Abstract
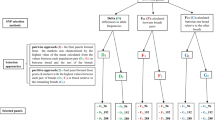
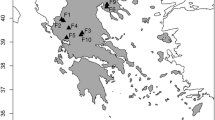
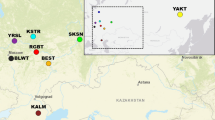
A comparative study of the informativeness of SNP and STR markers for interspecific and intraspecific differentiation of the two species of the genus Ovis, snow sheep (O. nivicola) and domestic sheep (O. aries), was conducted. Eleven STR loci combined into two multiplex panels were examined. SNP analysis was performed with the DNA microarray OvineSNP50K BeadChip featuring 54241 SNPs. The possibility of clear differentiation of the studied Ovis species with both types of genetic markers was demonstrated. The advantages of SNP markers for intraspecific differentiation of the O. aries breeds and O. nivicola geographical groups were revealed. The areas of application of the studied types of DNA markers are discussed.
Similar content being viewed by others
References
Khlestkina, E.K., Molecular markers in genetic studies and breeding, Russ. J. Genet.: Appl. Res., 2014, vol. 4 no. 2, pp. 236–244.
Putman, A.I. and Carbone, I., Challenges in analysis and interpretation of microsatellite data for population genetic studies, Ecol. Evol., 2014, vol. 4, no. 22, pp. 4399–4428.
Haasl, R.J. and Payseur, B.A., Multi-locus inference of population structure: a comparison between single nucleotide polymorphisms and microsatellites, J. Hered., 2011, vol. 106, pp. 158–171. doi 10.1038/hdy.2010.21
Smaragdov, M.G., Genomic selection as a possible accelerator of traditional selection, Russ. J. Genet., 2009, vol. 45, no. 6, pp. 633–636.
Kijas, J.W., Townley, D., Dalrymple, B.P., et al., A genome wide survey of SNP variation reveals the genetic structure of sheep breeds, PLoS One, 2009, vol. 4, no. 3. e4668. doi l0.1371/journal.pone.0004668
Morin, P.A., Luikart, G., and Wayne, R.K., The SNP workshop group: SNPs in ecology, evolution and conservation, Trends Ecol. Evol., 2004, vol. 19, pp. 208–216.
Vignal, A., Milan, D., Sancristobal, M., and Eggen, A., A review on SNP and other types of molecular markers and their use in animal genetics, Genet. Sel. Evol., 2002, vol. 34, pp. 275–305.
Schopen, G.C.B., Bovenhuis, H., Visker, M.H.P.W., and van Arendonk, J.A.M., Comparison of information content for microsatellites and SNPs in poultry and cattle, Anim. Gen., 2008, vol. 39, pp. 451–453. doi 10.1111/j.1365-2052.2008.01736.x
Gärke C., Ytournel, F. Bed’hom, B., et al., Comparison of SNPs and microsatellites for assessing the genetic structure of chicken populations, Anim. Gen., 2011, vol. 43, pp. 419–428. doi 10.1111/j.1365-2052.2011. 02284.x
Herraeza, D.L., Schafer, H., Mosner, J., Fries, H. R., and Wink, M., Comparison of microsatellite and single nucleotide polymorphism markers for the genetic analysis of a Galloway cattle population, J. Biosci., 2005, vol. 60, pp. 637–643.
Fernández, M.E., Goszczynski, D.E., Lirón, J.P., et al., Comparison of the effectiveness of microsatellites and SNP panels for genetic identification, traceability and assessment of parentage in an inbred Angus herd, Genet. Mol. Biol., 2013, vol. 36, no. 2, pp. 185–191.
Coates, B.S., Sumerford, D.V., Miller, N.J., et al., Comparative performance of single nucleotide polymorphism and microsatellite markers for population genetic analysis, J. Hered., 2009, vol. 100, no. 5, pp. 556–564. doi 10.1093/jhered/esp028
Seeb, J.E., Carvalho, G., Hauser, L., et al., Singlenucleotide polymorphism (SNP) discovery and applications of SNP genotyping in nonmodel organisms, Mol. Ecol. Res., 2011, vol. 11, no. 1, pp. 1–8.
Tokarska, M., Marshall, T., Kowalczyk, R., et al., Effectiveness of microsatellite and SNP markers for parentage and identity analysis in species with low genetic diversity: the case of European bison, J. Hered., 2009, vol. 103, pp. 326–332. doi 10.1038/hdy.2009.73
Haynes, G.D. and Latch, E.K., Identification of novel single nucleotide polymorphisms (SNPs) in deer (Odocoileus spp.) using the BovineSNP50 BeadChip, PLoS One, 2012, vol. 7, no. 5. e36536. doi 10.1371/journal.pone.0036536
Miller, J.M., Poissant, J., Kijas, J.W., et al., A genomewide set of SNPs detects population substructure and long-range linkage disequilibrium in wild sheep, Mol. Ecol. Res., 2011, vol. 11, no. 2, pp. 314–322. doi 10.1111/j.1755-0998.2010.02918.x
Miller, J.M., Kijas, J.W., Heaton, M.P., et al., Consistent divergence times and allele sharing measured from cross-species application of SNP chips developed for three domestic species, Mol. Ecol. Res., 2012, no. 12, pp. 1145–1150.
Peakall, R., GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research, Mol. Ecol. Notes, 2006, no. 6, pp. 288–295.
Purcell, S., Neale, B., Todd-Brown, K., et al., PLINK: a tool set for whole-genome association and population-based linkage analyses, Am. J. Hum. Genet., 2007, vol. 81, pp. 559–575.
R Development Core Team, R: A language and environment for statistical computing, Vienna: R Foundation for Statistical Computing, 2009. http://www.Rproject.org
Pritchard, J.K., Stephens, M., and Donnelly, P., Inference of population structure using multilocus genotype data, Genetics, 2000, vol. 155, pp. 945–959.
Alexander, D.H., Novembre, J., and Lange, K., Fast model-based estimation of ancestry in unrelated individuals, Genome Res., 2009, vol. 19, pp. 1655–1664.
Nasibov, Sh.N., Bagirov, V.A., Klenovitskii, P.M., et al., Conservation and management of the gene pool of the bighorn sheep, Dostizh. Nauki Tekh. Agrar. Prom. Kompleksa, 2010, no. 12, pp. 63–64.
Plakhina, D.A., Zvychainaya, E.Yu., Kholodova, M.V., and Danilkin, A.A., Identification of European (Capreolus capreolus L.) and Siberian (C. pygargus Pall.) roe deer hybrids by microsatellite marker analysis, Russ. J. Genet., 2014, vol. 50, no. 7, pp. 757–762.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © T.E. Deniskova, A.A. Sermyagin, V.A. Bagirov, I.M. Okhlopkov, E.A. Gladyr, R.V. Ivanov, G. Brem, N.A. Zinovieva, 2016, published in Genetika, 2016, Vol. 52, No. 1, pp. 90–96.
Rights and permissions
About this article
Cite this article
Deniskova, T.E., Sermyagin, A.A., Bagirov, V.A. et al. Comparative analysis of the effectiveness of STR and SNP markers for intraspecific and interspecific differentiation of the genus Ovis . Russ J Genet 52, 79–84 (2016). https://doi.org/10.1134/S1022795416010026
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795416010026