Abstract
Background
Microarray analyses allow the identification and assessment of molecular signatures in whole tissues undergoing pathological processes. To better understand cerebral malaria pathogenesis, we investigated intra-cerebral gene-expression profiles in well-defined genetically cerebral malaria-resistant (CM-R) and CM-susceptible (CM-S) mice, upon infection by Plasmodium berghei ANKA (PbA). We investigated mouse transcriptional responses at early and late stages of infection by use of cDNA microarrays.
Results
Through a rigorous statistical approach with multiple testing corrections, we showed that PbA significantly altered brain gene expression in CM-R (BALB/c), and in CM-S (CBA/J and C57BL/6) mice, and that 327 genes discriminated between early and late infection stages, between mouse strains, and between CM-R and CM-S mice. We further identified 104, 56, 84 genes with significant differential expression between CM-R and CM-S mice on days 2, 5, and 7 respectively. The analysis of their functional annotation indicates that genes involved in metabolic energy pathways, the inflammatory response, and the neuroprotection/neurotoxicity balance play a major role in cerebral malaria pathogenesis. In addition, our data suggest that cerebral malaria and Alzheimer's disease may share some common mechanisms of pathogenesis, as illustrated by the accumulation of β-amyloid proteins in brains of CM-S mice, but not of CM-R mice.
Conclusion
Our microarray analysis highlighted marked changes in several molecular pathways in CM-S compared to CM-R mice, particularly at early stages of infection. This study revealed some promising areas for exploration that may both provide new insight into the knowledge of CM pathogenesis and the development of novel therapeutic strategies.
Similar content being viewed by others
Background
Malaria is a disease affecting millions of people worldwide. Cerebral malaria (CM) is one of the most severe complications and is a major cause of death. Both host and parasite genetic factors play important roles in the outcome of malaria infection. Epidemiological data, candidate gene studies, and genetic linkage studies clearly support the existence of a genetic contribution to susceptibility to human malaria [1]. In parallel with human studies, malaria susceptibility genes have been mapped in mouse models, and the role of some genes has been demonstrated [2]. It is clear, however, that a number of malaria susceptibility genes remain to be identified. These include genes, whose expression is likely deregulated upon malaria infection.
Transcriptional profiling may provide new tools for identifying the key genes that govern host responses against pathogens. Recently, several reports have described gene expression changes that accompany the host response against Plasmodium spp. Microarrays have been analyzed from mice [3–5], rhesus monkey [6] and humans [7], upon infection by Plasmodium spp A parallel can be observed in the regulation of genes involved in immune responses, glycolysis, and erythropoiesis. These data suggested that variation in host gene expression may be associated with resistance or susceptibility to malaria.
Recently, we investigated brain gene expression patterns in well-defined genetically CM-resistant (CM-R) and CM-susceptible mice (CM-S) by use of cDNA microarrray [8]. We identified a set of genes that perfectly discriminates between CM-R and CM-S mice at the time of CM onset. This indicates that gene expression analysis using microarray tools may be useful for the identification of candidate genes that are potentially responsible for resistance or susceptibility to CM. Nevertheless, an important issue was to identify genes whose expression differ between CM-R and CM-S mice before the time of CM onset to identify early events that may participate in malaria pathogenesis. In this report, we present an analysis of genes differentially expressed in brains from CM-R and CM-S mice prior to infection, and at the early and late stages of infection with Plasmodium berghei ANKA (PbA). Data analysis reveals that molecules belonging to several biological processes were preferentially and differentially expressed between CM-R and CM-S mice, and that a number of gene expression changes occurred at the early and late stages of infection. Herein, we discuss new working hypotheses on this basis.
Results
Identification of genes regulated in brains by PbA infection
The ANOVA of microarray data revealed significant gene expression changes over the course of infection in BALB/c (n = 25) mice, CBA/J mice (n = 16), and in C57BL/6 mice (n = 20). We calculated empirical P values for each gene, and we considered P < 0.05 significant. On this basis, we selected 174, 210, and 342 genes for BALB/c mice, CBA/J mice, and C57BL/6 mice, respectively.
To further compare uninfected mice with infected mice, we performed a Welch t test and we applied a Bonferroni correction to account for multiple tests performed (Figure 1). Figure 2 shows the number of genes whose expression was significantly altered by PbA infection. The mouse strains displayed various patterns. Strikingly, the number of genes that showed significant expression changes was higher in C57BL/6 mice than in CBA/J mice on day 2 post-infection with PbA (Figure 2A). The number of genes that showed significant expression changes in CBA/J mice gradually increased during infection. In contrast, the number of genes that showed significant expression changes in both C57BL/6 mice and BALB/c mice on day 2 was similar to the number of genes identified on day 7; it was, nevertheless, lower on day 5 than on days 2 and 7. As shown in Figure 2, most of the genes identified in C57BL/6 mice (Figure 2B) and in CBA/J mice (Figure 2C) were under-expressed, while most of the genes identified in BALB/c mice (Figure 2D) were over-expressed.
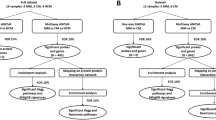
Schematic outline of data analysis. HCL: hierarchical clustering. a Brain gene expression prior to infection was compared with brain gene expression on days 2, 5 and 7. b Brain gene expression in CM-R mice was compared with that in CM-S mice at each time point. As represented by dashed arrows, we considered the whole data set (n = 2012 genes) to carry out multiple testing correction.
Distribution of genes regulated by PbA infection according to the time of infection. The genes whose expression was significantly altered by PbA infection were identified by using pair-wise Welch t tests with a Bonferroni correction.A. The number of genes with significant changes is shown at each time point for each mouse strain. B, C and D. The number of genes significantly up- (positive values) and down-regulated (negative values) is shown at each time point for each mouse strain.
A Venn diagram summarizes the number of overlapping genes with significant differential expression at different time points (Figure 3). Most of the genes identified on day 2 were no longer identified on day 7 in C57BL/6 mice and in BALB/c mice. As shown in Figure 3A–B and 3E–F, 42 of 53 and 31 of 36 genes with differential expression on day 2 did not show differential expression on days 5 and 7 in C57BL/6 and BALB/c mice, respectively. In contrast, 10 of 14 genes regulated by PbA infection on day 2 were also regulated on days 5 and 7 in CBA/J mice (Figure 3C–D), indicating that C57BL/6 mice and CBA/J mice partly differ in their transcriptional response over the course of infection.
Overlapping genes with significant differential expression at different time points during PbA infection. This Venn diagrams show the number of overlapping genes with significant expression changes on days 2, 5, and 7 post-infection with PbA, for C57BL/6 mice (A and B), CBA/J mice (C and D) and BALB/c mice (E and F). Genes that are either induced or suppressed by infection are distinguished for each strain: the number of genes induced is shown in A, C, and E, while the number of genes suppressed is shown in B, D, and F.
Microarray analysis discriminates between CM-R and CM-S mice according to the time after infection
We searched for genes showing differential expression between mouse strains before infection. The ANOVA identified 125 genes without multiple test correction (data not shown), and 7 genes with Bonferroni multiple test correction at the level of 5%: 1700023B02Rik, Acot8, Gpd2, Sec11c, Ngly1, Zfp346 and Tln2. These results revealed a minor natural variation in gene expression between mouse strains. Nevertheless, we took into account this natural variation in further analyses to search for transcriptional changes. Thus, the individual gene expression level in infected mice was adjusted for the gene expression level in uninfected mice for each mouse strain.
To focus on transcriptional changes associated with resistance or susceptibility, we searched for a set of genes that discriminated between CM-R mice (BALB/c) and CM-S mice (C57BL/6 and CBA/J). To this end, we used the multi-class Significant Analysis of Microarrays (SAM) procedure, and we applied a false discovery rate of 0% (Figure 1). The analysis yielded a set of 327 genes [see Additional file 1], which was used to perform unsupervised hierarchical clustering (Figure 4A). Interestingly, the expression of several genes was induced in CM-R mice or suppressed in CM-S mice as a result of infection, while the expression of other genes was suppressed in CM-R mice and induced in CM-S mice [see Additional file 1]. This led to successfully classify CM-R and CM-S mice according to the time after infection (Figure 4B) and to determine five clusters (Figure 4A). Clusters A and C fully grouped CM-R mice (BALB/c) versus CM-S mice (C57BL/6 and CBA/J) whatever the time after infection. Genes of cluster A were over-expressed in CM-S mice compared to CM-R mice, while genes of cluster C were under-expressed in CM-S mice compared to CM-R mice. Clusters B and D also grouped CM-R mice versus CM-S mice. Nevertheless, clusters B and D discriminated between C57BL/6 and CBA/J mice, indicating differential gene expression between the two CM-S strains at early time points of infection. Genes of clusters B and D were over-expressed in CBA/J mice on day 2 post-infection compared to C57BL/6 mice, and in C57BL/6 on days 2 and 5 post-infection compared to CBA/J mice, respectively. At late stage of infection, genes of clusters B and D showed similar gene expression between the two CM-S strains, and were under-expressed in CM-S mice compared to CM-R mice. Cluster E discriminated mice at early and late stage of infection. Nevertheless, genes of cluster E were over-expressed in CM-S mice compared to CM-R mice indicating an interval between the response of CM-S and the response of CM-R mice.
Hierarchical classification of mouse-strain-specific and CM-R/CM-S genes according to the time of infection. A. Hierarchical clustering of the 58 brain tissue samples, using expression levels of 327 significant genes differentially expressed between the mouse strains at early and late stages of infection. This set of genes was extracted from the full data set (n = 2012) by use of a SAM procedure and a false discovery rate of 0%. Each row represents a gene and each column represents a sample. Red and green indicate expression levels above and below the median, respectively. Grey indicates missing data. Dendograms of samples (above matrix) and genes (to the left of matrix) represent overall similarities in gene expression profiles. B. Dendogram of samples representing the results of the same global hierarchical clustering applied to the 58 brain tissue samples. Clustering of technical replicates (CBA/J-5, C57BL6-20 and BALB/c-13) is shown: the samples taken from a CBA/J mouse on day 2, a C57L/6 mouse on day 7, and a BALB/c mouse on day 7 were run on 5, 2, and 2 microarrays, respectively. Samples taken from mice on days 2, 5, 7, 9, and 15 post-infection are coded D2, D5, D7, D9, and D15, respectively.
To analyze functional annotations related to CM, we sought biological process Gene Ontology (GO) terms and KEGG pathways for the 327 genes that discriminated between CM-R mice (BALB/c) and CM-S mice (C57BL/6 and CBA/J). The analysis of biological process GO terms of gene clusters showed an over-representation of some GO terms. In particular, the GO terms related to the "defense response" category, such as the "immune response" or the "inflammatory response" GO terms were strongly over-represented in cluster E (Table 1). Similarly, most of the genes grouped in cluster E were found to be involved in KEGG pathways related to immune responses, such as "cytokine-cytokine receptor interaction" or "natural killer cell mediated cytotoxicity". The analysis of other clusters did not reveal, however, a strong over-representation of particular GO terms, suggesting that these clusters were heterogeneous. In particular, clusters B and C contained 38 and 207 genes with heterogeneous GO categories. This was further supported by the analysis of KEGG pathways, which pointed out a number of different pathways, such as metabolic energy pathways ("glycolysis/gluconeogenesis" and "oxidative phosphorylation"), immune responses ("cytokine-cytokine receptor interaction" and "antigen processing and presentation"), haematopoiesis, ("hematopoietic cell lineage"), cytoskeleton pathways ("regulation of active cytoskeleton"), or pathways related to brain function ("neurodegenerative disorders" and "axon guidance") (Figure 5).
View of biological functional annotation repartition of the genes grouped in clusters B and C. The KEGG pathways, in which the genes were known to be involved, are shown. Of the 245 genes of the clusters B and C, 80 were annotated. We represented only the KEGG pathways that contained at least three genes.
We specifically searched for genes with significant differential expression between CM-R and CM-S mice. Thus, we used the Welch t test to compare BALB/c mice (CM-R mice) on the one hand and CBA/J mice and C57BL/6 mice (CM-S mice) on the other hand (Figure 1). We tested the 327 genes identified by the SAM multi-class procedure, but we considered the whole data set (n = 2012 genes) to carry out multiple testing correction. Among the 327 genes, 177 genes were further found to be differentially expressed between CM-R mice and CM-S mice (Table 2). Among the 177 genes, 104, 56, and 84 genes showed significant differential expression on days 2, 5, and 7, respectively, and 11, 17, 138, and 11 were grouped in clusters A, B, C and E, respectively. The number of co-occurring genes with differential expression between CM-R and CM-S mice detected at different times is displayed in Figure 6. Eighteen of 177 genes were up-regulated in CM-S mice compared to CM-R mice, while 158 of 177 genes were up-regulated in CM-R mice compared to CM-S mice. Only one gene showed a complicated picture: Fyco1 was down-regulated in CBA/J and C57BL/6 mice (CM-S mice) mice on day 2, and in CBA/J mice on day 5, while it was up-regulated in C57BL/6 mice on day 5. Table 3 show the most represented KEGG pathways, which included pathways related to metabolism, erythropoiesis, immune responses, neuronal development, and neurodegenerative disorders.
We further analyzed the expression of genes involved in neuronal development or in neurodegenerative disorders using immunochemistry. Thus, we studied the expression of reelin (Reln), which is involved in neurogenesis, and we searched for the presence of β-amyloid protein, which is involved in Alzheimer's disease. We detected RELN only in brains of BALB/c mice at days 5 and 7, while we showed the presence of β-amyloid only in brains of CBA/J and C57BL/6 mice at days 5 and 7 (Table 4).
Overall, our gene expression analysis revealed marked changes in metabolic energy pathways, the inflammatory response, and pathways related to neurogenesis and neurodegenerative disorders in CM-S mice versus CM-R mice.
Discussion
In the present study, we have searched for genes and physiological pathways potentially involved in CM. To this aim, we performed a longitudinal analysis of differentially expressed genes in brains from well-defined genetically CM-R (BALB/c) and CM-S (C57BL/6, and CBA/J) mice at early and late stages of infection. The present study shows that PbA strongly altered gene expression in these mice. In particular, gene expression was deeply altered at the time of CM onset, confirming our previous study [8]. Here, we show that a number of genes were over-expressed in CM-R mice at the early stage of infection, suggesting that CM-R mice mount an early protective transcriptional response. In this way, we found an association of resistance to CM with an increase in the expression of a number of genes on day 2 post-infection (Figure 4 and Table 2). In contrast, a number of genes were found to be under-expressed in C57BL/6 mice at the early stage of infection, while few genes were found to be regulated in CBA/J mice at the same time (Figure 2). Also, CBA/J mice appeared to mount a gradual response that may be involved in CM pathogenesis, while C57BL/6 mice may mount two waves of transcriptional responses both of them potentially implicated in malaria pathogenesis. These observations are consistent with other reports suggesting that CM mediators partly differ between C57BL/6 and CBA/J mice [8–10]. Nevertheless, both CBA/J and C57BL/6 mice showed a pronounced up-regulation of genes involved in either interferon-associated response or in glycolysis at the late stage, and a down-regulation of genes involved in erythropoiesis both at early and late stages, as previously described [3].
To identify genes with significant transcriptional changes associated with CM, we performed, on the whole data set, a multi-class SAM procedure with a very stringent false discovery rate followed by a Welch t test with a Bonferroni correction. In other words, we applied a two-filtering procedure to decrease the number of "false positive" genes. To account for natural variation between mouse strains, we adjusted gene expression level in infected mice for gene expression level in uninfected mice. Thus, we identified a number of genes differentially regulated between CM-R and CM-S mice. The 327 most differentially expressed genes identified by the SAM analysis allowed the complete discrimination between CM-R and CM-S mice according to the time of infection. The same result was obtained with the subset of 177 genes identified by the Welch t test (data not shown). This further confirms our previous study that investigated gene expression at the time of CM onset [8].
EASE analysis of either the 327 genes or the 177 genes revealed that some of the most represented biological process categories were related to the "defense response", such as the "response to parasite" or the "inflammatory response" terms. This was further supported by the results of the KEGG pathway analysis. In addition, genes were found to be involved in KEGG pathways related to metabolism, such as "oxidative phosphorylation", "glycolysis/gluconeogenesis", or "tryptophan metabolism". The analysis of functional annotation also revealed GO terms and KEGG pathways related to brain, such as the "axon guidance" and the "neurodegenerative disorders" KEGG pathways.
Overall, the analysis of functional annotation is consistent with the view that mouse CM is characterized among others by the deregulation of both immune response and glucose metabolism [11, 12]. This leads to an abnormal increase in the inflammatory response and to hypoglycemia and acidosis in CM-S mice [11, 12]. In addition, our data provide evidence of an intrinsic deficiency in oxidative phosphorylation, and the functional annotations related to brain disease suggest the role of genes expressed by brain cells in resistance or susceptibility to CM. Thus, despite a number of genes identified by our microarray analysis, which may highlight highly complex interactions between the parasite and the host, several major features of the transcriptional profile can be deduced.
First, PbA infection affects the expression of genes involved in metabolic energy pathways. The expression of Uqcrq, Cox4i1, Ndufb8, Atp6v1g2 and Atp5b involved in oxidative phosphorylation was upregulated in CM-R mice on day 2 post-infection [see Additional file 2], suggesting that cerebral oxidative metabolism may be stimulated by PbA infection in CM-R mice. In contrast, these genes were downregulated in C57BL/6 mice on day 2 post-infection and in CBA/J mice on day 7 post-infection, while only Ndufb8 was downregulated in CBA/J mice on day 2 post-infection. These transcriptional changes were associated with CM. A similar pattern was observed for Hmgcs1 involved in ketone metabolism. In the same way, lower NAD+/NADH levels and decreased mitochondrial function have been observed in CM-S mice by others [12, 13]. These observations may be related to hypoxia and hypoglycemia, which reflect the low level of metabolic energy substrates. Alternatively, our data are consistent with the "cytopathic hypoxia" hypothesis, which rather proposes an adequate oxygen supply but an abnormal oxygen use [14]. Also, the low level of oxidative phosphorylation gene expression and ketone bodies pathway genes that we detected in the brain of CM-S mice suggests that cerebral oxidative metabolism may be inhibited even without oxygen delivery being impaired.
This metabolic disturbance also leads to lactate production and acidosis. In addition, this leads to an accumulation of ADP, which favors platelet aggregation [15]. Interestingly, platelet aggregation is known to be stimulated by PAF acether and inhibited by AMPc, the expression of which is inhibited by LIS1 (PAFAH1B1) and PDE4B, respectively [16, 17]. Overall, the lower expression of Pafah1b1 in CM-S mice compared to CM-R mice [8], the higher expression of Pde4b in CM-S mice compared to CM-R mice, and the metabolic disturbance leading to an accumulation of ADP may participate in the platelet aggregation process in the cerebral microvasculature of CM-S mice.
Second, it is likely that the inflammatory response plays a major role in CM pathogenesis. In particular, a surge of IFNγ production at 3 to 4 days p.i. was demonstrated to be essential for murine CM, and this may be due to the absence of regulation in IFNγ pathways at early stages in PbA infection [18–20]. IFNγ is a proinflammatory cytokine typically produced by Th1 lymphocytes, and it is thought that the Th2 response protects from CM [11]. In this way, our microarray analysis showed that IL4 and IL4R were over-expressed in CM-R mice from day 2. Similarly, Dnaja3, Foxo3a, and Ptpns1 that inhibit the activity of NF-kB [21–23] had lower expression levels in CM-S mice than in CM-R mice, while Nfkbia, a marker of the NF-kB signalling pathway involved in inflammation [24], was over-expressed in CM-S mice. In addition, C1qa and Pde4b that are involved in the inflammatory response [17, 25] were found to be over-expressed in CM-S mice compared to CM-R mice. S100a10 that inhibits the activity of phospholipase A2 [26], and Bcl6 that inhibits the production of MIP-alpha and IP-10 [27] were under-expressed in CM-S mice compared to CM-R mice. Blood cells are known to be involved in the inflammatory response due to malarial infection. Indeed, CD4+ and CD8+ T lymphocytes, platelets, monocytes have been shown to cooperate in the cerebral microvasculature, and this causes inflammation, endothelial cell damage, and hemorrhages [9, 20, 28–30]. In addition, Gzmb, whose expression was up-regulated in CM-S mice [8], encodes granzyme B in cytotoxic T lymphocytes, and is thought to be involved in the breakdown of the blood-brain barrier [31]. The influence of immune responses on the blood-brain barrier may be partly reflected by changes in expression of genes involved in either cytoskeletal and tight-junction pathways or cell adhesion pathways in CM-S mice.
It has been also suggested that glial cells actively participate in the local inflammatory response caused by malarial infection [32]. In this way, glial cells that have been shown to be activated on day 3 post-infection by PbA can produce C1q components [32, 33]. The NF-kB signalling pathway has been demonstrated in these cells [34]. Interestingly, Cd200 that is implicated in the control of the activation of glial cells was strongly under-expressed in the CBA/J CM-S mice [35]. Besides, the expression of IL4 and IL4R was shown to be higher in CM-R mice than in CM-S mice. Since IL-4 induces apoptosis in activated glial cells that express IL-4R [36], IL-4 may contribute to the down-regulation of brain inflammation in CM-R mice.
Third, genes involved in the neuroprotection/neurotoxicity balance and/or in neurogenesis may protect the host against CM. This hypothesis is based on changes in tryptophan metabolism caused by PbA infection. Sanni et al (1998) showed an increase of the activity of indoleamine 2,3-dioxygenase whose expression is induced by TNF and IFNγ [37]. This leads to an increase of the ratio quinolinic acid/kynurenic acid, and to neuro-excitotoxic damage associated with CM. In humans, high levels of quinolinic acid have been associated with CM [38]. Interestingly, genes involved in tryptophan metabolism, such as Ube3a, Prnt3, and Aldh1b1, were differentially regulated between CM-R and CM-S mice. The expression of these genes was enhanced by infection in CM-R mice. In addition, genes having a neuroprotective role, such Agtr2, Bag1, Csnk1a1 and Reln [39–42], were shown to be over-expressed in CM-R mice compared to CM-S mice. Indeed, Rtn3 and Creb1 that are markers of neuronal survival [43, 44], were also over-expressed in CM-R mice. Besides, Reln and Dab1, which were over-expressed in CM-R mice, are known to be involved in neurogenesis [45, 46]. Similarly, Pafah1b1 whose expression was associated with resistance to CM [8] is involved in the Reln pathway [46]. This suggests that CM-S mice are deficient in neurogenesis, and that they cannot repair neuronal damages. In contrast, CM-R mice might be able to repair such damage (Figure 7).
Schematic diagram showing the possible effects of the reelin pathway in protection from CM. Reelin (RELN) is an extracellular matrix serine protease expressed in some neurons, such as GABAergic interneurons, which inhibit excitotoxic neurotransmission [45]. RELN that is secreted into the extracellular space acts by paracrine and autocrine mechanisms. RELN interacts with very low-density lipoprotein receptors (VLDLR) and apolipoprotein E type 2 receptors (ApoER2) leading to tyrosine phosphorylation of the adaptor protein Disabled-1 (DAB1) by the SRC family kinases (SRC) [42]. DAB1 activation, in turn, activates PI3K/Akt signalling, which has been implicated in neuronal migration during development and adulthood. In addition, phosphorylated DAB1 interacts with LIS1, a protein encoded by Pafah1b1, which associates with microtubules and modulates neuronal migration [46]. LIS1 may be required for regulating crucial steps of reelin-dependent neuronal positioning. In parallel, phosphorylated DAB1 inhibits glycogen synthase kinase 3β (GSK3β), a kinase known to phosphorylate Tau protein at multiple sites. Therefore, the activation of RELN pathway diminishes the level of hyperphosphorylated Tau protein, which is a biomarker of brain injury. In particular, hyperphosphorylated Tau protein is a component of the neurofibrillary tangles involved in Alzheimer's disease. Reln, Dab1 and Pafah1b1 were shown to be over-expressed in CM-R mice compared to CM-S mice. The activation of RELN signalling may inhibit excitotoxic neurotransmission and Tau phosphorylation, and may activate neurogenesis in CM-R mice. This may lead to diminished brain injury and to increased brain injury repair. Solid arrows represent influences on the activity of proteins or physiological mechanisms. Dashed arrows represent impaired effects on the activity of proteins or physiological mechanisms. Negative signs indicate inhibition, and positive signs indicate activation.
So far, little research has been conducted on the issue of neuroprotective responses in CM. Interestingly, the Reln pathway that inhibits the phosphorylation of the protein Tau has been recently proposed as a protective mechanism against Alzheimer'disease [47]. In addition, decreased ribosomal RNA levels and decreased rates for protein synthesis have been recently described in Alzheimer'disease [48], while we showed the down-regulation of several genes encoding ribosomal proteins in CM-S mice. In the same way, Medana et al (2002) detected β-amyloid precursor protein in humans with CM [49], and we report here the β-amyloid protein in brains of CM-S mice but not in CM-R mice. This result is consistent with the down-regulation of Arc, Itm2b, Bsg, Rtn3, and Il4 in CM-S mice, and with the up-regulation of Pde4b in CM-S mice. Indeed, Itm2b, Bsg, Rtn3, and Il4 inhibit the production of the β-amyloid protein [50–53]. Besides, the β-amyloid protein inhibits the expression of Arc, while it increases the expression of Pde4b [25, 54]. These observations suggest that cerebral malaria and Alzheimer'disease share some common mechanisms of pathogenesis.
Conclusion
In this study, we have confirmed that gene-expression profiling discriminates between CM-R and CM-S mice, and we have identified genes whose expression showed consistent differential expression between CM-R and CM-S mice at early and late stages of infection. The analysis of gene functional annotation reveals several major features. First, it indicated that brain metabolic energy metabolism was early and deeply disturbed in CM-S mice, suggesting that high lactate production may be due rather to metabolic disturbance than to deficient oxygen supply. Second, the influence of inflammatory response on CM was also clearly detected, and our data are consistent with an active role of microglial cells in local inflammation. Third, the outcome of infection may critically depend on either cerebral tissue protective responses or brain repair capacity. Overall, our microarray analysis may give a global overview of critical events occurring in CM-S mice compared to CM-R mice. Searching for polymorphisms that alter the expression of genes identified should help in determining the genetic control of cerebral malaria. This analysis also revealed some promising areas for exploration that may both provide new insight into the key events that govern CM pathogenesis and the development of therapeutic strategies. In particular, novel neuroprotective therapies may be proposed as adjuncts to anti-malarial therapy.
Methods
Mouse strains and phenotyping
Six to 8 weeks old BALB/c (CM-R strain), C57BL/6J and CBA/J (CM-S strains) female, were obtained from IFFA CREDO (Ch. River Lab, France) and kept in our facilities. Three mice from each strain were not infected, and 22 BALB/c, 17 C57BL/6J, and 15 CBA/J were infected by i.p. injection of 106 PbA parasitized erythrocytes. The frozen stabilates were obtained with an uncloned line, and were prepared from CBA/J day 7-infected mice [30]. The parasite was conserved as stabilates of 107 parasitized erythrocytes stored in liquid nitrogen in Alsever's solution containing 10% glycerol. Parasitemia was monitored daily by blood smear. No difference was observed between mouse strains. Parasitemia was 0.8%±0.9, 4.5%±2.2, and 8.7%±5.3 on days 2, 5, and 7 post-infection, respectively. The CM-S mice developed a neurological syndrome (mono-, hemi-, para-, or tetraplegia, ataxia, deviation of the head, and convulsions), which occurred 6 to 7 days after parasite inoculation with a cumulative mortality of 100%. The CM-R mice did not present neurologic lesions and died during the 3rd or the 4th week of infection, with severe anaemia and hyperparasitemia [9]. The parasitemia of CM-R mice was 16.6%±8.9 and 62.8%±25.7 on days 9 and 15, respectively.
Organ sampling
Brains were taken from CM-R and CM-S mice before and after infection. Brains from three uninfected mice were taken for each strain. Three, 4, and 6 CBA/J were analyzed on days 2, 5, and 7 post-infection, respectively, while 4, 4, and 9 C57BL/6 were analyzed on days 2, 5, and 7 post-infection, respectively. Brains from 3, 4, 5, 5, and 5 BALB/c mice were taken on days 2, 5, 7, 9 and 15 post-infection. Brains were completely removed and were cut into two parts: one part was frozen in RNALater (Qiagen, TM) until RNA analysis, and the other part was embedded in Tissue Tek (Leica), snap frozen in liquid nitrogen, and kept at -80°C until histopathological analysis of cryosections.
Immunochemistry
For immunostaining, frozen sections were incubated for 45 minutes with primary monoclonal antibodies directed against murine Reelin and β-Amyloid peptide (Santa Cruz, Tebu Bio) after saturation with appropriate serum. After washing, sections were incubated for 45 minutes with biotinylated polyclonal antibodies, followed by the addition of HRPO-avidin (anti-rat or anti-hamster ABC kits; Vector, Peterborough, England). Color reaction was obtained by the addition of Novared (AbCys). Slides were counterstained with Mayer's hematoxylin before permanent mounting with Entellan (Merck, Brussels, Belgium). Slides were pictured at 200 magnification using an Eclipse 800 microscope (Nikon, Champignysur- Marne, France) and a digital camera; labelling was then analyzed by quantitative digitalized image analysis using Lucia software (Nikon). At least 3 brains were sampled for each time point and each mouse strain and image analysis was performed on an average of 25 microphotographs per mouse.
RNA isolation and cDNA microarray hybridizations
Total RNA from brains was extracted using TRIzol reagent (Gibco-BRL, Life Technologies). The quality of RNA was confirmed on a formaldehyde agarose gel, and the concentration of RNA was determined by reading absorbance at 260/280 nm. RNA from 2 CBA/J mice had an inadequate quality, and was not further processed. Each mRNA sample extracted from an individual brain was run on a single microarray. In addition, three samples were run on several microarrays, and were considered as technical replicates: samples from CBA/J, C57BL/6, and BALB/c mice were run on 5, 2, and 2 microarrays, respectively. All microarray procedures were done at our microarray core facility [55]. cDNA microarrays were designed and prepared as described [56]. Briefly, the microarrays used in this study were composed of 8388 sequences. The following cDNA libraries were used: the NIA Mouse 15 K cDNA clone set, 2NbMT (thymus), NbMLN (lymph node), and 3NbMS (spleen). Detailed descriptions of these cDNA libraries are available at the UniGene database website (2NbMT: Lib.544, 3NbMS: Lib.553, NbMLN: Lib.567, NIA 15 K: Lib.8622) [57]. PCR amplification was performed as previously described [56], and PCR products were spotted onto nylon membranes (Hybond-N+, Amersham) with a MicroGrid II arrayer (Affymetrix, Santa Clara, CA). About 10% of the genes included in this clone set are represented by two or more different cDNA clones, providing internal controls to assess the reproducibility of gene expression measurements. Microarrays were hybridized with 33P-labelled probes, first with an oligonucleotide sequence common to all spotted PCR products (5'-TCACACAGGAAACAGCTATGAC-3'), then after stripping, with complex probes made from 5 μg of retrotranscribed total RNA. Probe preparations, hybridizations and washes were carried out as described previously [56]. After 48 h hybridization, arrays were scanned with a FUJI BAS5000 machine at 25 μm resolution. Hybridization signals were quantified using ArrayGauge software (Fuji Ltd, Tokyo, Japan).
Microarray data analysis
All images were carefully inspected, and spots with overestimated intensities due to neighborhood effects were manually excluded. The data were filtered such that only spots with intensities that were two times greater than the median background in either microarray were used in the analysis, and the signal intensities were then corrected to take into account the amount of spotted DNA and the variability of experimental conditions, as described [58]. Of the 8388 spotted clones, we selected the clones that had detectable expression levels in at least 80% of the experiments (n = 2012). Unsupervised hierarchical clustering investigated relationships between samples and relationships between genes. It was applied to data log-transformed and median-centred using the Cluster and TreeView programs (average linkage clustering using Pearson's correlation as similarity metric) [59].
Microarray data were statistically analyzed using the TIGR MeV (MultiExperiment Viewer) v3.1 software [60]. Figure 1 shows an outline of data analysis. A one-way ANOVA and SAM (Significant Analysis of Microarrays) procedures were applied to look for time-, strain-, and CM-R/CM-S-specific variation in gene expression in the full data set. One-way ANOVA and Welch t-statistics were used to analyze gene expression changes upon infection for each mouse strain. The values on days 2, 5, 7, 9 and 15 post-infection were compared to values on day 0 before infection. To search for gene expression changes associated with CM, a multi-class SAM procedure and a Welch t test were performed on the log2 ratios of infected vs uninfected samples. For each gene, the level of gene expression in each sample taken from an infected mouse was divided by the median of gene expression levels in samples taken from three uninfected mice. This calculation was done for each mouse strain. Multiple test corrections were performed [61].
The Expression Analysis Systematic Explorer (EASE) was used to search for common biological themes within gene lists generated by our microarray analysis [62]. EASE assigns identified genes to Gene Ontology (GO) terms, and tests whether specific biological pathways were over-represented within specific gene clusters. A score based on Fisher Exact test reports the probability that the prevalence of a particular theme within a cluster is due to chance alone given the prevalence of that theme in the population of all genes under study. In addition, we checked whether the genes were included in a KEGG pathway [63].
All data are MIAME compliant and have been loaded into ArrayExpress database [64]. The ArrayExpress accession number of this experiment is E-MEXP-1018.
Abbreviations
- CM:
-
Cerebral Malaria
- CM-S:
-
Cerebral Malaria-Susceptible mice
- CM-R:
-
Cerebral Malaria-Resistant mice
- PbA:
-
Plasmodium berghei ANKA
- GO:
-
Gene Ontology
- SAM:
-
Significant Analysis of Microarrays
- RELN:
-
Reelin
References
Kwiatkowski DP: How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005, 77 (2): 171-192. 10.1086/432519.
Fortin A, Stevenson MM, Gros P: Complex genetic control of susceptibility to malaria in mice. Genes Immun. 2002, 3 (4): 177-186. 10.1038/sj.gene.6363841.
Sexton AC, Good RT, Hansen DS, D'Ombrain MC, Buckingham L, Simpson K, Schofield L: Transcriptional profiling reveals suppressed erythropoiesis, up-regulated glycolysis, and interferon-associated responses in murine malaria. J Infect Dis. 2004, 189 (7): 1245-1256. 10.1086/382596.
Schaecher K, Kumar S, Yadava A, Vahey M, Ockenhouse CF: Genome-wide expression profiling in malaria infection reveals transcriptional changes associated with lethal and nonlethal outcomes. Infect Immun. 2005, 73 (9): 6091-6100. 10.1128/IAI.73.9.6091-6100.2005.
Lovegrove FE, Pena-Castillo L, Mohammad N, Liles WC, Hughes TR, Kain KC: Simultaneous host and parasite expression profiling identifies tissue-specific transcriptional programs associated with susceptibility or resistance to experimental cerebral malaria. BMC Genomics. 2006, 7: 295-10.1186/1471-2164-7-295.
Ylostalo J, Randall AC, Myers TA, Metzger M, Krogstad DJ, Cogswell FB: Transcriptome profiles of host gene expression in a monkey model of human malaria. J Infect Dis. 2005, 191 (3): 400-409. 10.1086/426868.
Griffiths MJ, Shafi MJ, Popper SJ, Hemingway CA, Kortok MM, Wathen A, Rockett KA, Mott R, Levin M, Newton CR, Marsh K, Relman DA, Kwiatkowski DP: Genomewide analysis of the host response to malaria in Kenyan children. J Infect Dis. 2005, 191 (10): 1599-1611. 10.1086/429297.
Delahaye NF, Coltel N, Puthier D, Flori L, Houlgatte R, Iraqi FA, Nguyen C, Grau GE, Rihet P: Gene-Expression Profiling Discriminates between Cerebral Malaria (CM)-Susceptible Mice and CM-Resistant Mice. J Infect Dis. 2006, 193 (2): 312-321. 10.1086/498579.
Lou J, Lucas R, Grau GE: Pathogenesis of cerebral malaria: recent experimental data and possible applications for humans. Clin Microbiol Rev. 2001, 14 (4): 810-20, table of contents. 10.1128/CMR.14.4.810-820.2001.
Engwerda CR, Mynott TL, Sawhney S, De Souza JB, Bickle QD, Kaye PM: Locally up-regulated lymphotoxin alpha, not systemic tumor necrosis factor alpha, is the principle mediator of murine cerebral malaria. J Exp Med. 2002, 195 (10): 1371-1377. 10.1084/jem.20020128.
Hunt NH, Grau GE: Cytokines: accelerators and brakes in the pathogenesis of cerebral malaria. Trends Immunol. 2003, 24 (9): 491-499. 10.1016/S1471-4906(03)00229-1.
Rae C, McQuillan JA, Parekh SB, Bubb WA, Weiser S, Balcar VJ, Hansen AM, Ball HJ, Hunt NH: Brain gene expression, metabolism, and bioenergetics: interrelationships in murine models of cerebral and noncerebral malaria. Faseb J. 2004, 18 (3): 499-510. 10.1096/fj.03-0543com.
Sanni LA, Rae C, Maitland A, Stocker R, Hunt NH: Is ischemia involved in the pathogenesis of murine cerebral malaria?. Am J Pathol. 2001, 159 (3): 1105-1112.
Fink MP: Bench-to-bedside review: Cytopathic hypoxia. Crit Care. 2002, 6 (6): 491-499. 10.1186/cc1824.
Kunapuli SP, Dorsam RT, Kim S, Quinton TM: Platelet purinergic receptors. Curr Opin Pharmacol. 2003, 3 (2): 175-180. 10.1016/S1471-4892(03)00007-9.
Hattori M, Aoki J, Arai H, Inoue K: PAF and PAF acetylhydrolase in the nervous system. J Lipid Mediat Cell Signal. 1996, 14 (1-3): 99-102. 10.1016/0929-7855(96)00514-7.
Ariga M, Neitzert B, Nakae S, Mottin G, Bertrand C, Pruniaux MP, Jin SL, Conti M: Nonredundant function of phosphodiesterases 4D and 4B in neutrophil recruitment to the site of inflammation. J Immunol. 2004, 173 (12): 7531-7538.
Grau GE, Heremans H, Piguet PF, Pointaire P, Lambert PH, Billiau A, Vassalli P: Monoclonal antibody against interferon gamma can prevent experimental cerebral malaria and its associated overproduction of tumor necrosis factor. Proc Natl Acad Sci U S A. 1989, 86 (14): 5572-5574. 10.1073/pnas.86.14.5572.
Mitchell AJ, Hansen AM, Hee L, Ball HJ, Potter SM, Walker JC, Hunt NH: Early cytokine production is associated with protection from murine cerebral malaria. Infect Immun. 2005, 73 (9): 5645-5653. 10.1128/IAI.73.9.5645-5653.2005.
de Kossodo S, Grau GE: Profiles of cytokine production in relation with susceptibility to cerebral malaria. J Immunol. 1993, 151 (9): 4811-4820.
Cheng H, Cenciarelli C, Tao M, Parks WP, Cheng-Mayer C: HTLV-1 Tax-associated hTid-1, a human DnaJ protein, is a repressor of Ikappa B kinase beta subunit. J Biol Chem. 2002, 277 (23): 20605-20610. 10.1074/jbc.M201204200.
Lin L, Hron JD, Peng SL: Regulation of NF-kappaB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity. 2004, 21 (2): 203-213. 10.1016/j.immuni.2004.06.016.
Neznanov N, Neznanova L, Kondratov RV, Burdelya L, Kandel ES, O'Rourke DM, Ullrich A, Gudkov AV: Dominant negative form of signal-regulatory protein-alpha (SIRPalpha /SHPS-1) inhibits tumor necrosis factor-mediated apoptosis by activation of NF-kappa B. J Biol Chem. 2003, 278 (6): 3809-3815. 10.1074/jbc.M210698200.
Blais V, Rivest S: Inhibitory action of nitric oxide on circulating tumor necrosis factor-induced NF-kappaB activity and COX-2 transcription in the endothelium of the brain capillaries. J Neuropathol Exp Neurol. 2001, 60 (9): 893-905.
Sebastiani G, Morissette C, Lagace C, Boule M, Ouellette MJ, McLaughlin RW, Lacombe D, Gervais F, Tremblay P: The cAMP-specific phosphodiesterase 4B mediates Abeta-induced microglial activation. Neurobiol Aging. 2006, 27 (5): 691-701. 10.1016/j.neurobiolaging.2005.03.024.
Donato R: Functional roles of S100 proteins, calcium-binding proteins of the EF-hand type. Biochim Biophys Acta. 1999, 1450 (3): 191-231. 10.1016/S0167-4889(99)00058-0.
Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM: BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000, 13 (2): 199-212. 10.1016/S1074-7613(00)00020-0.
Hermsen C, van de Wiel T, Mommers E, Sauerwein R, Eling W: Depletion of CD4+ or CD8+ T-cells prevents Plasmodium berghei induced cerebral malaria in end-stage disease. Parasitology. 1997, 114 ( Pt 1): 7-12. 10.1017/S0031182096008293.
Yanez DM, Manning DD, Cooley AJ, Weidanz WP, van der Heyde HC: Participation of lymphocyte subpopulations in the pathogenesis of experimental murine cerebral malaria. J Immunol. 1996, 157 (4): 1620-1624.
Grau GE, Piguet PF, Engers HD, Louis JA, Vassalli P, Lambert PH: L3T4+ T lymphocytes play a major role in the pathogenesis of murine cerebral malaria. J Immunol. 1986, 137 (7): 2348-2354.
Potter S, Chan-Ling T, Ball HJ, Mansour H, Mitchell A, Maluish L, Hunt NH: Perforin mediated apoptosis of cerebral microvascular endothelial cells during experimental cerebral malaria. Int J Parasitol. 2006, 36 (4): 485-496. 10.1016/j.ijpara.2005.12.005.
Medana IM, Chaudhri G, Chan-Ling T, Hunt NH: Central nervous system in cerebral malaria: 'Innocent bystander' or active participant in the induction of immunopathology?. Immunol Cell Biol. 2001, 79 (2): 101-120. 10.1046/j.1440-1711.2001.00995.x.
Lynch NJ, Willis CL, Nolan CC, Roscher S, Fowler MJ, Weihe E, Ray DE, Schwaeble WJ: Microglial activation and increased synthesis of complement component C1q precedes blood-brain barrier dysfunction in rats. Mol Immunol. 2004, 40 (10): 709-716. 10.1016/j.molimm.2003.08.009.
Patrizio M: Tumor necrosis factor reduces cAMP production in rat microglia. Glia. 2004, 48 (3): 241-249. 10.1002/glia.20074.
Neumann H: Control of glial immune function by neurons. Glia. 2001, 36 (2): 191-199. 10.1002/glia.1108.
Park KW, Lee DY, Joe EH, Kim SU, Jin BK: Neuroprotective role of microglia expressing interleukin-4. J Neurosci Res. 2005, 81 (3): 397-402. 10.1002/jnr.20483.
Sanni LA, Thomas SR, Tattam BN, Moore DE, Chaudhri G, Stocker R, Hunt NH: Dramatic changes in oxidative tryptophan metabolism along the kynurenine pathway in experimental cerebral and noncerebral malaria. Am J Pathol. 1998, 152 (2): 611-619.
Dobbie M, Crawley J, Waruiru C, Marsh K, Surtees R: Cerebrospinal fluid studies in children with cerebral malaria: an excitotoxic mechanism?. Am J Trop Med Hyg. 2000, 62 (2): 284-290.
Li J, Culman J, Hortnagl H, Zhao Y, Gerova N, Timm M, Blume A, Zimmermann M, Seidel K, Dirnagl U, Unger T: Angiotensin AT2 receptor protects against cerebral ischemia-induced neuronal injury. Faseb J. 2005, 19 (6): 617-619. 10.1096/fj.04-2555com.
Liman J, Ganesan S, Dohm CP, Krajewski S, Reed JC, Bahr M, Wouters FS, Kermer P: Interaction of BAG1 and Hsp70 mediates neuroprotectivity and increases chaperone activity. Mol Cell Biol. 2005, 25 (9): 3715-3725. 10.1128/MCB.25.9.3715-3725.2005.
Chergui K, Svenningsson P, Greengard P: Physiological role for casein kinase 1 in glutamatergic synaptic transmission. J Neurosci. 2005, 25 (28): 6601-6609. 10.1523/JNEUROSCI.1082-05.2005.
Herz J, Chen Y: Reelin, lipoprotein receptors and synaptic plasticity. Nat Rev Neurosci. 2006, 7 (11): 850-859. 10.1038/nrn2009.
Di Scala F, Dupuis L, Gaiddon C, De Tapia M, Jokic N, Gonzalez de Aguilar JL, Raul JS, Ludes B, Loeffler JP: Tissue specificity and regulation of the N-terminal diversity of reticulon 3. Biochem J. 2005, 385 (Pt 1): 125-134.
Mantamadiotis T, Lemberger T, Bleckmann SC, Kern H, Kretz O, Martin Villalba A, Tronche F, Kellendonk C, Gau D, Kapfhammer J, Otto C, Schmid W, Schutz G: Disruption of CREB function in brain leads to neurodegeneration. Nat Genet. 2002, 31 (1): 47-54. 10.1038/ng882.
Won SJ, Kim SH, Xie L, Wang Y, Mao XO, Jin K, Greenberg DA: Reelin-deficient mice show impaired neurogenesis and increased stroke size. Exp Neurol. 2006, 198 (1): 250-259. 10.1016/j.expneurol.2005.12.008.
Assadi AH, Zhang G, Beffert U, McNeil RS, Renfro AL, Niu S, Quattrocchi CC, Antalffy BA, Sheldon M, Armstrong DD, Wynshaw-Boris A, Herz J, D'Arcangelo G, Clark GD: Interaction of reelin signaling and Lis1 in brain development. Nat Genet. 2003, 35 (3): 270-276. 10.1038/ng1257.
Deutsch SI, Rosse RB, Deutsch LH: Faulty regulation of tau phosphorylation by the reelin signal transduction pathway is a potential mechanism of pathogenesis and therapeutic target in Alzheimer's disease. Eur Neuropsychopharmacol. 2006, 16 (8): 547-551. 10.1016/j.euroneuro.2006.01.006.
Ding Q, Markesbery WR, Chen Q, Li F, Keller JN: Ribosome dysfunction is an early event in Alzheimer's disease. J Neurosci. 2005, 25 (40): 9171-9175. 10.1523/JNEUROSCI.3040-05.2005.
Medana IM, Day NP, Hien TT, Mai NT, Bethell D, Phu NH, Farrar J, Esiri MM, White NJ, Turner GD: Axonal injury in cerebral malaria. Am J Pathol. 2002, 160 (2): 655-666.
Matsuda S, Giliberto L, Matsuda Y, Davies P, McGowan E, Pickford F, Ghiso J, Frangione B, D'Adamio L: The familial dementia BRI2 gene binds the Alzheimer gene amyloid-beta precursor protein and inhibits amyloid-beta production. J Biol Chem. 2005, 280 (32): 28912-28916. 10.1074/jbc.C500217200.
Zhou S, Zhou H, Walian PJ, Jap BK: CD147 is a regulatory subunit of the gamma-secretase complex in Alzheimer's disease amyloid beta-peptide production. Proc Natl Acad Sci U S A. 2005, 102 (21): 7499-7504. 10.1073/pnas.0502768102.
He W, Lu Y, Qahwash I, Hu XY, Chang A, Yan R: Reticulon family members modulate BACE1 activity and amyloid-beta peptide generation. Nat Med. 2004, 10 (9): 959-965. 10.1038/nm1088.
Iribarren P, Chen K, Hu J, Zhang X, Gong W, Wang JM: IL-4 inhibits the expression of mouse formyl peptide receptor 2, a receptor for amyloid beta1-42, in TNF-alpha-activated microglia. J Immunol. 2005, 175 (9): 6100-6106.
Dickey CA, Gordon MN, Mason JE, Wilson NJ, Diamond DM, Guzowski JF, Morgan D: Amyloid suppresses induction of genes critical for memory consolidation in APP + PS1 transgenic mice. J Neurochem. 2004, 88 (2): 434-442.
TAGC website. [http://tagc.univ-mrs.fr/]
Puthier D, Joly F, Irla M, Saade M, Victorero G, Loriod B, Nguyen C: A general survey of thymocyte differentiation by transcriptional analysis of knockout mouse models. J Immunol. 2004, 173 (10): 6109-6118.
UniGene database website - Library browser. [http://www.ncbi.nlm.nih.gov/UniGene/lbrowse2.cgi?TAXID_10090]
Bertucci F, Salas S, Eysteries S, Nasser V, Finetti P, Ginestier C, Charafe-Jauffret E, Loriod B, Bachelart L, Montfort J, Victorero G, Viret F, Ollendorff V, Fert V, Giovaninni M, Delpero JR, Nguyen C, Viens P, Monges G, Birnbaum D, Houlgatte R: Gene expression profiling of colon cancer by DNA microarrays and correlation with histoclinical parameters. Oncogene. 2004, 23 (7): 1377-1391. 10.1038/sj.onc.1207262.
Eisen MB, Spellman PT, Brown PO, Botstein D: Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998, 95 (25): 14863-14868. 10.1073/pnas.95.25.14863.
TIGR MeV (MultiExperiment Viewer) v3.1 software. [http://www.tm4.org/mev.html]
Benjamini Y, Hochberg Y: Controlling the False Discovery Rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B. 1995, 57 (1): 289-300.
Expression Analysis Systematic Explorer (EASE). [http://david.abcc.ncifcrf.gov]
Kyoto Encyclopedia of Genes and Genomes (KEGG). [http://www.genome.jp/kegg]
ArrayExpress database. [http://www.ebi.ac.uk/arrayexpress]
Acknowledgements
We thank Béatrice Loriod and Geneviève Victorero for technical support and advice. We also thank Frédéric Foucault for helpful discussions, and Andrew Mitchell for critical reading of the manuscript. We acknowledge the technical support of Marseille-Nice genopole.
Financial support: French Ministry of Research and Technology (PAL+ Program); Fondation pour la Recherche Médicale; PACA Conseil Régional; Conseil Général des Bouches du Rhône. NFD was supported by a studentship from the Fondation pour la Recherche Médicale. MB and NC are supported by a studentship from the French Ministry of Research and Technology.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
NFD participated in the design of the study, the injection of parasitized erythrocytes, the sample preparation, and the analysis of the data, carried out microarray experiments, and prepared the figures. NC participated in sample preparation and carried out imunochemistry experiments. DP, FJ, and CN designed and produced the microarrays. DP participated in the statistical analysis, and CN contributed to study design. MB participated in microarray hybridizations. PB and GG participated in the interpretation of data that concerned genes involved in the inflammatory response. NFD and PR carried out a systematic analysis of the functional annotation of genes identified. FAI and GG participated in the design of animal studies. PR conceived and coordinated the study, and wrote the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12864_2007_1165_MOESM1_ESM.xls
Additional File 1: The full list of the 327 genes that discriminated between early and late infection stages, between mouse strains, and between CM-R and CM-S mice. (XLS 221 KB)
12864_2007_1165_MOESM2_ESM.pdf
Additional File 2: The graphical representation of expression profiles of genes involved in oxidative phosphorylation in CM-S and CM-R mice. (PDF 11 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Delahaye, N.F., Coltel, N., Puthier, D. et al. Gene expression analysis reveals early changes in several molecular pathways in cerebral malaria-susceptible mice versus cerebral malaria-resistant mice. BMC Genomics 8, 452 (2007). https://doi.org/10.1186/1471-2164-8-452
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-8-452