Abstract
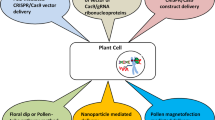
CRISPR/Cas genome editing of plants is realized in three basic variants, including knockout mutations as indels, insertion of alien DNA fragments, and base editing via deamination of nitrogenous bases. The most important stages of the CRISPR/Cas-based genome editing are the choice of a target site, design of guide RNAs, creation of genetically engineered constructions, and delivery of CRISPR/Cas components into a plant cell. Rapid developments in the field of plant genome editing with the use of CRISPR/Cas systems requires more detailed consideration of the last stage, so this review is dedicated to the description of the main ways to deliver CRISPR/Cas components into cells of higher plants. In the first studies on the genome editing of different plant species, these components were delivered to the target site mainly by Agrobacterium tumefaciens. This approach supposes integration of T-DNA into a genome and a stable expression of CRISPR/Cas components or their transient expression in the case of agroinfiltration. Another widespread approach included the use of plant viruses as delivery platforms; in this case, viruses were used mainly for production of an increased amount of guide RNAs that significantly improved the efficiency of genome editing. Another approach provides for the use of another bacterium, A. rhizogenes, as a platform for delivery of CRISPR/Cas components. This bacterium induces hairy root formation that may be an indirect confirmation of successful genome editing and assist in the selection of genetically modified forms. Other common ways to obtain genetically edited plants are the biolistic delivery of genetically engineered constructions into explants and various protoplast transformation technologies. The review also considers some issues transgenic and GM status of CRISPR/Cas-edited plants to transgenic and GM plants. There are a number of cases in which new organisms created by a CRISPR/Cas genome editing without any introduction of alien DNA were not considered as transgenic ones; it is quite possible that such plants will not fall under Russian legislation prohibiting GMO cultivation.

Similar content being viewed by others
REFERENCES
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J.A., and Charpentier, E., A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity, Science, 2012, vol. 337, pp. 816–821. https://doi.org/10.1126/science.1225829
Cong, L., Ran, F.A., Cox, D., Lin, S., Barretto, R., Habib, N., Hsu, P.D., Wu, X., Jiang, W., Marraffini, L.A., and Zhang, F., Multiplex genome engineering using CRISPR/Cas systems, Science, 2013, vol. 339, pp. 819–823. https://doi.org/10.1126/science.1231143
Puchta, H., Applying CRISPR/Cas for genome engineering in plants: the best is yet to come, Curr. Opin. Plant Biol., 2017, vol. 36, pp. 1–8. https://doi.org/10.1016/j.pbi.2016.11.011
Chemeris, D.A., Kir’yanova, O.Yu., Gerashchenkov, G.A., Kuluev, B.R., Rozhnova, N.A., Matniyazov, R.T., Baymiev, An.Kh., Baymiev, Al.Kh., Gubaidullin, I.M., and Chemeris, A.V., Bioinformatic resources for CRISPR/Cas genome editing, Biomics, 2017, vol. 9, no. 3, pp. 203–228.
Ma, X., Zhu, Q., Chen, Y., and Liu, Y.G., C-RISPR/Cas9 platforms for genome editing in plants: developments and applications, Mol. Plant, 2016, vol. 9, pp. 961–974. https://doi.org/10.1016/j.molp.2016.04.009
Zlobin, N.E., Ternovoi, V.V., Grebenkina, N.A., and Taranov, V.V., To make the complex easier: modern tools for editing the plant genome, Vavilov J. Genet. Breed., 2017, vol. 21, no. 1, pp. 104–111. https://doi.org/10.18699/VJ17.228
Kuluev, B.R., Gerashchenkov, G.A., Rozhnova, N.A., Baymiev, An.Kh., Vershinina, Z.R., Knyazev, A.V., Matniyazov, R.T., Gumerova, G.R., Mikhailova, E.V., Nikonorov, Yu.M., Chemeris, D.A., Baymiev, Al.Kh., and Chemeris, A.V., CRISPR/Cas genome editing of plants, Biomics, 2017, vol. 9, no. 3, pp. 155–182.
Liu, X., Xie, C., Si, H., and Yang, J., CRISPR/Cas9-mediated genome editing in plants, Methods, 2017, vols. 121–122, pp. 94–102. https://doi.org/10.1016/j.ymeth
Ran, Y., Liang, Z., and Gao, C., Current and future editing reagent delivery systems for plant genome editing, Sci. China Life Sci., 2017, vol. 60, pp. 490–505. https://doi.org/10.1007/s11427-017-9022-1
Jiang, W., Zhou, H., Bi, H., Fromm, M., Yang, B., and Weeks, D.P., Demonstration of CR-ISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice, Nucleic Acids Res., 2013, vol. 41: e188. https://doi.org/10.1093/nar/gkt780
Miao, J., Guo, D., Zhang, J., Huang, Q., Qin, G., Zhang, X., Wan, J., Gu, H., and Qu, L.J., Targeted mutagenesis in rice using CRISPR-Cas system, Cell Res., 2013, vol. 23, pp. 1233–1236. https://doi.org/10.1038/cr.2013.123
Feng, Z., Zhang, B., Ding, W., Liu, X., Yang, D.L., Wei, P., Cao, F., Zhu, S., Zhang, F., Mao, Y., and Zhu, J.K., Efficient genome editing in plants using a CRISPR/Cas system, Cell Res., 2013, vol. 23, pp. 1229–1232. https://doi.org/10.1038/cr.2013.114
Feng, Z., Mao, Y., Xu, N., Zhang, B., Wei, P., Yang, D.L., Wang, Z., Zhang, Z., Zheng, R., Yang, L., Zeng, L., Liu, X., and Zhu, J.K., Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis, Proc. Natl. Acad. Sci. USA, 2014, vol. 111, pp. 4632–4637. https://doi.org/10.1073/pnas.1400822111
Fauser, F., Schiml, S., and Puchta, H., Both CRI-SPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana, Plant J., 2014, vol. 79, pp. 348–359. https://doi.org/10.1111/tpj.12554
Schob, H., Kunc, C., and Meins, F., Silencing of transgenes introduced into leaves by agroinfiltration: a simple, rapid method for investigating sequence requirements for gene silencing, Mol. Gen. Genetics, 1997, vol. 256, pp. 581–585. https://doi.org/10.1007/s004380050604
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J.D., and Kamoun, S., Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease, Nat. Biotechnol., 2013, vol. 31, pp. 691–693. https://doi.org/10.1038/nbt.2655
Jia, H. and Wang, N., Xcc-facilitated agroinfiltration of citrus leaves: a tool for rapid functional analysis of transgenes in citrus leaves, Plant Cell Rep., 2014, vol. 33, pp. 1993–2001. https://doi.org/10.1007/s00299-014-1673-9
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., Wang, B., Yang, Z., Li, H., Lin, Y., Xie, Y., Shen, R., Chen, S., Wang, Z., Chen, Y., et al., A robust CR-ISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants, Mol. Plant, 2015, vol. 8, pp. 1274–1284. https://doi.org/10.1016/j.molp.2015.04.007
Pan, C., Ye, L., Qin, L., Liu, X., He, Y., Wang, J., Chen, L., and Lu, G., CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations, Sci. Rep., 2016, vol. 6: e24765. https://doi.org/10.1038/srep24765
Char, S.N., Neelakandan, A.K., Nahampun, H., Frame, B., Main, M., Spalding, M.H., Becraft, P.W., Meyers, B.C., Walbot, V., Wang, K., and Yang, B., An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize, Plant Biotechnol. J., 2017, vol. 15, pp. 257–268. https://doi.org/10.1111/pbi.12611
Nakajima, I., Ban, Y., Azuma, A., Onoue, N., Moriguchi, T., Yamamoto, T., Toki, S., and Endo, M., CR-ISPR/Cas9-mediated targeted mutagenesis in grape, PLoS One, 2017, vol. 12: e0177966. https://doi.org/10.1371/journal.pone.0177966
Odipio, J., Alicai, T., Ingelbrecht, I., Nusinow, D.A., Bart, R., and Taylor, N.J., Efficient CRISPR/Cas9 genome editing of Phytoene desaturase in cassava, Front. Plant Sci., 2017, vol. 8: e01780. https://doi.org/10.3389/fpls.2017.01780
Wang, M., Mao, Y., Lu, Y., Tao, X., and Zhu, J.K., Multiplex gene editing in rice using the CRISPR-Cpf1 system, Mol. Plant, 2017, vol. 10, pp. 1011–1013. https://doi.org/10.1016/j.molp.2017.03.001
Zhou, X., Jacobs, T.B., Xue, L.J., Harding, S.A., and Tsai, C.J., Exploiting SNPs for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4-coumarate:CoA ligase specificity and redundancy, New Phytol., 2015, vol. 208, pp. 298–301. https://doi.org/10.1111/nph.13470
Mikami, M., Toki, S., and Endo, M., Precision targeted mutagenesis via Cas9 paired nickases in rice, Plant Cell Physiol., 2016, vol. 57, pp. 1058–1068. https://doi.org/10.1093/pcp/pcw049
Mercx, S., Tollet, J., Magy, B., Navarre, C., and Boutry, M., Gene inactivation by CRISPR-Cas9 in N-icotiana tabacum BY-2 suspension cells, Front. Plant Sci., 2016, vol. 7: 40. https://doi.org/10.3389/fpls.2016.00040
Hanania, U., Ariel, T., Tekoah, Y., Fux, L., Sheva, M., Gubbay, Y., Weiss, M., Oz, D., Azulay, Y., Turbovski, A., Forster, Y., and Shaaltiel, Y., Establishment of a tobacco BY-2 cell line devoid of plant-specific xylose and fucose as a platform for the production of biotherapeutic proteins, Plant Biotechnol. J., 2017, vol. 15, pp. 1120–1129. https://doi.org/10.1111/pbi.12702
Murovec, J., Pirc, Ž. and Yang, B., New variants of CRISPR RNA-guided genome editing enzyme, Plant Biotechnol. J., 2017, vol. 15, pp. 917–926. https://doi.org/10.1111/pbi.12736
Li, S., Zhang, X., Wang, W., Guo, X., Wu, Z., Du, W., Zhao, Y., and Xia, L., Expanding the scope of CR-ISPR/Cpf1-mediated genome editing in rice, Mol. Plant, 2018, vol. 11, pp. 995–998. https://doi.org/10.1016/j.molp.2018.03.009
Wang, M., Lu, Y., Botella, J.R., Mao, Y., Hua, K., and Zhu, J.K., Gene targeting by homology-directed repair in rice using a geminivirus-based CRISPR/Cas9 system, Mol. Plant, 2017, vol. 10, pp. 1007–1010. https://doi.org/10.1016/j.molp.2017.03.002
Kang, B.C., Yun, J.Y., Kim, S.T., Shin, Y., Ryu, J., Choi, M., Woo, J.W., and Kim, J.S., Precision genome engineering through adenine base editing in plants, Nat. Plants, 2018, vol. 4, pp. 427–431. https://doi.org/10.1038/s41477-018-0178-x
Tian, S., Jiang, L., Cui, X., Zhang, J., Guo, S., Li, M., Zhang, H., Ren, Y., Gong, G., Zong, M., Liu, F., Chen, Q., and Xu, Y., Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing, Plant Cell Rep., 2018, vol. 37, pp. 1353–1356. https://doi.org/10.1007/s00299-018-2299-0
Tsutsui, H. and Higashiyama, T., pKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana, Plant Cell Physiol., 2017, vol. 58, pp. 46–56. https://doi.org/10.1093/pcp/pcw191
Baltes, N.J., Gil-Humanes, J., Cermak, T., Atkins, P.A., and Voytas, D.F., DNA replicons for plant genome engineering, Plant Cell, 2014, vol. 26, pp. 151–163. https://doi.org/10.1105/tpc.113.119792
Kumagai, M.H., Donson, J., della-Cioppa, G., Harvey, D., Hanley, K., and Grill, L.K., Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA, Proc. Natl. Acad. Sci. USA, 1995, vol. 92, pp. 1679–1683.
Ruiz, M.T., Voinnet, O., and Baulcombe, D.C., Initiation and maintenance of virus-induced gene silencing, Plant Cell, 1998, vol. 10, pp. 937–946. https://doi.org/10.1105/tpc.10.6.937
Yin, K., Han, T., Liu, G., Chen, T., Wang, Y., Yu, A.Y., and Liu, Y., A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing, Sci. Rep., 2015, vol. 5: 14926. https://doi.org/10.1038/srep14926
Čermák, T., Baltes, N.J., Čegan, R., Zhang, Y., and Voytas, D.F., High-frequency, precise modification of the tomato genome, Genome Biol., 2015, vol.16: 232. https://doi.org/10.1186/s13059-015-0796-9
Butler, N.M., Baltes, N.J., Voytas, D.F., and Douches, D.S., Geminivirus-mediated genome editing in potato (Solanum tuberosum L.) using sequence-specific nucleases, Front. Plant Sci., 2016, vol. 7: e01045. https://doi.org/10.3389/fpls.2016.01045
Gil-Humanes, J., Wang, Y., Liang, Z., Shan, Q., Ozuna, C.V., Sánchez-León, S., Baltes, N.J., Starker, C., Barro, F., Gao, C., and Voytas, D.F., High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9, Plant J., 2017, vol. 89, pp. 1251–1262. https://doi.org/10.1111/tpj.13446
Ali, Z., Abul-Faraj, A., Piatek, M., and Mahfouz, M.M., Activity and specificity of TRV-mediated gene editing in plants, Plant Signal. Behav., 2015, vol. 10: e1044191. https://doi.org/10.1080/15592324.2015.1044191
Ali, Z., Abul-Faraj, A., Li, L., Ghosh, N., Piatek, M., Mahjoub, A., Aouida, M., Piatek, A., Baltes, N.J., Voytas, D.F., Dinesh-Kumar, S., and Mahfouz, M.M., Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system, Mol. Plant, 2015, vol. 8, pp. 1288–1291. https://doi.org/10.1016/j.molp.2015.02.011
Ali, Z., Eid, A., Ali, S., and Mahfouz, M.M., Pea early-browning virus-mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis, Virus Res., 2018, vol. 244, pp. 333–337. https://doi.org/10.1016/j.virusres.2017.10.009
Ron, M., Kajala, K., Pauluzzi, G., Wang, D., Reynoso, M.A., Zumstein, K., Garcha, J., Winte, S., Masson, H., Inagaki, S., Federici, F., Sinha, N., Deal, R.B., Bailey-Serres, J., and Brady, S.M., Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model, Plant Physiol., 2014, vol. 166, pp. 455–469. https://doi.org/10.1104/pp.114.239392
Serbinov, I.L., Bacterial cancer of fruit trees, berry bushes and other garden and agricultural plants, Plodovodstvo, 1912, no. 9, pp. 787–795.
Curtin, S.J., Zhang, F., Sander, J.D., Haun, W.J., Starker, C., Baltes, N.J., Reyon, D., Dahlborg, E.J., Goodwin, M.J., Coffman, A.P., Dobbs, D., Joung, J.K., Voytas, D.F., and Stupar, R.M., Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases, Plant Physiol., 2011, vol. 156, pp. 466–473. https://doi.org/10.1104/pp.111.172981
Jacobs, T.B. and Martin, G.B., High-throughput CRISPR vector construction and characterization of DNA modifications by generation of tomato hairy roots, J. Vis. Exp., 2016, vol. 110: 53843. https://doi.org/10.3791/53843
Cai, Y., Chen, L., Liu, X., Sun, S., Wu, C., Jiang, B., Han, T., and Hou, W., CRISPR/Cas9-mediated genome editing in soybean hairy roots, PLoS One, 2015, vol. 10: e0136064. https://doi.org/10.1371/journal.pone.0136064
Jacobs, T.B., LaFayette, P.R., Schmitz, R.J., and Parrott, W.A., Targeted genome modifications in soybean with CRISPR/Cas9, BMC Biotechnol., 2015, vol. 15: 16. https://doi.org/10.1186/s12896-015-0131-2
Michno, J.M., Wang, X., Liu, J., Curtin, S.J., Kono, T.J., and Stupar, R.M., CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web-tool and a modified Cas9 enzyme, GM Crops Food, 2015, vol. 6, pp. 243–252. https://doi.org/10.1080/21645698.2015.1106063
Sun, X., Hu, Z., Chen, R., Jiang, Q., Song, G., Zhang, H., and Xi, Y., Targeted mutagenesis in soybean using the CRISPR-Cas9 system, Sci. Rep., 2015, vol. 5: 10342. https://doi.org/10.1038/srep10342
Du, H., Zeng, X., Zhao, M., Cui, X., Wang, Q., Yang, H., Cheng, H., and Yu, D., Efficient targeted mutagenesis in soybean by TALENs and CRI-SPR/Cas9, J. Biotechnol., 2016, vol. 217, pp. 90–97. https://doi.org/10.1016/j.jbiotec.2015.11.005
Wang, L., Wang, L., Tan, Q., Fan, Q., Zhu, H., Hong, Z., Zhang, Z., and Duanmu, D., Efficient inactivation of symbiotic nitrogen fixation related genes in Lotus japonicus using CRISPR-Cas9, Front. Plant Sci., 2016, vol. 7: e01333. https://doi.org/10.3389/fpls.2016.01333
Li, B., Cui, G., Shen, G., Zhan, Z., Huang, L., Chen, J., and Qi, X., Targeted mutagenesis in the medicinal plant Salvia miltiorrhiza, Sci. Rep., 2017, vol. 7: 43320. https://doi.org/10.1038/srep43320
Zhou, Z., Tan, H., Li, Q., Chen, J., Gao, S., Wang, Y., Chen, W., and Zhang, L., CRISPR/Cas9-mediated efficient targeted mutagenesis of RAS in Salvia miltiorrhiza, Phytochemistry, 2018, vol. 148, pp. 63–70. https://doi.org/10.1016/j.phytochem.2018.01.015
Iaffalando, B., Zhang, Y., and Cornish, K., C-RISPR/Cas9 genome editing of rubber producing dandelion Taraxacum kok-saghyz using Agrobacterium rhizogenes without selection, Ind. Crops Prod., 2016, vol. 89, pp. 356–362. https://doi.org/10.1016/j.indcrop.2016.05.029
Kirchner, T.W., Niehaus, M., Debener, T., Schenk, M.K., and Herde, M., Efficient generation of mutations mediated by CRISPR/Cas9 in the hairy root transformation system of Brassica carinata, PLoS One, 2017, vol. 12: e0185429. https://doi.org/10.1371/journal.pone.0185429
Nakayasu, M., Akiyama, R., Lee, H.J., Osakabe, K., Osakabe, Y., Watanabe, B., Sugimoto, Y., Umemoto, N., Saito, K., Muranaka, T., and Mizutani, M., Generation of [alpha]-solanine-free hairy roots of potato by CRISPR/Cas9 mediated genome editing of the St1-6DOX gene, Plant Physiol. Biochem., 2018, vol. 131, pp. 70–77. https://doi.org/10.1016/j.plaphy.2018.04.026
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., Zhang, K., Liu, J., Xi, J.J., Qiu, J.L., and Gao, C., Targeted genome modification of crop plants using a CRISPR-Cas system, Nat. Biotechnol., 2013, vol. 31, pp. 686–688. https://doi.org/10.1038/nbt.2650
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., and Qiu, J.L., Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew, Nat. Biotechnol., 2014, vol. 32, pp. 947–951. https://doi.org/10.1038/nbt.2969
Zhang, Y., Bai, Y., Wu, G., Zou, S., Chen, Y., Gao, C., and Tang, D., Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat, Plant J., 2017, vol. 91, pp. 714–724. https://doi.org/10.1111/tpj.13599
Sun, Y., Zhang, X., Wu, C., He, Y., Ma, Y., Hou, H., Guo, X., Du, W., Zhao, Y., and Xia, L., Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase, Mol. Plant, 2016, vol. 9, pp. 628–631. https://doi.org/10.1016/j.molp.2016.01.001
Zhang, Y., Liang, Z., Zong, Y., Wang, Y., Liu, J., Chen, K., Qiu, J.L., and Gao, C., Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA, Nat. Co-mmun., 2016, vol. 7: e12617. https://doi.org/10.1038/ncomms12617
Svitashev, S., Schwartz, C., Lenderts, B., Young, J.K., and Mark Cigan, A., Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes, Nat. Commun., 2016, vol. 7: e13274. https://doi.org/10.1038/ncomms13274
Martin-Ortigosa, S. and Wang, K., Proteolistics: a biolistic method for intracellular delivery of proteins, Transgenic Res., 2014, vol. 23, pp. 743–756. https://doi.org/10.1007/s11248-014-9807-y
Liang, Z., Chen, K., Li, T., Zhang, Y., Wang, Y., Zhao, Q., Liu, J., Zhang, H., Liu, C., Ran, Y., and Gao, C., Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes, Nat. Commun., 2017, vol. 8: e14261. https://doi.org/10.1038/ncomms14261
Liang, Z., Chen, K., Zhang, Y., Liu, J., Yin, K., Qiu, J.L., and Gao, C., Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins, Nat. Protoc., 2018, vol. 13, pp. 413–430. https://doi.org/10.1038/nprot.2017.145
Begemann, M.B., Gray, B.N., January, E., Gordon, G.C., He, Y., Liu, H., Wu, X., Brutnell, T.P., Mockler, T.C., and Oufattole, M., Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases, Sci. Rep., 2017, vol. 7: 11606. https://doi.org/10.1038/s41598-017-11760-6
Xie, K. and Yang, Y., RNA-guided genome editing in plants using a CRISPR-Cas system, Mol. Plant, 2013, vol. 6, pp. 1975–1983. https://doi.org/10.1093/mp/sst119
Butt, H., Eid, A., Ali, Z., Atia, M.A.M., Mokhtar, M.M., Hassan, N., Lee, C.M., Bao, G., and Mahfouz, M.M., Efficient CRISPR/Cas9-mediated genome editing using a chimeric single-guide RNA molecule, Front. Plant Sci., 2017, vol. 24: e01441. https://doi.org/10.3389/fpls.2017.01441
Malnoy, M., Viola, R., Jung, M.H., Koo, O.J., Kim, S., Kim, J.S., Velasco, R., and Nagamangala Kanchiswamy, C., DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins, Front. Plant Sci., 2016, vol. 7: e01904. https://doi.org/10.3389/fpls.2016.01904
Subburaj, S., Chung, S.J., Lee, C., Ryu, S.M., Kim, D.H., Kim, J.S., Bae, S., and Lee, G.J., Site-d-irected mutagenesis in Petunia × hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins, Plant Cell Rep., 2016, vol. 35, pp. 1535–1544. https://doi.org/10.1007/s00299-016-1937-7
Kim, D., Alptekin, B., and Budak, H., CRISPR/Cas9 genome editing in wheat, Funct. Integr. Genomics, 2018, vol. 18, pp. 31–41. https://doi.org/10.1007/s10142-017-0572-x
Hudzieczek, V., Cegan, R., Cermak, T., Bacovska, N., Machalkova, Z., Dolezal, K., Plihalova, L., Voytas, D., Hobza, R., and Vyskot, B., Agrobacterium rhizogenes-mediated transformation of a dioecious plant model S-ilene latifolia, New Biotechnol., 2018, vol. 48, pp. 20–28. https://doi.org/10.1016/j.nbt.2018.04.001
Andersson, M., Turesson, H., Nicolia, A., Fält, A.S., Samuelsson, M., and Hofvander, P., Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts, Plant Cell Rep., 2017, vol. 36, pp. 117–128. https://doi.org/10.1007/s00299-016-2062-3
Andersson, M., Turesson, H., Olsson, N., Fält, A.S., Ohlsson, P., Gonzalez, M.N., Samuelsson, M., and Hofvander, P., Genome editing in potato via CRI-SPR-Cas9 ribonucleoprotein delivery, Physiol. Plant., 2018, vol. 164, pp. 378–384. https://doi.org/10.1111/ppl.12731
Woo, J.W., Kim, J., Kwon, S.I., Corvalán, C., Cho, S.W., Kim, H., Kim, S.G., Kim, S.T., Choe, S., and Kim, J.S., DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins, Nat. Biotechnol., 2015, vol. 33, pp. 1162–1164. https://doi.org/10.1038/nbt.3389
Kim, H., Kim, S.T., Ryu, J., Kang, B.C., Kim, J.S., and Kim, S.G., CRISPR/Cpf1-mediated DNA-free plant genome editing, Nat. Commun., 2017, vol. 8: 14406. https://doi.org/10.1038/ncomms14406
Gao, J., Wang, G., Ma, S., Xie, X., Wu, X., Zhang, X., Wu, Y., Zhao, P., and Xia, Q., CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum, Plant Mol. Biol., 2015, vol. 87, pp. 99–110. https://doi.org/10.1007/s11103-014-0263-0
Kopertekh, L. and Schiemann, J., Marker removal in transgenic plants using Cre recombinase delivered with potato virus X, Methods Mol. Biol., 2017, vol. 1642, pp. 151–168. https://doi.org/10.1007/978-1-4939-7169-5_10
Sprink, T., Eriksson, D., Schiemann, J., and Hartung, F., Regulatory hurdles for genome editing: process- vs. product-based approaches in different regulatory contexts, Plant Cell Rep., 2016, vol. 35, pp. 1493–1506. https://doi.org/10.1007/s00299-016-1990-2
Podevin, N., Devos, Y., Davies, H.V., and Nielsen, K.M., Transgenic or not? No simple answer! New biotechnology-based plant breeding techniques and the regulatory landscape, EMBO Rep., 2012, vol. 13, pp. 1057–1061. https://doi.org/10.1038/embor.2012.168
Matveeva, T.V., Naturally transgenic plants as a model for the study of delayed environmental risks of cultivation of GMOs, Ecol. Genet., 2015, vol. 13, no. 2, pp. 118–126. https://doi.org/10.17816/ecogen132118-126
Matveeva, T.V. and Azarakhsh, M., Genetically modified organisms authorized for cultivation and breeding in Russia, Ecol. Genet., 2016, vol. 14, no. 4, pp. 32–40. https://doi.org/10.17816/ecogen14432-40
Baymiev, An.Kh., Kuluev, B.R., Vershinina, Z.R., Knyazev, A.V., Chemeris, D.A., Rozhnova, N.A., Gerashchenkov, G.A., Mikhailova, E.V., Baymiev, Al.Kh., and Chemeris, A.V., CRISPR/Cas genome editing (plants) and society, Biomics, 2017, vol. 9, no. 3, pp. 183–202.
Waltz, E., With a free pass, CRISPR-edited plants reach market in record time, Nat. Biotechnol., 2018, vol. 36, pp. 6–7. https://doi.org/10.1038/nbt0118-6b
Callaway, E., CRISPR plants now subject to tough GM laws in European Union, Nature, 2018, vol. 560: 16. https://doi.org/10.1038/d41586-018-05814-6
Urnov, F., Ronald, P.C., and Carroll, D., A call for science-based review of the European court’s decision on gene-edited crops, Nat. Biotechnol., 2018, vol. 36, pp. 800–802. https://doi.org/10.1038/nbt.4252
Gao, C., The future of CRISPR technologies in agriculture, Nat. Rev. Mol. Cell Biol., 2018, vol. 19, pp. 275–276. https://doi.org/10.1038/nrm.2018.2
Funding
The study was performed within the framework of the State Assignment (project nos. АААА-А16-116020350028-4 and AAAA-A19-119021190011-0) and also supported by the Russian Foundation for Basic Research (project nos. 18-04-00118, 19-016-00117, and 19-016-00139).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by N. Statsyuk
Rights and permissions
About this article
Cite this article
Kuluev, B.R., Gumerova, G.R., Mikhaylova, E.V. et al. Delivery of CRISPR/Cas Components into Higher Plant Cells for Genome Editing. Russ J Plant Physiol 66, 694–706 (2019). https://doi.org/10.1134/S102144371905011X
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S102144371905011X