Abstract
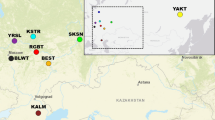
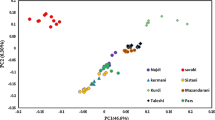
A potential application of single nucleotide polymorphisms (SNPs) in animal husbandry and production is identification of the animal breed. In this study, using chosen marker selection methods and genotypic data obtained with the use of Illumina Bovine SNP50 BeadChip for individuals belonging to ten cattle breeds, the reduced panels containing the most informative SNP markers were developed. The suitability of selected SNP panels for the effective and reliable assignment of the studied individuals to the breed of origin was checked by three allocation algorithms implemented in GeneClass 2. The studied breeds set included both Polish-native breeds under the genetic resources conservation programs and highly productive breeds with a global range. For all of the tested marker selection methods (“delta” and two FST-based variants), two separate methodological approaches of marker assortment were used and three marker panels were created with 96, 192, and 288 SNPs respectively, to determine the minimum number of markers required for effective differentiation of the studied breeds. Moreover, the usefulness of the most effective panels of markers to assess the population structure and genetic diversity of the analyzed breeds was examined. The conducted analyses showed the possibility of using SNP subsets from medium-density genotypic microarrays to distinguish breeds of cattle kept in Poland and to analyze their genetic structure.





Similar content being viewed by others
Data Availability
Raw genotyping data are available from corresponding author on reasonable request from.
References
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664. https://doi.org/10.1101/gr.094052.109
Baes CF, Makanjuola BO, Miglior F, Marras G, Howard JT, Fleming A, Maltecca C (2019) Symposium review: The genomic architecture of inbreeding: how homozygosity affects health and performance. J Dairy Sci 102:2807–2817. https://doi.org/10.3168/jds.2018-15520
Baudouin L, Lebrun P (2001) An operational Bayesian approach for the identification of sexually reproduced crossfertilized populations using molecular markers. Acta Hort 546:81–94. https://doi.org/10.17660/ActaHortic.2001.546.5
Bertolini F, Galimberti G, Calò DG, Schiavo G, Matassino D, Fontanesi L (2015) Combined use of principal component analysis and Random Forests identify population-informative single nucleotide polymorphisms: application in cattle breeds. Journal of Animal Breeding Genetics 132:346–356. https://doi.org/10.1111/jbg.12155
Bertolini F., Galimberti G., Schiavo G., Mastrangelo S., Di Gerlando R., Strillacci M.G., Bagnato A., Portolano B., Fontanesi L., 2017. Preselection statistics and Random Forest classification identify population informative single nucleotide polymorphisms in cosmopolitan and autochthonous cattle breeds. Animal: 1–8. https://doi.org/10.1017/S1751731117001355
Bjelland DW, Weigel KA, Vukasinovic N, Nkrumah JD (2013) Evaluation of inbreeding depression in Holstein cattle using whole-genome SNP markers and alternative measures of genomic inbreeding. J Dairy Sci 96:4697–4706. https://doi.org/10.3168/jds.2012-6435
Browett S, McHugo G, Richardson IW, Magee DA, Park SDE, Fahey AG, Kearney JF, Correia CN, Randhawa IAS, MacHugh DE (2018) Genomic characterisation of the indigenous Irish Kerry cattle breed. Front Genet 9:51. https://doi.org/10.3389/fgene.2018.00051
Cornuet JM, Piry S, Luikart G, Estoup A, Solignac M (1999) New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics 153:1989–2000
Dimauro C, Cellesi M, Steri R, Gaspa G, Sorbolini S, Stella A, Macciotta NPP (2013) Use of the canonical discriminant analysis to select SNP markers for bovine breed assignment and traceability purposes. Anim Genet 44:377–382. https://doi.org/10.1111/age.12021
Franklin IR (1977) The distribution of the proportion of the genome which is homozygous by descent in inbred individuals. Theor Popul Biol 11:60–80. https://doi.org/10.1016/0040-5809(77)90007-7
Gautier M, Laloe D, Moazami-Goudarzi K (2010) Insights into the genetic history of French cattle from dense SNP data on 47 worldwide breeds. PLoS One 5(9):e13038. https://doi.org/10.1371/journal.pone.0013038
Gurgul A, Rubiś D, Ząbek T, Żukowski K, Pawlina K, Semik E, Bugno-Poniewierska M (2013) The evaluation of the usefulness of pedigree verification-dedicated SNPs for breed assignment in three Polish cattle populations. Mol Biol Rep 40:6803–6809. https://doi.org/10.1007/s11033-013-2797-7
Gurgul A, Szmatoła T, Topolski P, Jasielczuk I, Żukowski K, Bugno-Poniewierska M (2016) The use of runs of homozygosity for estimation of recent inbreeding in Holstein cattle. J Appl Genet 57:527–530. https://doi.org/10.1007/s13353-016-0337-6
Gurgul A, Jasielczuk I, Szmatoła T, Sosin-Bzducha E, Majewska A, Litwińczuk Z (2020) Divergent selection signatures of phenotypic and production traits among conserved and commercial cattle breeds. Livest Sci 239:104174. https://doi.org/10.1016/j.livsci.2020.104174
Hill WG, Weir BS (2011) Variation in actual relationship as a consequence of Mendelian sampling and linkage. Genetics Research (camb) 93:47–64. https://doi.org/10.1017/S0016672310000480
Hulsegge B, Calus MP, Windig JJ, Hoving-Bolink AH, Maurice-van Eijndhoven MH, Hiemstra SJ (2013) Selection of SNP from 50K and 777K arrays to predict breed of origin in cattle. J Anim Sci 91:5128–5134. https://doi.org/10.2527/jas.2013-6678
Huston H, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23(2):254–267. https://doi.org/10.1093/molbev/msj030
Iso-Touru T, Tapio M, Vilkki J, Kiseleva T, Ammosov I, Ivanova Z, Popov R, Ozerov M, Kantanen J (2016) Genetic diversity and genomic signatures of selection among cattle breeds from Siberia, eastern and northern Europe. Anim Genet 47:647–657. https://doi.org/10.1111/age.12473
JASP Team (2020) JASP (Version 0.17.3)[Computer software]. https://jasp-stats.org/
Kelleher M, Berry D, Kearney J, McParland S, Buckley F, Purfield D (2017) Inference of population structure of purebred dairy and beef cattle using high-density genotype data. Animal 11(1):15–23. https://doi.org/10.1017/S1751731116001099
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15(5):1179–1191. https://doi.org/10.1111/1755-0998.12387
Majewska A., 2006. Ochrona zasobów genetycznych bydła w ramach Programu Rozwoju Obszarów Wiejskich. Wiadomości Zootechniczne, R. XLIV, 4: 3-8
Makina SO, Muchadeyi FC, van Marle-Köster E, MacNeil MD, Maiwashe A (2014) Genetic diversity and population structure among six cattle breeds in South Africa using a whole genome SNP panel. Front Genet 5:333. https://doi.org/10.3389/fgene.2014.00333
Mateus JC, Russo-Almeida PA (2015) Traceability of 9 Portuguese cattle breeds with PDO products in the market using microsatellites. Food Control 47:487e492. https://doi.org/10.1016/j.foodcont.2014.07.038
Matukumalli LK, Lawley CT, Schnabel RD, Taylor JF, Allan MF, Heaton MP, O’Connell J, Moore SS, Smith TPL, Sonstegard TS, Van Tassell CP (2009) Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE 4(4):e5350. https://doi.org/10.1371/journal.pone.0005350
McKay SD, Schnabel RD, Murdoch BM, Matukumalli LK, Aerts J, Coppieters W, Crews D, Dias E, Gill CA, Gao C, Mannen H, Wang Z, Van Tassell CP, Williams JL, Taylor JF, Moore SS (2008) An assessment of population structure in eight breeds of cattle using a whole genome SNP panel. BMC Genet 9:9. https://doi.org/10.1186/1471-2156-9-37
Negrini R, Milanesi E, Colli L, Pellecchia M, Nicoloso L, Crepaldi P, Lenstra JA, Ajmone-Marsan P (2007) Breed assignment of Italian cattle using biallelic AFLP markers. Anim Genet 38:147–153. https://doi.org/10.1111/j.1365-2052.2007.01573.x
Negrini R, Nicoloso L, Crepaldi P, Milanesi E, Marino R, Perini D, Pariset L, Dunner S, Leveziel H, Williams JL, Ajmone MP (2008) Traceability of four European protected geographic indication (PGI) beef products using single nucleotide polymorphisms (SNP) and Bayesian statistics. Meat Sci 80:1212–1217. https://doi.org/10.1016/j.meatsci.2008.05.021
Negrini R, Nicoloso L, Crepaldi P, Milanesi E, Colli L, Chegdani F, Pariset L, Dunner S, Leveziel H, Williams JL, Ajmone MP (2009) Assessing SNP markers for assigning individuals to cattle populations. Anim Genet 40:18–26. https://doi.org/10.1111/j.1365-2052.2008.01800.x
Nishimura S, Watanabe T, Ogino A, Shimizu K, Morita M, Sugimoto Y, Takasuga A (2013) Application of highly differentiated SNPs between Japanese Black and Holstein to a breed assignment test between Japanese Black and F1 (Japanese Black x Holstein) and Holstein. Anim Sci J 84:1–7. https://doi.org/10.1111/j.1740-0929.2012.01036.x
Paetkau D, Calvert W, Stirling I, Strobeck C (1995) Microsatellite analysis of population structure in Canadian polar bears. Mol Ecol 4:347–354. https://doi.org/10.1111/j.1365-294x.1995.tb00227.x
Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A (2004) GENECLASS 2, a software for genetic assignment and first-generation migrant detection. J Hered 95:536–539. https://doi.org/10.1093/jhered/esh074
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. https://doi.org/10.1007/s13353-019-00495-x
Putnová L, Štohl R (2019) Comparing assignment-based approaches to breed identification within a large set of horses. J Appl Genet 60:187–198. https://doi.org/10.1007/s13353-019-00495-x
Ramos AM, Megens HJ, Crooijmans RPMA, Schook LB, Groenen MAM (2011) Identification of high utility SNPs for population assignment and traceability purposes in the pig using high-throughput sequencing. Anim Genet 42:613–620. https://doi.org/10.1111/j.1365-2052.2011.02198.x
Rannala B, Mountain JL (1997) Detecting immigration by using multilocus genotypes. Proc Natl Acad Sci USA 94:9197–9221. https://doi.org/10.1073/pnas.94.17.9197
Sermyagin AA, Dotsev AV, Gladyr EA, Traspov AA, Deniskova TE, Kostyunina OV, Reyer H, Wimmers K, Barbato M, Paronyan IA, Plemyashov KV, Sölkner J, Popov RG, Brem G, Zinovieva NA (2018) Whole-genome SNP analysis elucidates the genetic structure of Russian cattle and its relationship with Eurasian taurine breeds. Genet Sel Evol 50:37. https://doi.org/10.1186/s12711-018-0408-8
Sosin-Bzducha E (2012) Charakterystyka bydła polskiego czerwono-białego objętego programem ochrony zasobów genetycznych w latach 2008–2011*. Roczniki Naukowe Zootechniki 39(1):3–16
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370. https://doi.org/10.2307/2408641
Wilkinson S, Wiener P, Archibald AL, Law A, Schnabel RD, McKay SD, Taylor JF, Ogden R (2011) Evaluation of approaches for identifying population informative markers from high density SNP chips. BMC Genet 12:45. https://doi.org/10.1186/1471-2156-12-45
Wright S (1965) The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19(3):395–420. https://doi.org/10.2307/2406450
Yaro M, Munyard KA, Stear MJ, Groth DM (2017) Molecular identification of livestock breeds: a tool for modern conservation biology. Biol Rev Camb Philos Soc 92(2):993–1010. https://doi.org/10.1111/brv.12265
Zhang Q, Calus MPL, Guldbrandtsen B, Lund MS, Sahana G (2015) Estimation of inbreeding using pedigree, 50k SNP chip genotypes and full sequence data in three cattle breeds. BMC Genet 16:88. https://doi.org/10.1186/s12863-015-0227-7
Funding
The study was financed from funds of the project: “Directions for use and conservation of livestock genetic resources in sustainable development” co-financed by the National Research and Development Center (Poland) under the Strategic Research and Development Program: “Environment, Agriculture and Forestry” – BIOSTRATEG, the decision number BIOSTRATEG2/297267/14/NCBR/2016.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Communicated by: Maciej Szydlowski
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file2 (DOCX 1.39 MB)
Supplementary file3 (DOCX 18.3 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jasielczuk, I., Gurgul, A., Szmatoła, T. et al. The use of SNP markers for cattle breed identification. J Appl Genetics (2024). https://doi.org/10.1007/s13353-024-00857-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13353-024-00857-0