Abstract
Background
Tachykinins (TK), such as substance P, and their neurokinin receptors which are ubiquitously expressed in the human urinary tract, represent an endogenous system regulating bladder inflammatory, immune responses, and visceral hypersensitivity. Increasing evidence correlates alterations in the TK system with urinary tract diseases such as neurogenic bladders, outflow obstruction, idiopathic detrusor instability, and interstitial cystitis. However, despite promising effects in animal models, there seems to be no published clinical study showing that NK-receptor antagonists are an effective treatment of pain in general or urinary tract disorders, such as detrusor overactivity. In order to search for therapeutic targets that could block the tachykinin system, we set forth to determine the regulatory network downstream of NK1 receptor activation. First, NK1R-dependent transcripts were determined and used to query known databases for their respective transcription regulatory elements (TREs).
Methods
An expression analysis was performed using urinary bladders isolated from sensitized wild type (WT) and NK1R-/- mice that were stimulated with saline, LPS, or antigen to provoke inflammation. Based on cDNA array results, NK1R-dependent genes were selected. PAINT software was used to query TRANSFAC database and to retrieve upstream TREs that were confirmed by electrophoretic mobility shift assays.
Results
The regulatory network of TREs driving NK1R-dependent genes presented cRel in a central position driving 22% of all genes, followed by AP-1, NF-kappaB, v-Myb, CRE-BP1/c-Jun, USF, Pax-6, Efr-1, Egr-3, and AREB6. A comparison between NK1R-dependent and NK1R-independent genes revealed Nkx-2.5 as a unique discriminator. In the presence of NK1R, Nkx2-5 _01 was significantly correlated with 36 transcripts which included several candidates for mediating bladder development (FGF) and inflammation (PAR-3, IL-1R, IL-6, α-NGF, TSP2). In the absence of NK1R, the matrix Nkx2-5_02 had a predominant participation driving 8 transcripts, which includes those involved in cancer (EYA1, Trail, HSF1, and ELK-1), smooth-to-skeletal muscle trans-differentiation, and Z01, a tight-junction protein, expression. Electrophoretic mobility shift assays confirmed that, in the mouse urinary bladder, activation of NK1R by substance P (SP) induces both NKx-2.5 and NF-kappaB translocations.
Conclusion
This is the first report describing a role for Nkx2.5 in the urinary tract. As Nkx2.5 is the unique discriminator of NK1R-modulated inflammation, it can be imagined that in the near future, new based therapies selective for controlling Nkx2.5 activity in the urinary tract may be used in the treatment in a number of bladder disorders.
Similar content being viewed by others
Background
Substance P belongs to the tachykinins (TKs) family of peptides involved in the peripheral and central regulation of urinary functions [1] through the stimulation of neurokinin (NK) NK1, NK2, and NK3 receptors [2, 3]. At the urinary system level, TKs stimulate smooth muscle tone, ureteric peristalsis and bladder contractions, initiate neurogenic inflammation, and trigger local and spinal reflexes [4] aimed to maintain organ functions in emergency conditions [2]. The most studied effects produced by TKs in these systems are smooth muscle contraction [5–9], modulation of inflammation [10, 11], mucus secretion, and recruitment/activation of immune cells [12]. At least in the mouse bladder, TKs are spontaneously released and their levels maintained low by the activity of neutral-endopeptidase [13]. Indeed, null deletion of NEP in mice leads to spontaneous plasma extravasation in the urinary bladder that was reversed by a recombinant of NK1 and bradykinin B2 receptors antagonists [14].
In the urinary tract, the major recognized sources of TKs are the primary afferent neurons expressing transient receptor potential vanilloid-1 receptors, which have the unique property of releasing transmitters both in the periphery (efferent function) and the spinal cord (afferent function) upon stimulation [2].
NK1R are the predominant subtype involved in inflammation in general [3] and may underlie persistent pain, such as that observed during chronic bladder inflammation [15]. SP activation of NK1R [3] induces a sequential activation of signaling pathways leading to the production of pro-inflammatory mediators [10, 16, 17] and pro-inflammatory cytokines such as macrophage migration inhibitory factor (MIF) that plays a major role in bladder inflammation [18].
The use of NK1R-/- mice confirmed a central role for SP in models of bladder inflammation [19]. Indeed, NK1R-/- mice do not mount bladder inflammatory response to antigen-complex stimulation and that NK1Rs are required in cystitis [19]. In this context, an up-regulation of NK1R was found in bladder inflammation [20] and bladder biopsies from cystitis patients present an increase in NK1R density [21], nerves [22], and SP-containing fibers [23]. Furthermore, the finding that sensory C fibers desensitization decreases urinary bladder hyperreflexia further supports a role for sensory peptides in this disorder [24]. In fact, NK1R antagonists reduce detrusor hyperreflexia caused by chemical [25] and bacterial cystitis [26], and decrease cyclophosphamide-induced inflammation [27]. In addition, changes in SP expression following cystitis may contribute to the altered visceral sensation (allodynia) and/or urinary bladder hyperreflexia in the clinical syndrome, interstitial cystitis [4].
The bulk of data obtained in experimental animal models suggests that TKs could contribute to the genesis of symptoms accompanying various diseases of the urinary tract, which includes cystitis and incontinence [28]. Indeed, a significant increase in the density of suburothelial, SP-containing nerves was found in patients with idiopathic detrusor overactivity, compared with stable controls [29, 30]. Therefore, it cannot be excluded that peripheral tachykinins may be involved in pathophysiologic afferent signaling associated with detrusor overactivity [28]. However, despite promising effects in animal models, there seems to be no published clinical study showing that NK-receptor antagonists are an effective treatment of pain [31] or overactive bladder disease [28]. In addition, despite the known existence of NK2 receptors in the human detrusor, NK2 receptor antagonist does not block the non-cholinergic contraction in unstable human bladder [32].
Therefore, in order to search for putative therapeutic targets that could be manipulated to reduce the influence of the tachykinin system, we set forth to determine the regulatory network downstream of NK1R activation. This network is composed of genes and the transcriptional regulatory elements (TREs) that are putative binding sites for the transcription factors. In this way, we could define not only genes downstream of NK1R activation but also the regulators of their expression. This is based on the fact that when active transcription factors associate with TREs of their target genes, they can function to specifically repress (down-regulate) or induce (up-regulate) synthesis of the corresponding RNA. The overall hypothesis is that genes sharing the same TREs can be associated in a molecular network that would represent key pharmacological targets for modulating the influence of tachykinins in bladder diseases.
For this purpose, we used a combination of cDNA array and in silico analysis of TREs, as described previously [33]. cDNA array analysis defined the interactome of NK1-dependent genes by querying a web-based entry tool developed by Ingenuity Systems Inc [34]. Next, we uploaded the sequence of NK1-dependent genes into PAINT software and the respective TREs were identified using MATCH® tool in the TRANSFAC Professional database. Genes and TREs were assembled in regulatory networks and selected TREs were confirmed by EMSA.
Methods
Animals
All animal experimentation described here was performed in conformity with the "Guiding Principles for Research Involving Animals and Human Beings (OUHSC Animal Care & Use Committee protocol #00-109 and #00-108). Groups of ten to twelve-week old female mice were used in these experiments. NK1R-/- and wild type (WT, C57BL6) littermate control mice were generated by Dr. Norma P. Gerard. The colonies at OUHSC were genotyped as described previously [35].
Antigen sensitization protocol
All mice in this study were sensitized with 1 μg DNP4-human serum albumin (HSA) in 1 mg alum on days 0, 7, 14, and 21, intraperitoneally (i.p.). In normal mice, this protocol induces sustained levels of IgE antibodies up to 56 days post-sensitization [36]. One week after the last sensitization, cystitis was induced. Briefly, sensitized WT and NK1R-/- mice were anesthetized (ketamine 40 mg/kg and xylazine 2.5 mg/kg, i.p.), then transurethrally catheterized (24 Ga.; 3/4 in; Angiocath, Becton Dickson, Sandy, Utah), and the urine was drained by applying slight digital pressure to the lower abdomen. The urinary bladders were instilled with 200 μl of pyrogen-free saline or DNP4-OVA (1 μg/ml). One, four, and twenty-four hours after instillation, mice were sacrificed with pentobarbital (100 mg/kg, i.p.) and bladders were removed rapidly.
Alterations at histological level
Previous results from our laboratory demonstrated a mandatory role of NK1R on antigen-induced cystitis [19, 37]. In the present work, we also investigated whether NK1Rs are important for both SP- and LPS-induced cystitis. For this purpose, an additional group of NK1R-/- and wild type (WT, C57BL6) were anesthetized as described above and challenged intravesically with 200 μl of pyrogen-free saline, SP (10 μM), or Escherichia coli LPS strain 055:B5 (Sigma, St. Louis, MO; 100 μg/ml). Twenty-four hours after instillation, mice were euthanized with pentobarbital (200 mg/kg, i.p.), and the bladders were removed rapidly for evaluation of inflammatory cell infiltrates and the presence of interstitial edema. A semi-quantitative score using defined criteria of inflammation severity was used to evaluate cystitis [37]. A cross-section of bladder wall was fixed in formalin, dehydrated in graded alcohol and xylene, embedded in paraffin, and cut serially into four 5-μm sections (8 μm apart) to be stained with hematoxylinand eosin (H&E) and Giemsa. H&E stained sections were visualized under microscope (Eclipse E600, Nikon, Lewisville, TX). All tissues were photographed at room temperature by a digital camera (DXM1200; Nikon). Exposure times were held constant when acquiring images from different groups. Images were analyzed with Image-Pro Analyzer® (Media Cybernetics Inc.; Silver Spring, MD 20910). The severity of lesions in the urinary bladder was graded as follows: 1+, mild (infiltration of 0–10 neutrophils/cross-section in the lamina propria, and little or no interstitial edema); 2+, moderate(infiltration of 10–20 neutrophils/cross-section in the lamina propria, and moderate interstitial edema); 3+, severe (diffuse infiltration of >20 neutrophils/cross-section in the lamina propria and severe interstitial edema) [19, 37, 38]. Identification of mast cells was performed in Giemsa-stained sections [37].
Minimum information about microarray experiments – MIAME [39]
a. Objective
To determine the time course of gene-expression in control and antigen-inflamed wild type and NK1R-/- mice.
b. Array design
Mouse 5K Arrays (Clontech, Palo Alto, CA, Cat. #GPL151), for a complete list of genes in this array, please access Gene Expression Omnibus, GEO [40].
c. Animal numbers
Female WT and NK1R-/- mice were instilled with antigen (in sensitized mice), or saline. At 1, 4, and 24 hours following stimulation, the urinary bladders were randomly distributed into the following groups: a) RNA extraction (n = 3), b) replicate of RNA extraction (n = 3), and c) morphological analysis (n = 6).
d. Sample preparation for cDNA expression arrays
Three bladders from each group were homogenized together in Ultraspec RNA solution (Biotecx Laboratories Inc. Houston, TX) for isolation and purification of total RNA. Mouse bladders were pooled to ensure enough RNA for gene array analysis. The justification for this approach is that there is not enough RNA in a single mouse bladder for performing cDNA array experiments, and the step of purification reduces the amount of total RNA. RNA was DNase-treated according to manufacturer's instructions (Clontech Laboratories, Palo Alto, CA), and the quality of 10 μg was evaluated by denaturing formaldehyde/agarose gel electrophoresis.
d. Mouse cDNA expression arrays
cDNA probes were prepared from DNase-treated RNAs obtained from each of the experiments. Five μg of DNase-treated RNA was reverse-transcribed to cDNA and labeled with [α-32P]dATP, according to the manufacturer's protocol (Clontech, Palo Alto, CA). The radioactively labeled complex cDNA probes were hybridized overnight to Atlas™ Mouse 5K Arrays (Clontech, Palo Alto, CA) using ExpressHyb™ hybridization solution with continuous agitation at 68°C. After two high-stringency washes, the hybridized membranes were exposed (at room temperature) to an ST Cyclone phosphor screen overnight. Spots on the arrays were quantified by BD AtlasImage™ 2.7 software (Clontech, Palo Alto, CA). The results were placed in an Excel spreadsheet.
f. Data normalization and analysis
Data was normalized by linear regression analysis using only genes expressed above background, as described [11, 41], and the ratio of gene-expression between antigen- and saline-challenge was obtained. NK1R-dependent genes were selected according to the following criterion: a. In tissues isolated from WT mice, the expression of a particular gene should be up-regulated (ratio between antigen- and saline-treated >3.0) in at least one of the time points (1, 4, and 24 hours post challenged); b. in tissues isolated from NK1R-/- mice, the expression of same gene should not be altered by antigen-challenge in any of the time points.
g. Database submission of microarray data
The microarray data was prepared according to "minimum information about a microarray experiment" (MIAME) recommendations [39], has been deposited in the Gene Expression Omnibus (GEO) database and can be retrieved with GEO accession number GSE2821 [42].
Ingenuity pathways analysis
We used a novel approach [34] to fully annotate and represent NK1R-dependent genes by using the Ingenuity Pathways Analysis tool [43]. Using Ingenuity knowledge base network, we identified specific and canonical pathways downstream of NK1R activation.
Analysis of transcriptional regulatory elements (TREs)
We employed a bioinformatics approach to hypothesize functionally relevant transcriptional regulatory elements (TREs) of NK1R-dependent and -independent genes. The regulatory network was determined by a combination of micro array-selected transcripts and PAINT 3.3 [44], available online [45], to query the transcription factor database (TRANSFAC) [46]. PAINT 3.3 was employed to examine 2000 base pairs of regulatory regions upstream of the transcriptional start site of each differentially expressed gene detected with the microarray. PAINT is a suite of bioinformatics and computational tools that integrate functional genomics information, as is the case of our microarray-based gene expression data, with genomic sequence and TRE data to derive hypotheses on the TREs relevant to the biological function under study.
Genbank accession numbers were used as the gene identifiers in PAINT test files. Over-representation of TREs in the matrix was calculated at levels of 0 < p <= 0.01 and 0.01 < p < 0.05 when compared to the reference (TREs regulating all genes in the original array). Employing the microarray accounts for any 'bias' present in the genes on the microarray relative to entire genome, nd guards from incorrectly concluding that certain TREs are relevant to the current experiment. The TRE hypotheses were generated from statistical enrichment analysis and were defined as those TREs that are significantly enriched such as NK1R-dependent and -independent genes, over random occurrence in the gene groups [44].
Electrophoretic mobility shift assays (EMSA)
Anesthetized C57BL6 female mice were instilled with 200 μl of saline or SP (10 μM) and bladders were removed 2, 6, and 24 hours after instillation. In one additional group (zero hours), the urinary bladders were removed without instillation. Urinary bladders were placed in cold phosphate buffered saline (0°C), containing peptidase inhibitors (aprotinin, pepstatin, leupeptin at 0.01 mg/ml,) and the mucosa was dissected away from the muscle, as described previously [33]. Nuclear proteins were extracted and used for electrophoretic mobility shift assay for Nkx-2.5 and NF-kappaB.
NF-kappaB EMSA
The NF-kappaB probe was constructed by annealing complementary synthetic oligonucleotides (5' GAT CAT GGG GAA TCC CCA 3'). Annealed probes were end-labeled with [α-32P] ATP (3000 Ci/mmole; GE Healthcare) and T4 polynucleotide kinase (New England Biolabs), and then purified using a G-50 column (GE Healthcare). Nuclear extracts (10 μg) were incubated with 1 ng of [32P] NF-kappaB double-stranded probe in 20 μl 20 mM HEPES, 70 mM KCl, 2 mM DTT, 0.01% NP-40, 4% Ficoll, 1 mg/ml BSA, and 1.4 μg poly d(I-C). For competition reactions, a 50-fold excess of unlabelled NF-kappaB probe was added to the reaction mixture, and it was incubated at room temperature for 5 minutes before the addition of the [32P] NF-kappaB double-stranded probe. Reaction mixtures were incubated for 20 minutes at room temperature. DNA-protein complexes were resolved on a non-denaturing 6% polyacrylamide gel at 200 V for 3 hours in 0.5 TBE (45 mM Tris-borate and 1 mM EDTA). Gels were vacuum-dried and visualized on Kodak Biomax MS Film and quantified using ImageJ Software (NIH) and statistical differences were determined using GraphPad Prism Software (GraphPad Software, San Diego, CA).
Nkx-2.5 EMSA
Electrophoretic mobility shift assays for Nkx-2.5 were performed using a non-radioactive Gel Shift Kit (Panomics, Redwood City, CA) according to the experimental procedures provided by the manufacturer. Briefly, nuclear extracts (5 μg) were incubated with biotinylated Nkx-2.5 probe in Binding Buffer with poly d(I-C). For competition reactions, an excess of unlabelled Nkx-2.5 probe was added to the reaction mixture and it was incubated at room temperature for 5 minutes before the addition of the biotinylated Nkx-2.5 probe. Reaction mixtures were incubated for 30 minutes at 20°C. DNA-protein complexes were resolved on a non-denaturing 6% polyacrylamide gel at 200 V for 2 hours in 0.5× TBE (45 mM Tris-borate and 1 mM EDTA). The gel was then transferred onto a 0.45 μm Nytran SuperCharge membrane (Schleicher & Schuell, Keene, NH) at 300 mA for 30 minutes in 0.5× TBE. The blot was dried for 60 minutes at 80°C and then UV cross-linked for 3 minutes. After blocking, the blot was incubated with Streptavidin-HRP and developed using the substrate solutions included in the kit. Blots were visualized on Pierce CL-XPosure film (Pierce, Rockford, IL). Bands were quantified using ImageJ Software (NIH) and statistical differences were determined using GraphPad Prism Software (GraphPad Software, San Diego, CA). The sequence for the NKx-2.5 motif-containing probe, not provided by the manufacturer, is 5' AAA CAA GTC ATA ATA GGA AGC A 3'.
Results
Essential role of NK1R in cystitis
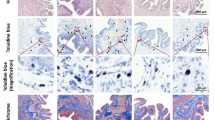
Previous results from our laboratory demonstrated a mandatory role of NK1R on antigen-induced cystitis [19, 37]. In the present work we also investigated whether NK1Rs are important for both SP- and LPS-induced cystitis. In contrast to mice treated with saline (Figure 1A), instillation of LPS into the bladder of wild type mice leads to inflammation characterized by edema (Figure 1B; black line delimits the area of sub-epithelial edema) and infiltration of inflammatory cells (Figure 1D; black arrows indicate PMNs). In contrast, the urinary bladders of NK1R-knockout mice failed to mount an inflammatory response to LPS (Figures 1C and 1E) despite the capacity of LPS to induce urothelial cell injury, as indicated by intra-cytoplasmatic vacuolization [47] (Figure 1E; black circles). Similar results were obtained with WT mice that mounted an inflammatory reaction to intravesicall instillation of SP as seem in H&E (Figure 2A and 2C) and Giemsa stained (Figure 2E) sections. NK1R-knockout failed to mount an inflammatory response to SP (Figures 2B, 2D, and 2F) with a reduced number of migrating inflammatory cells but presenting visible resident mast cells (Figure 2F; black line). Figure 3 presents the quantification of the results here described. In conclusion, these results extend our previous observation of a mandatory role for NK1R in antigen-induced cystitis to other pro-inflammatory mediators.
A-E. Comparison of bladder inflammatory responses in wild type (C57BL6) and NK1-R knockout mice. Representative photomicrographs of bladder inflammation in mice treated intravesically with 200 μl of pyrogen-free saline (1A), LPS (100 μg/ml; 1B, 1C, 1D, and 1E), or SP (10 μM; 2A, 2B, 2C, 2D, 2E, and 2F). The urinary bladders were removed 24 hours after bladder instillation and processed for histological stains: H&E (1 A-E and 2A-D) and Giemsa (2E and 2F), (n = 6).
A-F. Comparison of bladder inflammatory responses in wild type (C57BL6) and NK1-R knockout mice. Representative photomicrographs of bladder inflammation in mice treated intravesically with 200 μl of pyrogen-free saline (1A), LPS (100 μg/ml; 1B, 1C, 1D, and 1E), or SP (10 μM; 2A, 2B, 2C, 2D, 2E, and 2F). The urinary bladders were removed 24 hours after bladder instillation and processed for histological stains: H&E (1 A-E and 2A-D) and Giemsa (2E and 2F), (n = 6).
Quantification of inflammation. Wild type (C57BL6) and NK1-R knockout mice were treated intravesically with 200 μl of pyrogen-free saline, LPS (100 μg/ml), or SP (10 μM). The urinary bladders were removed 24 hours after bladder instillation and processed for histological stains for quantification of inflammation. A = Grade of edema and B = Grade of polymorphonuclear [PMNs] leukocytes (see material and methods).
NK1-dependent genes
Two hundred and nine genes fulfilled the established criteria (see material and methods) and were considered NK1R-dependent. In contrast, 236 genes were found to be up-regulated secondary to antigen challenge in tissues isolated from WT and NK1R-/- mice and, therefore, were considered to be NK1R-independent.
NK1-dependent transcriptional regulatory elements
PAINT 3.3 was employed to examine 2000 base pairs of regulatory regions upstream of the transcriptional start site of each differentially expressed gene from the microarray expression data. Genbank accession numbers were used as gene identifiers in PAINT input files. For the NK1R-dependent genes, out of a total of 209 genes from the expression analysis, only 153 genes had corresponding upstream sequence information owing to the incomplete nature of genomic annotation. A total of 87 TREs were identified on these sequences using MATCH® tool in the TRANSFAC Professional database. Similarly, for the NK1R-independent genes, only 188 promoters were retrieved for the 236 genes from expression analysis. A total of 88 TREs were identified on these sequences using MATCH®. These TREs were examined in the enrichment analysis to derive regulatory network hypotheses. The resulting candidate interaction network can be visualized as an interaction matrix where the individual elements of the matrix are color-coded based on the p-values for statistical enrichment. A p-value threshold of 0.07 was used to filter the enrichment analysis results to derive the regulatory network hypotheses. These results are shown in Figure 4 for NK1R-dependent genes and in Figure 5 for NK1R-independent genes.
PAINT 3.3 representation of an interaction matrix for NK 1 R-dependent genes and respective TREs. PAINT 3.3 was employed to examine 2000 base pairs of regulatory regions upstream of the transcriptional start site of each differentially expressed gene from the microarray expression data. Genbank accession numbers were used as the gene identifiers in PAINT input files. Individual elements of the matrix are colored by the significance of the p-values: over-representation in the matrix is indicated in red, under-representation is indicated in cyan, and the TREs that are neither significantly over nor under-represented in the matrix are colored in gray.
PAINT 3.3 representation of an interaction matrix for NK 1 R-independent genes and respective TREs. PAINT 3.3 was employed to examine 2000 base pairs of regulatory regions upstream of the transcriptional start site of each differentially expressed gene from the microarray expression data. Genbank accession numbers were used as the gene identifiers in PAINT input files. Individual elements of the matrix are colored by the significance of the p-values: over-representation in the matrix is indicated in red, under-representation is indicated in cyan, and the TREs that are neither significantly over nor under-represented in the matrix are colored in gray.
Regulatory network downstream of NK1R activation
Figure 6 depicts the hypothesized regulatory network corresponding to the NK1R-dependent genes whose TRE were found to be over-represented (0.01 < p < 0.05) in this set when compared to a reference (all genes in the array). cRel was the predominant TRE, driving 22% of the NK1R-dependent genes. The order of predominance for the different TREs was: cRel, v-Myb, CRE-BP1/c-Jun, USF, AP-1_Q2, Pax-6, AP-1_C, NF-kappaB_Q6, Efr-1, Egr-3, and AREB6 (Figure 6 and Table 1 [additional file 1]).
Regulatory Network downstream NK 1 receptor activation. Hypothesized regulatory network corresponding to the NK1R-dependent genes and respective TREs. A complete list of genes and TREs is summarized on Table 1 (Additional file 1).
Ingenuity pathways analysis
In order to make sense of the vast information generated by cDNA array expression, we used the recently developed Ingenuity Pathways Knowledge Base [34] to design pathways of NK1R-dependent genes and to query canonic pathways regarding the relative importance of each of the transcripts (Figure 7). As a result, genes were localized to different compartments depending on the predominant expression of the protein they encoded (Figure 7). In this way, four compartments are depicted: extracellular space, plasma membrane, cytoplasm, and nucleus. According to their primary function, ingenuity grouped NK1-dependent genes into 4 different networks: cell morphology, cell cycle, inflammation, and cell death. However, a strong degree of overlapping between cell morphology, cell cycle, cell death, and cancer was observed. In contrast, genes involved in inflammation were also listed to be involved in embryonic development. The NK1R-dependent genes were significantly correlated with the following canonical signaling pathways: p38 MAPK, NF-kappaB, PPAR, IL-6, death receptor, apoptosis, and SAPK/JNK (Table 2, [additional file 2]).
Overall NK1R-dependent genes were classified following the biological processes with which they are involved (GO anthology): apoptosis (GZMA, TNFRSF1B, TNFRSF1A, TRAF3, NOS2A, and BID); cell adhesion/hyaluronic acid binding (CD44 and AGC1); cell cycle (CCND2 and CCNG1); cell-cell signaling (FGF11 and GJA7); cytokinesis (CDC42 and KIF1B); development (FMR2); extracellular transporters & carriers (APOE); G-protein coupled receptors (GNA13 and PTGIR); growth factors (MXD1); heat shock proteins (PRNP, HSPH1, and HSPD1); immune response (BST-1, CTSW, and IL1R1); interferons (INFGR1) intracellular kinases (WBP6); intracellular transducers (MAP3K7); kinase activators & inhibitors (YWHAH); membrane channels (KCNAB1, KCNJ12, KCNQ1, and SLC30A4); nucleotide metabolism (PCSK1); oncogenes & tumor suppressors (BRCA1, MAP3K8, RET, VIL2, FLI1, MET, NF2, and VEGFR1); receptor mediated endocytosis (DAB2); receptor tyrosine kinase (EPHA2); regulation of transcription (NEUROD6); serotonin biosynthesis (YTPH1); symporters & antiporters (SLC16A1 and SLC1A1); transcription activators & repressors (FOXA1, HSF1, IER2, and NR1H2); and synaptic transmission (GRID1).
Comparing NK1R-dependent and -independent genes
The whole set of genes that were up-regulated at least 3-fold during inflammation including NK1R-dependent and NK1R-independent genes were analyzed in PAINT. A p-value threshold of 0.05 was used to derive statistically enriched regulatory elements in these two gene groups (Figure 8). The enrichment in this case was obtained by using the interaction matrix for the combined gene list as a reference. This enabled us to contrast the two gene groups relative to each other and characterize those regulatory elements that are specific to one group or the other. We observed differential enrichment of several regulatory elements including over-expression of AP1 specific in the NK1R-dependent genes, as we previously suggested [41] (Figure 8). However, the most striking finding of this analysis was the revelation that Nkx-2.5, a murine homeo box gene, is a unique discriminator of NK1R-dependent genes. Nkx-2.5 matrix _01 was found over represented in the set of NK1R-independent transcripts (colored in red) and under-represented or suppressed in the set of NK1R-dependent TREs (colored in cyan). In contrast, Nkx-2.5 matrix _02 had the completely opposite behavior since it was over-represented in NK1R-dependent and under-represented in the NK1R-independent set. Table 3 [additional file 3] lists all genes under the control of both Nkx-2.5 matrix _01 and matrix _02. With the exception of A10 (L21027) and EGF (J00380) that were associated with both matrixes, a unique set of genes is correlated with Nkx-2.5 matrix _01 and Nkx-2.5 matrix _02.
Direct comparison of TREs driving NK 1 R-dependent and-independent genes. The whole set of genes that were up-regulated at least 3-fold during bladder inflammation were analyzed in PAINT 3.3. These genes were divided into two groups: NK1R-dependent and NK1R-independent. A p-value threshold of 0.05 was used to derive statistically enriched regulatory elements in these two gene groups. Note that AP-1 is over represented in the set of NK1R-dependent genes, whereas it is not significantly correlated with NK1R-independent transcripts. Also note that Nkx-2.5 matrix _01 was found over represented in the set of NK1R-independent transcripts (colored in red) and under-represented or suppressed in the set of NK1R-dependent TRs (colored in cyan). In contrast, Nkx-2.5 matrix _02 had the completely opposite behavior.
Intravesical instillation of SP stimulates NF-kappaB and Nkx-2.5 translocation in the bladder mucosa
To determine whether activation of NK1R increases the transcriptional activities of NF-kappaB and Nkx-2.5, an EMSA was performed. Results show that instillation of SP into the mouse bladder increased the amount of shifted Nkx-2.5 and NF-kappaB probes that peaked at 24 hours post stimulation (Figure 9 A-D). Because of the high level of expression of both NKx-2.5 and NF-kappaB in the bladders pre-treated with saline, in an additional group, urinary bladders were removed without insertion of the catheter or fluid instillation. Interestingly, in this group (0 hours) almost the same degree of constitutive transcriptional activities was observed.
Intravesical SP stimulates NKx-2.5 and NF-kappaB activity. Anesthetized mice were instilled with 200 μl of saline or SP (10 μM) and bladders were removed 2, 6, and 24 hours after instillation. In one additional group (zero hours) the urinary bladders were removed without instillation. Urinary bladders were placed in cold phosphate buffered saline (0°C) containing protease inhibitors, and the mucosa was dissected away from the muscle. Nuclear proteins were extracted and used for electrophoretic mobility shift assay for NKx-2.5 (A) and NF-kappaB (C). Amount of shifted Nkx-2.5 (B) and NF-kappaB (D) probes were quantified as described in materials and methods.
Discussion
It has to be taken into consideration that the approach of this study permitted the generation of new testable hypothesis rather than the more traditional hypothesis-driven research. Indeed, the major question being answered by this work is what TREs can be therapeutically targeted for reducing the influence of tachykinins in bladder disorders? The method introduced here, supplements the standard procedure of multiple paired comparisons used in microarray analysis by associating the expression level of each gene in the experimental group with a family transcription regulatory elements[33] and to compare with the occurrence of each TRE in a reference file (all genes in the array). This bottom-up approach builds mechanistic models for each individual case, e.g., identifying the binding sites for selected genes and their respective TREs [48], then specifies the role of each TRE in the network generating a testable hypothesis for the network downstream of NK1R activation. Next, we used EMSA to confirm that selected TREs (NF-kappaB and NKx-2.5) are indeed part of the molecular network downstream of NK1R activation.
An extended family of TREs was significantly correlated with NK1R-dependent genes. Those included c-Rel, NF-kappaB_Q6, PAX-6, CREB_01, CRE-BP1/c-Jun, and v-Myb (Figure 4). However, most of the studies on transcription regulatory elements in urology are related to oncology, which makes it difficult to further illustrate the clinical relevance of our findings. Therefore, we are discussing only the most relevant TREs that modulate bladder inflammatory responses to SP.
AP1 was among the NK1R-dependent TREs. We have provided evidence for a predominant role for AP1 controlling highly expressed NK1R-dependent genes [41]. In the present work, we confirmed the regulatory relationships between AP1 (AP1_Q2 and AP1_C) and NK1R-dependent genes. It is known that the activation of MAPK (JNK, p38) and NF-kappaB signaling pathways leads to the activation of AP-1 and, consequently, the inflammation [49]. The present results extend these findings to the urinary bladder where these pathways can be explored as potential therapeutic targets to decrease the symptoms of cystitis.
NF-kappaB is believed to trigger both the onset and the resolution of inflammation. NF-kappaB activity is correlated with bladder cancer [50–52] and bladder urothelial cells respond to insults with a translocation of NF-kappaB [53] leading ultimately to an increased NK1R expression [54]. Our present work confirms previous indication that Tachykinins, such as SP, activate NF-kappaB translocation [55, 56]. Indeed, in the urinary bladder, activation of NK1R by SP induces NF-kappaB translocation, as seen by EMSA results (Figure 9), and up-regulation of pro-inflammatory genes, such as the encoding prostaglandin I2 receptor (Figure 6).
Another TRE over-expressed in the NK1R-dependent cluster was the upstream stimulatory factor (USF). Although widely expressed, USF can mediate tissue-specific transcripts. USF is stimulated by glucose in murine mesangial cells, binds to TGF-β1 promoter, contributes to TGF-β1 expression, and may play a role in diabetes-related gene regulation in the kidney [57].
However, the most impressive switch between NK1R-dependent and independent transcripts was the one observed with two different matrixes of Nkx-2.5 (_01 and _02). Both Nkx-2.5_01 and _02 are binding sites derived from mouse sequences [58, 59]. Nkx-2.5 is a murine homeobox named tinman homeodomain factor and is considered to be a new member of the sub-family of homeobox genes related to the Drosophila [58]. Nkx-2.5 is proposed as a valuable marker in the analysis of mesoderm development [59]. It was first described as an essential transcription factor for normal heart morphogenesis, myogenesis, and function [60]. However, more recently it was shown that Nkx-2.5 is required for the expression of atrial natriuretic peptide [61] and, along with NF-kappaB, is part of the brain natriuretic peptide promoter [62]. Outside of the heart, this element is important in vessel remodeling [63], skeletal myogenesis [64], and pyloric sphincter development [65]. Other sites of Nkx-2.5 expression include pharyngeal endoderm and its derivatives, branchial arch epithelium, stomach, spleen, pancreas and liver [66].
To our knowledge, this is the first report describing a role for Nkx-2.5 in the urinary tract. In the presence of NK1R, Nkx-2.5 _01 was significantly correlated with 36 transcripts which included several candidates for mediating bladder development (FGF) and inflammation (PAR-3, IL-1R, IL-6, NGF, TSP2) (Table 3, [additional file 3]). In the absence of NK1R, the matrix _02 had a predominant participation driving 8 transcripts, which includes those involved in cancer (EYA1, Trail, HSF, and ELK-1), smooth-to-skeletal muscle trans-differentiation, and Z01, a tight-junction protein, expression (Table 3, [additional file 3]).
An interesting finding was the constitutive translocation of NKx-2.5 and NF-kappaB in the bladder mucosa. One possible explanation was that mechanical stimulation caused by instillation of saline caused the shift. Therefore, an additional control group was added in which the bladder was removed without instillation. This group (0 hours) also presented a certain amount of shifted NKx-2.5 and NF-kappaB probes. An alternative explanation for these results is that mechanical isolation of the bladder mucosa caused the translocation of both transcription factors. We, therefore, generated preliminary results using an urothelial cell line (J82) which indicated a constitutive activation of both NF-kappaB and NKx-2.5 in the absence of overt stimulation (data not shown). Therefore, we suggest that the bladder mucosa/urothelium might present a constitutive activation of both transcription factors. The similarity of basal translocation of NKx-2.5 and NF-kappaB translocation in the urinary bladder may be related to an overlap of binding motifs in some genes. Indeed, others have shown an overlap of conserved DNA binding motifs including AP-1 sites, NF-kappaB, GATA, and Nkx-2.5 in promoter regions of genes, such as MMP13 [67].
Conclusion
This work indicates an overriding participation of NK1R in bladder inflammation, provides a working model for the involvement of transcription regulators such as NF-kappaB, and Nkx-2.5, and evokes testable hypotheses regarding a role for tachykinins in the urinary tract pathology. It remains to be determined whether the control of Nkx-2.5 activity by gene silencing or double mutant negative blockers will ameliorate the clinical manifestations of cystitis.
Abbreviations
- AP-1:
-
activator protein-1
- ELK-1:
-
Elk-1 ets-related proto-oncogene
- EMSA:
-
electrophoretic mobility shift assay
- EYA1:
-
eyes absent homolog 1
- FGF:
-
fibroblast growth factor
- HSF1:
-
transcription factor 1 for heat shock gene
- IL-1R:
-
interleukin-1 receptor
- IL-6:
-
Interleukin-6
- LPS:
-
lipopolysaccharide
- PAR-3:
-
proteinase-activated receptor 3
- NGF:
-
nerve growth factor
- Trail:
-
TNF-related apoptosis inducing ligand
- TSP2:
-
thrombospondin 2
- SP:
-
substance P.
References
Lecci A, Maggi CA: Tachykinins as modulators of the micturition reflex in the central and peripheral nervous system. Regul Pept. 2001, 101 (1–3): 1-18. 10.1016/S0167-0115(01)00285-3.
Candenas L, Lecci A, Pinto FM, Patak E, Maggi CA, Pennefather JN: Tachykinins and tachykinin receptors: effects in the genitourinary tract. Life Sci. 2005, 76 (8): 835-862. 10.1016/j.lfs.2004.10.004.
Pennefather JN, Lecci A, Candenas ML, Patak E, Pinto FM, Maggi CA: Tachykinins and tachykinin receptors: a growing family. Life Sci. 2004, 74 (12): 1445-1463. 10.1016/j.lfs.2003.09.039.
Vizzard MA: Alterations in neuropeptide expression in lumbosacral bladder pathways following chronic cystitis. Journal of chemical neuroanatomy. 2001, 21 (2): 125-138. 10.1016/S0891-0618(00)00115-0.
Jerde TJ, Saban R, Bjorling DE, Nakada SY: NK-2 is the predominant tachykinin receptor subtype in the swine ureter. BJU Int. 1999, 83 (3): 312-318. 10.1046/j.1464-410x.1999.00943.x.
Saban R, Keith IM, Nielsen KT, Christensen MM, Rhodes PR, Bruskewitz RC: In vitro effects of bladder mucosa and an enkephalinase inhibitor on tachykinin induced contractility of the dog bladder. J Urol. 1992, 147 (3): 750-755.
Jerde TJ, Saban R, Bjorling DE, Steinberg H, Nakada SY: Distribution of neuropeptides, histamine content, and inflammatory cells in the ureter. Urology. 2000, 56 (1): 173-178. 10.1016/S0090-4295(00)00559-8.
Jerde TJ, Saban R, Nakada SY: Evaluation of ureteric contraction: a comparison among ring, spiral-cut and longitudinal segments. BJU Int. 1999, 83 (1): 95-100. 10.1046/j.1464-410X.1999.00865.x.
Nakada SY, Jerde TJ, Bjorling DE, Saban R: In vitro contractile effects of neurokinin receptor blockade in the human ureter. J Urol. 2001, 166 (4): 1534-1538. 10.1016/S0022-5347(05)65826-0.
Bjorling DE, Beckman M, Saban R: Neurogenic inflammation of the bladder. Adv Exp Med Biol. 2003, 539 (Pt B): 551-583.
Dozmorov I, Saban MR, Knowlton N, Centola M, Saban R: Connective molecular pathways of experimental bladder inflammation. Physiol Genomics. 2003, 15 (3): 209-222.
Busser BW, Hammond TG, Bjorling DE, Saban R: Lipopolysaccharide upregulates bradykinin 1 receptors in the isolated mouse bladder. J Urol. 1998, 160 (6 Pt 1): 2267-2273.
Saban R, Franz J, Bjorling DE: Spontaneously released substance P and bradykinin from isolated guinea-pig bladder. Br J Urol. 1997, 79 (4): 516-524.
Lu B, Figini M, Emanueli C, Geppetti P, Grady EF, Gerard NP, Ansell J, Payan DG, Gerard C, Bunnett N: The control of microvascular permeability and blood pressure by neutral endopeptidase. Nat Med. 1997, 3 (8): 904-907. 10.1038/nm0897-904.
Abbadie C, Brown JL, Mantyh PW, Basbaum AI: Spinal cord substance P receptor immunoreactivity increases in both inflammatory and nerve injury models of persistent pain. Neuroscience. 1996, 70: 201-209. 10.1016/0306-4522(95)00343-H.
Saban M, Nguyen N-B, Hammond T, Saban R: Gene Expression profiling of mouse bladder inflammatory responses to LPS, substance P, and antigen-stimulation. Am J Pathol. 2002, 160: 2095-2110.
Saban MR, Saban R, Bjorling DE: Kinetics of peptide-induced release of inflammatory mediators by the urinary bladder. Br J Urol. 1997, 80 (5): 742-747.
Meyer-Siegler KL, Vera PL: Substance P induced changes in CD74 and CD44 in the rat bladder. J Urol. 2005, 173 (2): 615-620. 10.1097/01.ju.0000143188.02802.f3.
Saban R, Saban MR, Nguyen NB, Lu B, Gerard C, Gerard NP, Hammond TG: Neurokinin-1 (NK-1) receptor is required in antigen-induced cystitis. Am J Pathol. 2000, 156 (3): 775-780.
Hammond TG, Saban R, Bost KL, Harris HW, Kaysen JH, Goda FO, Wang XC, Lewis FC, Navar GL, Campbell WC, Bjorling DE, Saban M, Zeidel ML: Substance P dependence of endosomal fusion during bladder inflammation. Am J Physiol Renal Physiol. 2000, 278 (3): F440-451.
Marchand JE, Sant GR, Kream RM: Increased expression of substance P receptor-encoding mRNA in bladder biopsies from patients with interstitial cystitis. Br J Urol. 1998, 81: 224-228.
Hohenfellner M, Nunes L, Schmidt RA, Lampel A, Thuroff JW, Tanagho EA: Interstitial cystitis: increased sympathetic innervation and related neuropeptide synthesis. J Urol. 1992, 147 (3): 587-591.
Pang X, Marchand J, Sant GR, Kream RM, Theoharides TC: Increased number of substance P positive nerve fibres in interstitial cystitis. Br J Urol. 1995, 75 (6): 744-750.
Cruz F, Guimaraes M, Silva C, Reis M: Suppression of bladder hyperreflexia by intravesical resiniferatoxin. Lancet. 1997, 350 (9078): 640-641. 10.1016/S0140-6736(05)63330-2.
Lecci A, Giuliani S, Santicioli P, Maggi CA: Involvement of spinal tachykinin NK1 and NK2 receptors in detrusor hyperreflexia during chemical cystitis in anaesthetized rats. Eur J Pharmacol. 1994, 259 (2): 129-135. 10.1016/0014-2999(94)90501-0.
Lecci A, Tramontana M, Giuliani S, Criscuoli M, Maggi CA: Effect of tachykinin NK2 receptor blockade on detrusor hyperreflexia induced by bacterial toxin in rats. J Urol. 1998, 160 (1): 206-209. 10.1016/S0022-5347(01)63091-X.
Alfieri A, Gardner C: The NK1 antagonist GR203040 inhibits cyclophosphamide-induced damage in the rat and ferret bladder. Gen Pharmacol. 1997, 29 (2): 245-250.
Andersson KE: Tachykinins: role in detrusor overactivity?. Eur Urol. 2006, 49 (3): 423-425. 10.1016/j.eururo.2005.11.021.
Moore KH, Gilpin SA, Dixon JS, Richmond DH, Sutherst JR: Increase in presumptive sensory nerves of the urinary bladder in idiopathic detrusor instability. Br J Urol. 1992, 70 (4): 370-372.
Smet PJ, Moore KH, Jonavicius J: Distribution and colocalization of calcitonin gene-related peptide, tachykinins, and vasoactive intestinal peptide in normal and idiopathic unstable human urinary bladder. Lab Invest. 1997, 77 (1): 37-49.
Herbert MK, Holzer P: [Why are substance P(NK1)-receptor antagonists ineffective in pain treatment?]. Der Anaesthesist. 2002, 51 (4): 308-319. 10.1007/s00101-002-0296-7.
Moore KH, Lam DS, Lynch W, Burcher E: The tachykinin NK-2 receptor antagonist SR48968 does not block noncholinergic contractions in unstable human bladder. Peptides. 2002, 23 (6): 1155-1160. 10.1016/S0196-9781(02)00049-9.
Saban MR, Hellmich HL, Turner M, Nguyen NB, Vadigepalli R, Dyer DW, Hurst RE, Centola M, Saban R: The inflammatory and normal transcriptome of mouse bladder detrusor and mucosa. BMC Physiol. 2006, 6 (1): 1-10.1186/1472-6793-6-1.
Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF: A network-based analysis of systemic inflammation in humans. Nature. 2005, 437 (7061): 1032-1037. 10.1038/nature03985.
Bozic CR, Lu B, Hopken UE, Gerard C, Gerard NP: Neurogenic amplification of immune complex inflammation. Science. 1996, 273 (5282): 1722-1725. 10.1126/science.273.5282.1722.
Haak-Frendscho M, Saban R, Shields RL, Jardieu PM: Anti-immunoglobulin E antibody treatment blocks histamine release and tissue contraction in sensitized mice. Immunology. 1998, 94 (1): 115-121. 10.1046/j.1365-2567.1998.00478.x.
Saban MR, Nguyen NB, Hammond TG, Saban R: Gene expression profiling of mouse bladder inflammatory responses to LPS, substance P, and antigen-stimulation. Am J Pathol. 2002, 160 (6): 2095-2110.
D'Andrea MR, Saban MR, Gerard NP, Wershil BK, Saban R: Lack of neurokinin-1 receptor expression affects tissue mast cell numbers but not their spatial relationship with nerves. Am J Physiol Regul Integr Comp Physiol. 2005, 288 (2): R491-500.
Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M: Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001, 29 (4): 365-371. 10.1038/ng1201-365.
[http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GPL151]
Dozmorov I, Saban MR, Gerard NP, Lu B, Nguyen NB, Centola M, Saban R: Neurokinin 1 receptors and neprilysin modulation of mouse bladder gene regulation. Physiol Genomics. 2003, 12 (3): 239-250.
Ingenuity. [http://www.ingenuity.com]
Vadigepalli R, Chakravarthula P, Zak DE, Schwaber JS, Gonye GE: PAINT: a promoter analysis and interaction network generation tool for gene regulatory network identification. Omics. 2003, 7 (3): 235-252. 10.1089/153623103322452378.
Biobase. [http://www.biobase.de/pages/]
Saban MR, Saban R, Hammond TG, Haak-Frendscho M, Steinberg H, Tengowski MW, Bjorling DE: LPS-sensory peptide communication in experimental cystitis. Am J Physiol Renal Physiol. 2002, 282 (2): F202-210.
Wang W, Cherry JM, Nochomovitz Y, Jolly E, Botstein D, Li H: Inference of combinatorial regulation in yeast transcriptional networks: a case study of sporulation. Proc Natl Acad Sci USA. 2005, 102 (6): 1998-2003. 10.1073/pnas.0405537102.
Pahl HL: Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999, 18 (49): 6853-6866. 10.1038/sj.onc.1203239.
Kang S, Kim YB, Kim MH, Yoon KS, Kim JW, Park NH, Song YS, Kang D, Yoo KY, Kang SB, Lee HP: Polymorphism in the nuclear factor kappa-B binding promoter region of cyclooxygenase-2 is associated with an increased risk of bladder cancer. Cancer Lett. 2005, 217 (1): 11-16. 10.1016/j.canlet.2004.06.053.
Horiguchi Y, Kuroda K, Nakashima J, Murai M, Umezawa K: Antitumor effect of a novel nuclear factor-kappa B activation inhibitor in bladder cancer cells. Expert Rev Anticancer Ther. 2003, 3 (6): 793-798. 10.1586/14737140.3.6.793.
Karashima T, Sweeney P, Kamat A, Huang S, Kim SJ, Bar-Eli M, McConkey DJ, Dinney CP: Nuclear factor-kappaB mediates angiogenesis and metastasis of human bladder cancer through the regulation of interleukin-8. Clin Cancer Res. 2003, 9 (7): 2786-2797.
Saban MR, Memet S, Jackson DG, Ash J, Roig AA, Israel A, Saban R: Visualization of lymphatic vessels through NF-kB activity. Blood. 2004, 104 (10): 3228-3230. 10.1182/blood-2004-04-1428.
Wang XC, Saban R, Kaysen JH, Saban MR, Allen PL, Benes EN, Hammond TG: Nuclear factor kappa B mediates lipopolysaccharide-induced inflammation in the urinary bladder. J Urol. 2000, 163 (3): 993-998. 10.1016/S0022-5347(05)67870-6.
Quinlan KL, Naik SM, Cannon G, Armstrong CA, Bunnett NW, Ansel JC, Caughman SW: Substance P activates coincident NF-AT- and NF-kappa B-dependent adhesion molecule gene expression in microvascular endothelial cells through intracellular calcium mobilization. J Immunol. 1999, 163 (10): 5656-5665.
Lieb K, Fiebich BL, Berger M, Bauer J, Schulze-Osthoff K: The neuropeptide substance P activates transcription factor NF-kappa B and kappa B-dependent gene expression in human astrocytoma cells. J Immunol. 1997, 159 (10): 4952-4958.
Zhu Y, Casado M, Vaulont S, Sharma K: Role of Upstream Stimulatory Factors in Regulation of Renal Transforming Growth Factor-{beta}1. Diabetes. 2005, 54 (7): 1976-1984. 10.2337/diabetes.54.7.1976.
Chen CY, Schwartz RJ: Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, nkx-2.5. J Biol Chem. 1995, 270 (26): 15628-15633. 10.1074/jbc.270.26.15628.
Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP: Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993, 119 (2): 419-431.
Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP: Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995, 9 (13): 1654-1666.
Small EM, Krieg PA: Transgenic analysis of the atrialnatriuretic factor (ANF) promoter: Nkx2-5 and GATA-4 binding sites are required for atrial specific expression of ANF. Dev Biol. 2003, 261 (1): 116-131. 10.1016/S0012-1606(03)00306-3.
Ma KK, Banas K, de Bold AJ: Determinants of inducible brain natriuretic peptide promoter activity. Regul Pept. 2005, 128 (3): 169-176. 10.1016/j.regpep.2004.12.025.
Ponticos M, Partridge T, Black CM, Abraham DJ, Bou-Gharios G: Regulation of collagen type I in vascular smooth muscle cells by competition between Nkx2.5 and deltaEF1/ZEB1. Mol Cell Biol. 2004, 24 (14): 6151-6161. 10.1128/MCB.24.14.6151-6161.2004.
Riazi AM, Lee H, Hsu C, Van Arsdell G: CSX/Nkx2.5 modulates differentiation of skeletal myoblasts and promotes differentiation into neuronal cells in vitro. J Biol Chem. 2005, 280 (11): 10716-10720. 10.1074/jbc.M500028200.
Theodosiou NA, Tabin CJ: Sox9 and Nkx2.5 determine the pyloric sphincter epithelium under the control of BMP signaling. Dev Biol. 2005, 279 (2): 481-490. 10.1016/j.ydbio.2004.12.019.
Stanley EG, Biben C, Elefanty A, Barnett L, Koentgen F, Robb L, Harvey RP: Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3'UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int J Dev Biol. 2002, 46 (4): 431-439.
Campbell SE, Sood A, Argyle DJ, Nasir L, Argyle SA, Bennett D: The cloning and functional analysis of canine matrix metalloproteinase-13 gene promoter. Gene. 2002, 286 (2): 233-240. 10.1016/S0378-1119(02)00436-5.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2490/7/7/prepub
Acknowledgements
Supported by National Institutes of Health grants DK 55828-01 and DK066101-01 (R.S.).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
RS conceived of the study and drafted the manuscript. CS performed the EMSAs. RV consulted RS regarding PAINT and TRANSFAC analysis. SM introduced EMSA experiments in RS's laboratory and helped CS with the interpretation on EMSA results. ID normalized and analyzed the gene-array results. MRS participated in the experimental design, carried out the animal experiments, extracted the RNA and performed gene array results.
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Saban, R., Simpson, C., Vadigepalli, R. et al. Bladder inflammatory transcriptome in response to tachykinins: Neurokinin 1 receptor-dependent genes and transcription regulatory elements. BMC Urol 7, 7 (2007). https://doi.org/10.1186/1471-2490-7-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2490-7-7