Abstract—
Neurexins are a family of synaptic adhesion proteins that play a key role in synapse formation and maintenance. Neurexins undergo extensive alternative splicing at six sites (SS1–SS6) resulting in expression of multiplicity of different isoforms. Alternative splicing regulates the functional activity of neurexins in different types of tissues and cells and presumably plays a key role in determining the specificity of the interaction of various neurons. In this study, we have investigated the pattern of tissue expression of neurexin-1α mRNA isoforms including an insert in the recently discovered splice site SS6 using TaqMan Real-Time PCR in different organs of Wistar rats. The isoform containing the insert in the SS6 site was found only in neural tissues suggesting its potential functional importance. Position of the SS6 insert in the hinge region between the LNS5 and LNS6 domains increases variability of possible conformations of the molecule which may represent an additional mechanism for regulating functional activity of the neurexin-1α in the brain.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Neurexins (Nrxn1–3) are a family of synaptic adhesion proteins that play a key role in the formation and stabilization of synapses [1]. Neurexins and their ligands form complex interacting networks, mediating many regulatory functions. Disfunction of neurexins and proteins interacting with them causes autism spectrum disorder (ASD), schizophrenia and mental retardation [2]. Nrxns are type 1-membrane proteins and were originally discovered as receptors for α-latrotoxin [3]. Neurexins are encoded by three homologous genes Nrxn1–3. Each gene, in turn, is transcribed from two independent promoters (α and β), leading to the formation of long (α) and short (β) forms of neurexins [4, 5]. The extracellular region of α-neurexins consists of three modules, containing two LNS domains (laminin, neurexin, sex hormone binding protein domain, also called as laminin G domain), between which there is an EGF-like domain. The extracellular moiety of β-neurexins contains only one LNS domain [4].
Neurexins undergo extensive alternative splicing, resulting in the expression of multiplicity of different isoforms. Alternative splicing of neurexins is regulated differently in various brain regions and depends on the developmental stage and synaptic activity [6, 7]. It is believed that alternative splicing regulates the functional activity of neurexins and, presumably, plays a key role in determining the specificity of the interaction of various neurons.
Initially, five splicing sites (SS1–SS5) in α-neurexins were known, two of which (SS4 and SS5) are also found in β-neurexins. Later, a sixth splicing site (SS6) of neurexin-1α (Nrxn1α) was identified, which is located between the fifth LNS and the third EGF-like domains. The insert in this region corresponds to the 9-amino acid peptide VALMKADLQ, which is conserved in animals [8, 9]. Nrxn genes were initially found to be expressed in the brain, but later studies have shown Nrxn expression not only in neural tissues.In smaller amounts Nrxn1 mRNA expression was detected in the kidneys, liver, heart, stomach, and lungs [10].
Neurexins were also found in the vascular system [11] and in the β-cells of the pancreas [12], where they play a functional role. Nrxn1α is a component of the mechanism of regulation of exocytosis; and is essential for the docking of insulin granules with the membrane in pancreatic β-cells [13]. Nrxn3 mRNA was found in the lungs, pancreas, heart, placenta, liver, and kidneys. Moreover, specific splicing variants of Nrxn3 have been identified in the heart [14]. It is not clear what exactly triggers the expression of neurexin isoforms in different tissues.
The aim of this work is to analyze the expression of neurexin-1α with alternative splicing at the SS6 site using TaqMan real-time PCR in various rat organs to reveal the potential role of alternative splicing in the regulation of the functional activity of neurexins in different tissues.
RESULTS AND DISCUSSION
Since neurexins are found not only in neural tissues we investigated the expression of Nrxn1α with an insert in the recently discovered SS6 splicing site in various organs of Wistar rats using TaqMan real-time PCR assay. Organs were isolated from four female rats: the liver, kidneys, heart, pancreas, and brain. Total RNA was isolated from each extracted rat organ. The brain was divided into three parts: the cerebral cortex, cerebellum, and the remaining part. Total RNA was isolated from each part of the brain separately. Using the reverse transcription reaction, cDNA samples were obtained from various rat organs and were further analyzed by TaqMan Real-Time PCR.
To detect the isoforms of Nrxn1α (SS6+) and (SS6–), we used the same forward primer and probe, but different reverse primers (Table 1). One of them hybridized at the boundary of exons 17 and 18, which allowed us to amplify only a cDNA fragment containing the insert at the SS6 site. Another primer was annealed at the boundary of exons 16 and 18, which allowed us to obtain a cDNA fragment lacking the SS6 insert. Household gene Gapdh was used as a reference gene.
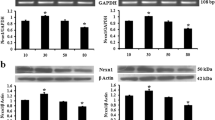
The receptor isoform containing the SS6 insert was found only in the brain, while the splicing variant lacking the insert was found in all five analyzed organs (Fig. 1). In the brain, the expression level of the neurexin isoform without the SS6 insert significantly exceeds the expression level of the isoform containing the insert (Fig. 1). The ratio of the expression of these two isoforms in various parts of the brain was also different. The largest amount of Nrxn1α (SS6–) was found in the cerebellum, while the maximum expression of Nrxn1α (SS6+) was observed in the cerebral cortex. If the total amount of Nrxn1α found in each part of the brain is represented as 100%, then the values of expression of isoforms (SS6–) and (SS6+) would be as follows: 87 and 13% in the cerebral cortex, 96 and 4% in the cerebellum, 94 and 6% in the remaining part of the brain.
Analysis of the expression of Nrxn1α isoforms (SS6+) and (SS6–) in organs of Wistar rats (n = 4) using TaqMan Real-Time PCR. Bars denote the standard error of the mean (SEM). PCR was performed in triplicate with each cDNA sample. * p < 0.001. Numbers indicate organs: (1) cerebral cortex, (2) cerebellum, (3) the remaining part of the brain, (4) heart, (5) kidney, (6) liver, (7) pancreas.
Unlike the Nrxn1α isoform (SS6+), which was expressed only in the brain, the (SS6–) isoform was found in other tissues, such as the heart, kidneys, liver, and pancreas, but in much smaller quantities. Among the latter, the greatest amount of the isoform (SS6–) was observed in the pancreas.
The expression of the Nrxn1 gene that we found in non-neuronal tissues is confirmed by modern literature data on RNA sequencing from various organs. According to the data of single-cell RNA sequencing, the Nrxn1 gene is expressed in the islets of the human pancreas [15], the expression of Nrxn1 was detected in alpha, beta, delta, acinar cells, and the pancreatic duct cells (Table 2). There is also evidence of Nrxn1 expression in rare pancreatic gamma cells [16]. Mouse kidney RNA sequencing revealed Nrxn1 expression in cells of cortical collecting duct and thick ascending limbs of the loop of Henle. The mean expression of the Nrxn1 gene in the collecting duct is 1.8 transcripts per million (TPM) [17].
Alternative splicing is differentially regulated among neurexin genes despite their homology. Each Nrxn isoform displays a unique expression profile in a region-, cell type- and sensory system-specific manner [18]. The mechanism of regulation of the neurexin isoform expression is poorly understood. Recent studies indicate that alternative splicing at the SS3 and SS4 sites is regulated by neuronal activity [1]. We found an Nrxn1α isoform with an inclusionin the SS6 site only in the brain, while a splicing variant lacking an insert in the SS6 was detected in all organs tested. The largest amount of Nrxn1α (SS6–) was found in the cerebellum, while the maximum expression of Nrxn1α (SS6 +) was observed in the cerebral cortex. It can be assumed that alternative splicing at the SS6 splice site represents a mechanism that controls the functional activity of Nrxn1α in various tissues.
Recent studies utilizing a combination of individual particle electron tomography (IPET), X-ray crystallography, and small-angle X-ray scattering (SAXS) have shown that the Nrxn1α ectodomain adopts several discrete conformations [19]. Diversity of observed conformations is provided by two main hinges within Nrxn1α. One of the hinges is located between the LNS1 and LNS2 domains, and the other, between the LNS5 and LNS6 domains [19]. It has been shown that an insertion in the SS6 site, located directly in the molecular hinge between the LNS5 and LNS6 domains, increases the variability of the molecular conformations [19].
It can be assumed that the inclusion of the SS6 insert can affect the binding of protein partners in the synaptic cleft, changing the actual binding sites or the accessibility to these binding sites. The SS6 insert is reported to be sensitive to proteolysis. Thus, the probable function of the SS6 insertion may be to make the neurexin-1α molecule susceptible to proteolysis, allowing the L1–L5 region to be cleaved [19] with the formation of a secreted form of the receptor. A similar soluble secreted form was found for the Nrxn3 receptor [20, 21]. The functional significance of the soluble form of Nrxn3 is evidenced by the reduced expression and ratio of transmembrane and soluble isoforms of Nrxn3 in Alzheimer’s disease post mortem brains [22].
Tissue-specific expression of various Nrxn1α isoforms indicates a potential role of SS6 alternative splicing in regulating the functional activity of Nrxn1α.
EXPERIMENTAL
Experiments with animals. Female Wistar rats (n = 4) aged 3–4 months (“Stolbovaya” Nursery of Laboratory Animals, Russia) were used in the experiments. The animals were healthy and kept under standard conditions. Before surgical removal of organs, the rats were anesthetized with zoletil (20 mg/kg body weight) and xylazine (5 mg/kg body weight) in 0.9% NaCl solution. The liver, kidneys, pancreas, heart, and brain were excised from rats. Whole organs were placed in separate IKA DT-20-M tubes (IKA, Germany) with TRIzol Reagent (Invitrogen, USA) and immediately afterwards homogenized using a homogenizer IKA Ultra-Turrax Tube Drive. The brain was divided into three parts: the cerebral cortex, cerebellum, and the remaining part of the brain. Each part of the brain was homogenized separately.
Preparation of cDNA. Total RNA was isolated from rat organs using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s protocol. To remove contaminants of genomic DNA total RNA samples were treated with DNase I (Thermo Scientific, USA) according to the manufacturer’s protocol. cDNA was obtained by a reverse transcription reaction using a RevertAid RT Reverse Transcription Kit (Thermo Scientific, USA) and random hexamer primers included in the kit according to the manufacturer’s protocol.
TaqMan real-time PCR assay. Primers and a specifically annealing probe for the each gene were selected using the Gene Runner (http://www.generunner.net/), Oligo Explorer (https://www.genelink.com/tools/gl-oe.asp) and Oligo Analyzer (https://eu.idtdna.com/pages/ tools/oligoanalyzer) programs. The specificity of primers and probes was confirmed using BLAST (https://blast.ncbi.nlm.nih.gov/). The Gapdh gene was used as a reference gene. Primers were synthesized by Evrogen (Russia), probes by Lumiprob RUS (Russia). To detect the isoforms Nrxn1α (SS6+) and (SS6–), we used the same forward primer and probe, but different reverse primers (Table 1). The probe contained a FAM fluorescence label (Fluorescein amidites) at the 5'-end, and a BHQ1 quencher (Black Hole Quencher 1) at the 3'-end.
For each pair of primers, the reaction efficiency (E) was calculated using the formula: E = 10–1/k, where k is taken from the straight-line equation: CT = klogP0 + b, in which P0 is the concentration of cDNA, CT is the number of cycles in which the fluorescence reaches the threshold level, and the values of k and b are obtained from a linear approximation of the experimental data. The efficiency of the reaction for the target and reference genes was 2, which corresponds to the maximum efficiency of the reaction.
TaqMan real-time PCR with each cDNA sample was performed in three replicas using a DTprime instrument (DNA-Technology, Russia) in 20 μL of the reaction mixture containing 1 unit of Hot Start Taq DNA polymerase (Evrogen, Russia). Primer concentration was 0.4 µM, and the probe concentration was 0.2 µM. The reaction parameters were as follows: preliminary denaturation at 95°С for 5 min; 45 amplification cycles: 95°C, 15 s; 60°C, 10 s; 72°C, 10 s.
A mean of normalized expression (NE) was calculated using the formula [23]:
where ER is the reaction efficiency for the reference gene, ET is the reaction efficiency for the target gene, CTR is the number of cycles in which fluorescence reaches the threshold level for the reference gene, CTT is the number of cycles in which the fluorescence reaches the threshold level for the target gene.
Real-time PCR data were statistically processed applying Prism 6 software (GraphPad Software, USA) using Student’s t-test. Differences were considered statistically significant at p <0.05.
CONCLUSIONS
We performed a detailed analysis of the expression of neurexin-1α isoforms (Nrxn1α) with alternative splicing in the SS6 site using TaqMan real-time PCR in various organs of Wistar rats. Tissue-specific expression of Nrxn1α isoforms has been shown. An isoform containing an insert at the SS6 site was found only in neuronal tissues, indicating its potential functional importance. The position of the SS6 insert in the hinge region between the LNS5 and LNS6 domains increases the variability of the possible conformations of the molecule, which may represent an additional mechanism for regulating the functional activity of Nrxn1α in the brain.
REFERENCES
Sudhof, T.C., Cell, 2017, vol. 171, pp. 745–769. https://doi.org/10.1016/j.cell.2017.10.024
Rudenko, G., Curr. Opin. Struct. Biol., 2019, vol. 54, pp. 112–121. https://doi.org/10.1016/j.sbi.2019.01.009
Petrenko, A.G., Kovalenko, V.A., Shamotienko, O.G., Surkova, I.N., Tarasyuk, T.A., Ushkaryov, Yu.A., and Grishin, E.V., EMBO J., 1990, vol. 9, pp. 2023–2027.
Ushkaryov, Y.A., Petrenko, A.G., Geppert, M., and Sudhof, T.C., Science, 1992, vol. 257, pp. 50–56. https://doi.org/10.1126/science.1621094
Geppert, M., Ushkaryov, Y.A., Hata, Y., Davletov, B., Petrenko, A.G., and Sudhof, T.C., Cold Spring Harb. Symp. Quant. Biol., 1992, vol. 57, pp. 483–490. https://doi.org/10.1101/sqb.1992.057.01.053
Ullrich, B., Ushkaryov, Y.A., and Sudhof, T.C., Neuron, 1995, vol. 14, pp. 497–507. https://doi.org/10.1016/0896-6273(95)90306-2
Schreiner, D., Nguyen, T.M., Russo, G., Heber, S., Patrignani, A., Ahrne, E., and Scheiffele, P., Neuron, 2014, vol. 84, pp. 386–398. https://doi.org/10.1016/j.neuron.2014.09.011
Serova, O.V., Radionov, N.V., Shayahmetova, D.M., Deyev, I.E., and Petrenko, A.G., Dokl. Biochem. Biophys., 2015, vol. 463, pp. 239–242. https://doi.org/10.1134/S1607672915040110
Treutlein, B., Gokce, O., Quake, S.R., and Sudhof, T.C., Proc. Natl. Acad. Sci. U. S. A., 2014, vol. 111, pp. E1291–E1299. https://doi.org/10.1073/pnas.1403244111
Saito, A., Miyauchi, N., Hashimoto, T., Karasawa, T., Han, G.D., Kayaba, M., Sumi, T., Tomita, M., Ikezumi, Y., Suzuki, K., Koitabashi, Y., Shimizu, F., and Kawachi, H., Am. J. Physiol. Regul. Integr. Comp. Physiol., 2011, vol. 300, pp. R340–R348. https://doi.org/10.1152/ajpregu.00640.2009
Bottos, A., Destro, E., Rissone, A., Graziano, S., Cordara, G., Assenzio, B., Cera, M.R., Mascia, L., Bussolino, F., and Arese, M., Proc. Natl. Acad. Sci. U. S. A., 2009, vol. 106, pp. 20782–20787. https://doi.org/10.1073/pnas.0809510106
Suckow, A.T., Comoletti, D., Waldrop, M.A., Mosedale, M., Egodage, S., Taylor, P., and Chessler, S.D., Endocrinology, 2008, vol. 149, pp. 6006–6017.
Mosedale, M., Egodage, S., Calma, R.C., Chi, N.W., and Chessler, S.D., J. Biol. Chem., 2012, vol. 287, pp. 6350–6361.
Occhi, G., Rampazzo, A., Beffagna, G., and Antonio Danieli, G., Biochem. Biophys. Res. Commun., 2002, vol. 298, pp. 151–155.
Li, J., Klughammer, J., Farlik, M., Penz, T., Spittler, A., Barbieux, C., Berishvili, E., Bock, C., and Kubicek, S., EMBO Rep., 2016, vol. 17, pp. 178–187. https://doi.org/10.15252/embr.201540946
Segerstolpe, A., Palasantza, A., Eliasson, P., Andersson, E.M., Andreasson, A.C., Sun, X., Picelli, S., Sabirsh, A., Clausen, M., Bjursell, M.K., Smith, D.M., Kasper, M., Ämmälä, C., and Sandberg, R., Cell Metab., 2016, vol. 24, pp. 593–607. https://doi.org/10.1016/j.cmet.2016.08.020
Chen, L., Lee, J.W., Chou, C.L., Nair, A.V., Battistone, M.A., Paunescu, T.G., Merkulova, M., Breton, S., Verlander, J.W., Wall, S.M., Brown, D., Burg, M.B., and Knepper, M.A., Proc. Natl. Acad. Sci. U. S. A., 2017, vol. 114, pp. E9989–E9998. https://doi.org/10.1073/pnas.1710964114
Uchigashima, M., Cheung, A., Suh, J., Watanabe, M., and Futai, K., J. Comp. Neurol., 2019, vol. 527, pp. 1940–1965. https://doi.org/10.1002/cne.24664
Liu, J., Misra, A., Reddy, M., White, M.A., Ren, G., and Rudenko, G., J. Mol. Biol., 2018, vol. 430, pp. 4325–4343. https://doi.org/10.1016/j.jmb.2018.08.026
Ushkaryov, Y.A. and Sudhof, T.C., Proc. Natl. Acad. Sci. U. S. A., 1993, vol. 90, pp. 6410–6414. https://doi.org/10.1073/pnas.90.14.6410
Hishimoto, A., Liu, Q.R., Drgon, T., Pletnikova, O., Walther, D., Zhu, X.G., Troncoso, J.C., and Uhl, G.R., Hum. Mol. Genet., 2007, vol. 16, pp. 2880–2891. https://doi.org/10.1093/hmg/ddm247
Hishimoto, A., Pletnikova, O., Lang, D.L., Troncoso, J.C., Egan, J.M., and Liu, Q.R., Alzheimer’s Res. Ther., 2019, vol. 11, p. 28. https://doi.org/10.1186/s13195-019-0475-2
Simon, P., Bioinformatics, 2003, vol. 19, pp. 1439–1440. https://doi.org/10.1093/bioinformatics/btg157
Funding
The study was financially supported by the Russian Foundation for Basic Research (project nos. 19-04-01042, 19-34-90177 and 19-34-51034).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
COMPLIANCE WITH ETHICAL STANDARDS
All experiments with animals were performed according to the protocol of the Institutional Animal Care and Use Committee (IACUC) certified by the Bioethics Commission of the Shemyakin–Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences (IBCh RAS).
Conflict of Interests
The authors declare they have no conflict of interests.
Rights and permissions
Open Access. This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Serova, O.V., Gantsova, E.A., Deyev, I.E. et al. Tissue-Specific Expression of Neurexin-1α Isoforms in Rat Organs. Russ J Bioorg Chem 48, 321–325 (2022). https://doi.org/10.1134/S1068162022020194
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1068162022020194