Abstract
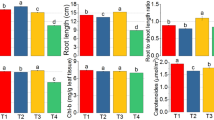
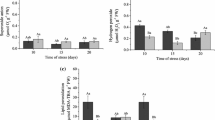
Salt stress as a major limiting factor negatively affects many physiological processes in plants. Salinity promotes the generation of reactive oxygen species and subsequently oxidative damage of cellular components. Plant salt stress tolerance requires activation of antioxidative pathways to prevent plant cell from injurious effects. In this study real-time quantitative reverse transcription–polymerase chain reaction was used to determine the protective role of two antioxidant genes, i.e. iron-superoxide dismutase (Fe-SOD) and catalase (CAT) in Cuminum cyminum L. after their treatment with 50, 100, 150 and 200 mM NaCl. Enzymatic activities were assayed spectrophotometrically for three antioxidants. Moreover, growth parameters, protein content and proline accumulation were measured. In comparison with the control plants, those plants which were exposed to 50 and 100 mM NaCl concentration accumulated higher levels of proline. At 50, 100 and 150 mM of NaCl plants showed higher superoxide dismutase, ascorbate peroxidase and catalase activities. The same condition also induced expression of the Fe-SOD and CAT genes at mRNA level. Protein content of the treated plants was significantly decreased at 50 mM NaCl and remained constant at other concentrations. Whereas, the growth parameters, with one exception in case of shoot length, did not change at plants receiving low and mild salt concentrations of up to 150 mM NaCl, 200 mM of NaCl affected these parameters negatively. From these details, it can be concluded that C. cyminum respond to salt stress by antioxidant system efficiency and proline accumulation.
Similar content being viewed by others
Abbreviations
- APX:
-
ascorbate peroxidase
- CAT:
-
catalase
- DR:
-
dehydroascorbatereductase
- GPX:
-
glutathione peroxidase
- GR:
-
glutathione reductase
- H2O2 :
-
hydrogen peroxide
- MR:
-
monodehydroascorbate reductase
- NBT:
-
nitro blue tetrazolium
- O2 •− :
-
superoxide radical
- OH• :
-
hydroxyl radical
- 1O2 :
-
singlet oxygen
- SOD:
-
superoxide dismutase
References
Pal, M., Singh, D.K., Rao, L.S., and Singh, K.P., Photosynthetic characteristics and activity of antioxidant enzymes in salinity tolerant and sensitive rice cultivars, Indian J. Plant Physiol., 2004, vol. 9, pp. 407–412.
Bartels, D. and Sunkar, R., Drought and salt tolerance in plants, Crit. Rev. Plant Sci., 2005, vol. 24, pp. 23–58.
Mittova, V., Guy, M., Tal, M., and Volokita, M., Salinity up-regulates the antioxidative system in root mitochondria and peroxisomes of the wild salt-tolerant tomato species Lycopersicon pennellii, J. Exp. Bot., 2004, vol. 55, pp. 1105–1113.
Attia, H., Arnaud, N., Karray, N., and Lachaâl, M., Long-term effects of mild salt stress on growth, ion accumulation and superoxide dismutase expression of Arabidopsis rosette leaves, Physiol. Plant., 2008, vol. 132, pp. 293–305.
Mittler, R., Oxidative stress, antioxidants and stress tolerance, Trends Plant Sci., 2002, vol. 7, pp. 405–410.
Mittler, R., Vanderauwera, S., Gollery, M., and van Breusegem, F., Reactive oxygen gene network of plants, Trends Plant Sci., 2004, vol. 9, pp. 490–498.
Pignocchi, C., Fletcher, J.M., Wilkinson, J.E., Barnes, J.D., and Foyer, C.H., The function of ascorbate oxidase in tobacco, Plant Physiol., 2003, vol. 132, pp. 1631–1641.
Thippeswamy, N.B. and Naidu, K.A., Antioxidant potency of cumin varieties—cumin, black cumin and bitter cumin—on antioxidant systems, Eur. Food Res. Technol., 2005, vol. 220, pp. 472–476.
Rebey, I.B., Jabri-Karoui, I., Hamrouni-Sellami, I., Bourgou, S., Limam, F., and Marzouk, B., Effect of drought on the biochemical composition and antioxidant activities of cumin (Cuminum cyminum L.) seeds, Ind. Crop. Prod., 2012, vol. 36, pp. 238–245.
Oroojalian, F., Kasra-Kermanshahi, R., Azizi, M., and Bassami, M.R., Phytochemical composition of the essential oils from three Apiaceae species and their antibacterial effects on food-borne pathogens, Food Chem., 2010, vol. 120, pp. 765–770.
Johri, R.K., Cuminum cyminum and Carum carvi: an update, Pharmacogn. Rev., 2011, vol. 5, pp. 63–72.
De Martino, L., De Feo, V., Fratianni, F., and Nazzaro, F., Chemistry, antioxidant, antibacterial and antifungal activities of volatile oils and their components, Nat. Prod. Commun., 2009, vol. 4, pp. 1741–1750.
Bates, L., Waldren, R., and Teare, I., Rapid determination of free proline for water stress studies, Plant Soil, 1973, vol. 39, pp. 205–207.
Bradford, M.M., A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding, Anal. Biochem., 1976, vol. 72, nos. 1–2, pp. 248–254.
Beauchamp, C. and Fridovich, I., Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels, Anal. Biochem., 1971, vol. 44, no. 1, pp. 276–287.
Nakano, Y. and Asada, K., Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts, Plant Cell Physiol., 1981, vol. 22, pp. 867–880.
Aebi, H., Catalase in vitro, Methods Enzymol., 1984, vol. 105, pp. 121–126.
Livak, K.J. and Schmittgen, T.D., Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method, Methods, 2001, vol. 25, pp. 402–408.
Gill, S.S. and Tuteja, N., Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants, Plant Physiol. Biochem., 2010, vol. 48, pp. 909–930.
Rubio, M.C., Bustos-Sanmamed, P., Clemente, M.R., and Becana, M., Effects of salt stress on the expression of antioxidant genes and proteins in the model legume Lotus japonicus, New Phytol., 2009, vol. 181, pp. 851–859.
Pandey, S., Patel, M.K., Mishra, A., and Jha, B., Physio-biochemical composition and untargeted metabolomics of cumin (Cuminum cyminum L.) make it promising functional food and help in mitigating salinity stress, PLoS One, 2015, vol. 10, p. e0144469
Shoor, M., Afrousheh, M., Rabeie, J., and Vahidi, M., The effect of salinity priming on germination and growth stage of cumin (Cuminum cyminum L), Res. J. Agric. Environ. Manage., 2014, vol. 3, pp. 340–352.
Szabados, L. and Savouré, A., Proline: a multifunctional amino acid, Trends Plant Sci., 2010, vol. 15, pp. 89–97.
Torabi, S. and Niknam, V., Effects of iso-osmotic concentrations of NaCl and mannitol on some metabolic activity in calluses of two Salicornia species, In Vitro Cell. Dev. Biol.: Plant, 2011, vol. 47, pp. 734–742.
Tavakkoli, E., Fatehi, F., Coventry, S., Rengasamy, P., and McDonald, G.K., Additive effects of Na+ and Cl–ions on barley growth under salinity stress, J. Exp. Bot., 2011, vol. 62, pp. 2189–2203.
Aghaleh, M., Niknam, V., Ebrahimzadeh, H., and Razavi, K., Salt stress effects on growth, pigments, proteins and lipid peroxidation in Salicornia persica and S. europaea, Biol. Plant., 2009, vol. 53, pp. 243–248.
Panda, S.K. and Khan, M.H., Growth, oxidative damage and antioxidant responses in greengram (Vigna radiata L.) under short-term salinity stress and its recovery, J. Agron. Crop Sci., 2009, vol. 195, pp. 442–454.
Liu, Z.J., Guo, Y.K., and Bai, J.G., Exogenous hydrogen peroxide changes antioxidant enzyme activity and protects ultrastructure in leaves of two cucumber ecotypes under osmotic stress, J. Plant Growth Regul., 2010, vol. 29, pp. 171–183.
Hu, L., Li, H., Pang, H., and Fu, J., Responses of antioxidant gene, protein and enzymes to salinity stress in two genotypes of perennial ryegrass (Lolium perenne) differing in salt tolerance, J. Plant Physiol., 2012, vol. 169, pp. 146–156.
Sekmen, A.H., Turkan, I., Tanyolac, Z.O., Ozfidan, C., and Dinc, A., Different antioxidant defense responses to salt stress during germination and vegetative stages of endemic halophyte Gypsophila oblanceolata Bark, Environ. Exp. Bot., 2012, vol. 77, pp. 63–76.
Author information
Authors and Affiliations
Corresponding author
Additional information
The article is published in the original.
Rights and permissions
About this article
Cite this article
Soleimani, Z., Afshar, A.S. & Nematpour, F.S. Responses of antioxidant gene and enzymes to salinity stress in the Cuminum cyminum L.. Russ J Plant Physiol 64, 361–367 (2017). https://doi.org/10.1134/S1021443717030177
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1021443717030177