1 kits were purchased from IBL international (Hamburg, Germany) and the Human Vitamin D binding–protein from USCN Inc. (Wuhan, China). Protocols were performed as recommended by the manufacturers. For each test a standard curve was established with serial dilution of calibrator in order to determine the protein concentration. Statistical analysis were performed using the Student t test and the GraphPadPrism 4.03 software.
References
Bedin F: [Brazil, land of choice for arbovirosis?]. Med Trop (Mars) 2007, 67: 281–287.
Kittigul L, Pitakarnjanakul P, Sujirarat D, Siripanichgon K: The differences of clinical manifestations and laboratory findings in children and adults with dengue virus infection. J Clin Virol 2007, 39: 76–81. 10.1016/j.jcv.2007.04.006
WHO: Guidelines for diagnosis, treatment, prevention and control. Geneva: World Health Organization; 2009.
Falconar AK: The dengue virus nonstructural-1 protein (NS1) generates antibodies to common epitopes on human blood clotting, integrin/adhesin proteins and binds to human endothelial cells: potential implications in haemorrhagic fever pathogenesis. Arch Virol 1997, 142: 897–916. 10.1007/s007050050127
Halstead SB, Suaya JA, Shepard DS: The burden of dengue infection. Lancet 2007, 369: 1410–1411. 10.1016/S0140-6736(07)60645-X
Lin CF, Chiu SC, Hsiao YL, Wan SW, Lei HY, Shiau AL, Liu HS, Yeh TM, Chen SH, Liu CC, Lin YS: Expression of cytokine, chemokine, and adhesion molecules during endothelial cell activation induced by antibodies against dengue virus nonstructural protein 1. J Immunol 2005, 174: 395–403.
Malasit P: Complement and dengue haemorrhagic fever/shock syndrome. Southeast Asian J Trop Med Public Health 1987, 18: 316–320.
Figueiredo MA, Rodrigues LC, Barreto ML, Lima JW, Costa MC, Morato V, Blanton R, Vasconcelos PF, Nunes MR, Teixeira MG: Allergies and diabetes as risk factors for dengue hemorrhagic fever: results of a case control study. PLoS Negl Trop Dis 2010, 4: e699. 10.1371/journal.pntd.0000699
Koraka P, Suharti C, Setiati TE, Mairuhu AT, Van Gorp E, Hack CE, Juffrie M, Sutaryo J, Van Der Meer GM, Groen J, Osterhaus AD: Kinetics of dengue virus-specific serum immunoglobulin classes and subclasses correlate with clinical outcome of infection. J Clin Microbiol 2001, 39: 4332–4338. 10.1128/JCM.39.12.4332-4338.2001
Avirutnan P, Punyadee N, Noisakran S, Komoltri C, Thiemmeca S, Auethavornanan K, Jairungsri A, Kanlaya R, Tangthawornchaikul N, Puttikhunt C, et al.: Vascular leakage in severe dengue virus infections: a potential role for the nonstructural viral protein NS1 and complement. J Infect Dis 2006, 193: 1078–1088. 10.1086/500949
de Kruif MD, Setiati TE, Mairuhu AT, Koraka P, Aberson HA, Spek CA, Osterhaus AD, Reitsma PH, Brandjes DP, Soemantri A, van Gorp EC: Differential gene expression changes in children with severe dengue virus infections. PLoS Negl Trop Dis 2008, 2: e215. 10.1371/journal.pntd.0000215
Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M: Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat Methods 2010, 7: 383–385. 10.1038/nmeth.1446
Lottspeich F, Kellermann J: ICPL labeling strategies for proteome research. Methods Mol Biol 2011, 753: 55–64. 10.1007/978-1-61779-148-2_4
Albuquerque LM, Trugilho MR, Chapeaurouge A, Jurgilas PB, Bozza PT, Bozza FA, Perales J, Neves-Ferreira AG: Two-dimensional difference gel electrophoresis (DiGE) analysis of plasmas from dengue fever patients. J Proteome Res 2009, 8: 5431–5441. 10.1021/pr900236f
Devignot S, Sapet C, Duong V, Bergon A, Rihet P, Ong S, Lorn PT, Chroeung N, Ngeav S, Tolou HJ, et al.: Genome-wide expression profiling deciphers host responses altered during dengue shock syndrome and reveals the role of innate immunity in severe dengue. PLoS One 2010, 5: e11671. 10.1371/journal.pone.0011671
Loke P, Hammond SN, Leung JM, Kim CC, Batra S, Rocha C, Balmaseda A, Harris E: Gene expression patterns of dengue virus-infected children from nicaragua reveal a distinct signature of increased metabolism. PLoS Negl Trop Dis 2010, 4: e710. 10.1371/journal.pntd.0000710
Thayan R, Huat TL, See LL, Tan CP, Khairullah NS, Yusof R, Devi S: The use of two-dimension electrophoresis to identify serum biomarkers from patients with dengue haemorrhagic fever. Trans R Soc Trop Med Hyg 2009, 103: 413–419. 10.1016/j.trstmh.2008.12.018
Brasier AR, Garcia J, Wiktorowicz JE, Spratt HM, Comach G, Ju H, Recinos A 3rd, Soman K, Forshey BM, Halsey ES, et al.: Discovery proteomics and nonparametric modeling pipeline in the development of a candidate biomarker panel for dengue hemorrhagic fever. Clin Transl Sci 2012, 5: 8–20. 10.1111/j.1752-8062.2011.00377.x
Issaq HJ, Xiao Z, Veenstra TD: Serum and plasma proteomics. Chem Rev 2007, 107: 3601–3620. 10.1021/cr068287r
Omenn GS: Exploring the human plasma proteome. Proteomics 2005, 5: 3223–3225. 10.1002/pmic.200590056
Jochim N, Gerhard R, Just I, Pich A: Impact of clostridial glucosylating toxins on the proteome of colonic cells determined by isotope-coded protein labeling and LC-MALDI. Proteome Sci 2011, 9: 48. 10.1186/1477-5956-9-48
Kellermann J: ICPL–isotope-coded protein label. Methods Mol Biol 2008, 424: 113–123. 10.1007/978-1-60327-064-9_10
Roche S, Tiers L, Provansal M, Seveno M, Piva MT, Jouin P, Lehmann S: Depletion of one, six, twelve or twenty major blood proteins before proteomic analysis: the more the better? J Proteomics 2009, 72: 945–951. 10.1016/j.jprot.2009.03.008
Levin Y, Bahn S: Quantification of proteins by label-free LC-MS/MS. Methods Mol Biol 2010, 658: 217–231. 10.1007/978-1-60761-780-8_13
Liu C, Zhang N, Yu H, Chen Y, Liang Y, Deng H, Zhang Z: Proteomic analysis of human serum for finding pathogenic factors and potential biomarkers in preeclampsia. Placenta 2011, 32: 168–174. 10.1016/j.placenta.2010.11.007
Pan S, Chen R, Crispin DA, May D, Stevens T, McIntosh MW, Bronner MP, Ziogas A, Anton-Culver H, Brentnall TA: Protein alterations associated with pancreatic cancer and chronic pancreatitis found in human plasma using global quantitative proteomics profiling. J Proteome Res 2011, 10: 2359–2376. 10.1021/pr101148r
O'Donnell LC, Druhan LJ, Avalos BR: Molecular characterization and expression analysis of leucine-rich alpha2-glycoprotein, a novel marker of granulocytic differentiation. J Leukoc Biol 2002, 72: 478–485.
Cummings C, Walder J, Treeful A, Jemmerson R: Serum leucine-rich alpha-2-glycoprotein-1 binds cytochrome c and inhibits antibody detection of this apoptotic marker in enzyme-linked immunosorbent assay. Apoptosis 2006, 11: 1121–1129. 10.1007/s10495-006-8159-3
Codina R, Vanasse A, Kelekar A, Vezys V, Jemmerson R: Cytochrome c-induced lymphocyte death from the outside in: inhibition by serum leucine-rich alpha-2-glycoprotein-1. Apoptosis 2010, 15: 139–152. 10.1007/s10495-009-0412-0
Watson CJ, Ledwidge MT, Phelan D, Collier P, Byrne JC, Dunn MJ, McDonald KM, Baugh JA: Proteomic analysis of coronary sinus serum reveals leucine-rich alpha2-glycoprotein as a novel biomarker of ventricular dysfunction and heart failure. Circ Heart Fail 2011, 4: 188–197. 10.1161/CIRCHEARTFAILURE.110.952200
Saito K, Tanaka T, Kanda H, Ebisuno Y, Izawa D, Kawamoto S, Okubo K, Miyasaka M: Gene expression profiling of mucosal addressin cell adhesion molecule-1+ high endothelial venule cells (HEV) and identification of a leucine-rich HEV glycoprotein as a HEV marker. J Immunol 2002, 168: 1050–1059.
Head JF, Swamy N, Ray R: Crystal structure of the complex between actin and human vitamin D-binding protein at 2.5 A resolution. Biochemistry 2002, 41: 9015–9020. 10.1021/bi026054y
Otterbein LR, Cosio C, Graceffa P, Dominguez R: Crystal structures of the vitamin D-binding protein and its complex with actin: structural basis of the actin-scavenger system. Proc Natl Acad Sci U S A 2002, 99: 8003–8008. 10.1073/pnas.122126299
McLeod JF, Cooke NE: The vitamin D-binding protein, alpha-fetoprotein, albumin multigene family: detection of transcripts in multiple tissues. J Biol Chem 1989, 264: 21760–21769.
Seckback J: Ferreting out the secrets of plant ferritin - A review. Journal of Plant Nutrition 1982, 5: 369–394. 10.1080/01904168209362966
Orino K, Lehman L, Tsuji Y, Ayaki H, Torti SV, Torti FM: Ferritin and the response to oxidative stress. Biochem J 2001, 357: 241–247. 10.1042/0264-6021:3570241
Ong DS, Wang L, Zhu Y, Ho B, Ding JL: The response of ferritin to LPS and acute phase of Pseudomonas infection. J Endotoxin Res 2005, 11: 267–280.
Larade K, Storey KB: Accumulation and translation of ferritin heavy chain transcripts following anoxia exposure in a marine invertebrate. J Exp Biol 2004, 207: 1353–1360. 10.1242/jeb.00872
Chaiyaratana W, Chuansumrit A, Atamasirikul K, Tangnararatchakit K: Serum ferritin levels in children with dengue infection. Southeast Asian J Trop Med Public Health 2008, 39: 832–836.
Schmidt A, Kellermann J, Lottspeich F: A novel strategy for quantitative proteomics using isotope-coded protein labels. Proteomics 2005, 5: 4–15. 10.1002/pmic.200400873
Shevchenko A, Wilm M, Vorm O, Mann M: Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 1996, 68: 850–858. 10.1021/ac950914h
Luecke N, Templin C, Muetzelburg MV, Neumann D, Just I, Pich A: Secreted proteome of the murine multipotent hematopoietic progenitor cell line DKmix. Rapid Commun Mass Spectrom 2010, 24: 561–570. 10.1002/rcm.4412
Dupierris V, Masselon C, Court M, Kieffer-Jaquinod S, Bruley C: A toolbox for validation of mass spectrometry peptides identification and generation of database: IRMa. Bioinformatics 2009, 25: 1980–1981. 10.1093/bioinformatics/btp301
Acknowledgements
We would like to thank Dr Y. Ataman-Önal for helpful discussions and manuscript reading, Dr M. Cao-Lormeau (Institut Louis Malardé, Papeete, Tahiti) for providing dengue patient plasma specimens and Dr N. Piga (BioMerieux SA, Lyon, France) for her precious support in getting plasma samples from healthy donors.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare to be employed by bioMérieux SA.
Authors’ contributions
Conception and design of the experiments: RF, FB. Realization of the experiments : RF, FB, Writing of the paper: RF, FB, JYM. Review of the paper: FB, JYM, AP. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Fragnoud, R., Yugueros-Marcos, J., Pachot, A. et al. Isotope Coded Protein Labeling analysis of plasma specimens from acute severe dengue fever patients. Proteome Sci 10, 60 (2012). https://doi.org/10.1186/1477-5956-10-60
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1477-5956-10-60