Abstract
In this paper, we consider the course of the coronavirus disease (COVID-19) in human patients. We investigate anamnesis, examination, and clinical analysis data, as well as other features that can affect the severity and mortality of COVID-19. Based on these features, we develop a set of machine learning and statistical models that can predict the severity of the coronavirus disease and its outcome for inpatients and outpatients.
The main contribution of this work is the development of the CT Calculator service, which is integrated in the Moscow city medical information system. This service allows one to assesses the degree of changes in the lung tissue of COVID-19 patients in an express mode without computed tomography (CT) scan, as well as predict the degree of lung damage.
The developed machine learning models make it possible to determine the degree of risk for mild and severe forms of the coronavirus disease depending on various factors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 INTRODUCTION
On March 11, 2020, the World Health Organization declared the coronavirus disease (COVID-19) caused by the SARS-CoV-2 virus a global pandemic. Since the beginning of the pandemic, the Center for Systems Science and Engineering (CSSE) at the Johns Hopkins UniversityFootnote 1 registered more than 10 million COVID-19 cases and more than 292 thousand coronavirus-related deaths in the Russian Federation as of December 20, 2021.
The daily growth in the number of infected people causes the increase in the workload on medical personnel and medical equipment, deterioration in the quality of assessment of the condition of patients, and overall growth of healthcare expenditures.
The pandemic dramatically accelerated the introduction of digital services in Moscow medical organizations. Since March 2019, Moscow polyclinics and hospitals accumulated a large amount of medical history data on more than two million people.
A patient with confirmed COVID-19 undergoes a complex clinical examination.
First, an anamnesis is collected, including the individual characteristics of the patient (chronic diseases, gender, age, etc.). Then, the doctor conducts a physical examination (respiratory rate, saturation, body temperature, and severity of the decease). Finally, laboratory data are collected: clinical analysis of blood and urine, as well as blood biochemistry test.
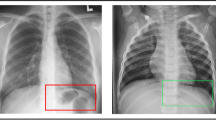
Computed tomography (CT) is widely employed to assess the state of lung tissue. CT results can serve as predictors of hospitalization or help to predict an adverse outcome in the intensive care unit. Figure 1 matches the severity of COVID-19 forms with the degree of lung damage on CT scans.
However, the CT-based approach to assessing lung damage has certain disadvantages, e.g., the risk of creating artificial epidemic foci, inefficient operation of ambulance services, and high cost of the CT procedure. In addition, there are problems associated with the radiation safety of patients and doctors.
The development of predictive models for assessing lung damage based on collected data, as well as identifying the most important features that affect the development of pulmonary pneumonia, is very important. As an alternative diagnostic tool for analyzing the state of lung tissue in COVID-19 patients, we design and investigate several data mining methods to assess the degree of lung damage in the express mode based on the physical and clinical features of the patient.
It should be noted that, in this work, we use real-world data from several sources: CT centers, laboratories, clinics, and hospitals. When collecting data from several sources, a number of problems arise: input errors and contradicting features, nonuniform filling of certain features (in particular, clinical and biochemical data are rarely available for ordinary patients), as well as different volumes and completeness of collected data. High accuracy of predictive models cannot be achieved without the construction and detailed investigation of algorithms for solving these problems.
The developed predictive models can be used as a medical decision support system, which reduces both the number of examinations and radiation exposure of the patients. In addition, this system reduces the workload on medical equipment (CT centers) and can be employed in the regions where access to CT scanners is limited or this equipment is not available.
This paper is organized as follows. Section 2 overviews some related works on the use of machine learning methods to analyze COVID-19 data. Section 3 discusses the specifics of the problem under consideration and investigates the possibility of using machine learning models for its solution. Section 4 describes the datasets and quality metrics used in this work and considers a complete algorithm for data collection and preprocessing, including elimination of inconsistencies (artifacts) and unification of feature values. In addition, the section conducts an experimental investigation of the quality of the proposed methods. Section 5 is devoted to the implementation of the CT Calculator service, interaction with the service, and its integration into external environment. Section 6 presents the main results of this work.
2 OVERVIEW OF RELATED LITERATURE
This section considers several types of research papers devoted to the use of machine learning methods for COVID-19 data analysis: the analysis of the spread of COVID-19 [1], analysis of CT images [2], and analysis of clinical data [3–5].
2.1 Analysis of the Spread of COVID-19
Currently, there are several types of research papers devoted to COVID-19 prediction.
First, there are papers on predicting daily morbidity and mortality. In [1], the problem of analyzing and predicting the time series of cumulative incidence of COVID-19 over a 14-day period was addressed. In that study, the authors considered LSTM and GRU neural network models, as well as AR and ARIMA statistical models.
As a dataset, mobility data collected in Spain using the GMD tool (Google)Footnote 2 were used. This dataset contained aggregated and anonymized data obtained using Google products (in particular, Google Maps). The main features were citizen mobility trends in places of different types: parks, supermarkets, public transport, workplaces, and residences.
The best short-term prediction result was shown by an ensemble of the considered methods (0.93R2, \(4.16RMSE\), and \(1.08MAE\)).
2.2 Analysis of CT Images
There are papers devoted to analyzing CT images and predicting the severity of the coronavirus disease. In [2], the binary classification of chest X-rays based on the presence of COVID-19 was carried out. It was proposed to extract features from the chest X-rays by using orthogonal fractional-order exponent moments (FrMEMs).
Then, the MRFODE feature selection algorithm was used to generate a set of solutions and compute a fitness value for each feature by using a trainable k‑nearest neighbors (k-NN) classifier with the determination of the best feature. The selection of the best feature was carried out until certain termination criteria were met. Based on differential evolution, binary vectors were generated using the selected features, and the k-NN classifier was trained.
Two different datasets were considered. The first dataset, collected from pediatric patients (one to five years) from the Guangzhou Medical Center, contained images of common and viral pneumonia. This dataset contained 216 positive and 1675 negative COVID-19 images. The second dataset was an extract from the database of the Italian Society of Medical and Interventional Radiology (SIRM). This dataset contained 219 positive and 1341 negative COVID-19 images.
The experimental assessment of the quality of the methods was carried out using the accuracy metric and included a comparison of the proposed method with a trained MobileNet deep neural network. The proposed algorithm showed the best result, reaching the accuracy rates of 0.9609 and 0.9809 on the first and second datasets, respectively.
2.3 Analysis of Clinical Data
Finally, there are papers devoted to the analysis of clinical data of hospitalized patients and the prediction of COVID-19 mortality. In [3], the problem of predicting 7-day survival for patients hospitalized with COVID-19 was addressed. The proposed method was based on the preselection of the most relevant features by using LASSO regression and the prediction of 7‑day survival based on Bayes’ theorem.
The dataset used contained the data of the patients hospitalized between March 1 and May 6, 2020, to 13 New York health facilities. This dataset covered over 11 000 patients with the average age of 65 years and the overall 7-day survival rate of 89%. Based on electronic health records, 42 features were extracted, including the demographic, laboratory, and clinical data of the patients.
The developed Northwell COVID-19 (NOCOS) calculator was based on six most relevant features: serum urea nitrogen, age, absolute neutrophil count, erythrocyte distribution width, oxygen saturation, and sodium; on the test sample, it reached an AUC value of 0.86.
In [4], the problem of predicting the mortality risk of COVID-19 was addressed. The proposed approach was based on the generation of a set of Cox proportional-hazards models on the subsamples grouped with respect to location and age (groups of 18–44, 45–74, and 75+ ages were considered). Based on the estimated risks, a logistic regression model was also constructed for each location.
As a dataset, the data of the patients insured under the National Health Insurance Scheme from June 7, 2020 to October 1, 2020 were used. This dataset covered more than 4.1 million patients across 259 U.S. Counties, as well as more than 15 features, including anamnesis, physical examination, and clinical data. The proposed method reached an AUC value of 0.895 on the test sample.
In [5], the problem of predicting the mortality of hospitalized patients diagnosed with COVID-19 was addressed. The following machine learning models were considered: logistic regression, support vector machine, random forest, and XGBoost.
For research purposes, the data from the Mount Sinai Health System (New York, USA) were used. The dataset contained more than 3800 observations and more than 20 features, including anamnesis (age, gender, and chronic diseases) and physical examination (saturation, arterial blood pressure, and body temperature).
The best result in terms of the AUC metric was shown by the XGBoost method (\(0.91\) AUC), which is based on the idea of gradient boosting and focuses on more difficult-to-predict subsets of training data. The following features were considered the most significant ones: saturation, age, and arterial blood pressure.
3 CONSTRUCTION AND INVESTIGATION OF THE SOLUTION TO THE PROBLEM AT HAND
The problem of predicting the degree of lung damage based on CT scans can be represented as two binary classification problems:
(1) determining the probability of a mild damage (CT 0–1): the patient does not require hospitalization and can be treated at home;
(2) determining the probability of a severe damage (CT 3–4): the patient should be immediately admitted to the hospital for intensive treatment without an intermediate visit to the CT center or clinic.
3.1 Specifics of the Solution
It should be noted that, in this work, we use real-world medical data, which causes several significant problems.
The first problem is the complexity of the data collection procedure: to develop a complete treatment regimen, it is required to process data from several sources (CT center, polyclinic, hospital, and laboratories for clinical analyzes and tests). These data sources provide different amounts of data and can contain different input errors.
When aggregating data from different sources, there can be data inconsistencies and missings in patient characteristics. For correct operation of predictive models, it is required to preliminary eliminate artifacts and inconsistencies, as well as investigate approaches for processing missing values.
Due to different amounts of data provided by these sources, there is also a problem of nonuniform filling of certain features. For instance, features from CT centers are filled for all patients because the target feature is the degree of lung damage on CT scans. In contrast, clinical or biochemical features may not be complete (due to the absence of tests or loss of data) or be outdated.
In addition, there is a problem of data completeness because access to certain data is limited due to the complexity of the collection procedure. In particular, the significant influence of the SpO2 feature (saturation) on the course of the coronavirus disease was noted in [6]. However, in open access, saturation data are available only for a small number of patients. To improve the filling of features, we can use additional data sources or carry out approximation based on available features.
Finally, there is a problem of an imbalanced dataset with respect to the degree of lung damage on CT scans. The dominant majority of patients have the mild degree of damage (CT-1), whereas the critical degrees (CT-3 and CT-4) are present in a small percentage of samples. Thus, multi-class classification with class balancing uses only a part of the available data. In this paper, we develop binary classifiers of severe and mild degrees, each of which is constructed based on samples balanced with respect to the degree of lung damage.
This section describes some basic supervised machine learning models [7, 8], in particular, binary classifiers: random forest [9, 10], neural networks [11–14], and gradient boosting [15, 16].
These models are selected due to the presence of complex non-linear dependencies in data. However, with the nonuniform filling of features in a dataset, these basic models can exhibit instability and overfitting.
To improve the stability of the predictions provided by the basic models, as well as to take into account the nonuniform filling of features, this section also considers ensembles of the basic predictive models.
3.2 Random Forest (RF)
The RF algorithm proposed in [9] is based on the idea of constructing an ensemble of decision trees [10] and aggregating their predictions.
1. From the original sample, \(N\) bootstrap samples (without replacement) are extracted. Each bootstrap sample excludes, on average, 37% of the data, which are called out-of-bag (OOB) data.
2. On each bootstrap sample, a decision tree is constructed, and \(P\) random features are selected at each node of the tree to find the best split. The split that maximizes the difference between child nodes (in particular, maximizes the logrank statistics) is selected.
3. Decision trees are constructed until the bootstrap sample is exhausted.
Classification is carried out by voting: each tree of the ensemble attributes an object to be classified to one of the classes, and the class with the maximum number of votes wins.
3.3 Neural Networks (NNs)
The NN algorithm proposed in [11] imitates the operation of the human brain’s neural system. Using neural networks, one can approximate any continuous function as accurately as possible and imitate any continuous automaton.
A neuron is an information processing unit in the neural network. This model has three main components. The first one is a set of synapses xj connected to neurons k, which are characterized by their weights \({{w}_{k}}j\). The second component is an adder that sums input signals weighted with respect to the corresponding synapses of the neuron and introduces bias \({{b}_{k}}\). The third component is activation function \(\varphi (.)\), which is applied to the resulting sum to generate the output signal \({{y}_{k}}\) of the neuron.
Thus, in mathematical notation, the operation of neuron k is described by the equation
Since the neuron model implements a function of its inputs, neurons can be interconnected based on the rules of superposition of functions to obtain more complex models called perceptrons [12], or direct-propagation artificial neural networks.
A multilayer perceptron [13] has several distinctive features: each neuron has a non-linear activation function and the whole network has one or more layers of hidden neurons.
To train a multilayer perceptron, the back-propagation method is used, which is a learning technique based on computing the gradient of an error function. In the process of training, the weights of the neurons at each layer of the neural network are adjusted taking into account the signals received from the previous layer, and the residual (deviation) of each layer is calculated recursively from the last layer to the first one.
For binary classification, cross-entropy is most commonly employed as an error function, and the logistic function is used as an activation function [17].
To avoid overfitting, a regularization layer can be added to the neural network architecture in order to limit the size of the weights. In practice, dropout layers, which null some of the weights before proceeding to the next layer, are most widely used. When adding the dropout layer, the neural network is trained on partially filled data to prevent the occurrence of global dependencies on a small number of features. Complete information on available features improves the stability of the neural network architecture on real-world data.
3.4 Gradient Boosting Machines
Gradient boosting is an alternative approach to constructing ensembles of decision trees, which was presented in [15].
Unlike a random forest, the gradient boosting algorithm is based on iterative training of the next decision tree on the errors of the previous tree, rather than on independent construction of decision trees and averaging their predictions. Aggregation of tree predictions is based on the weights computed when a new decision tree is added to the ensemble.
The gradient boosting algorithm minimizes the loss function based on which the ensemble error is computed. By default, the logarithmic loss function (log loss) [17] is used. Suppose that \(\left\{ {({{x}_{i}},{{y}_{i}})} \right\}_{{i = 1}}^{n}\) is the training set, L is the loss function, and M is the size of the ensemble. The algorithm consists of the following steps.
1. The model is initialized with a constant value α:
2. Pseudo-residuals are computed for all observations in the training set (\(i = 1,\; \ldots ,\;n\)):
3. Based on training sample \(\left\{ {({{x}_{i}},{{r}_{{im}}})} \right\}_{{i = 1}}^{n}\), decision tree \({{h}_{m}}(x)\) is constructed.
4. Weight \({{{v}}_{m}}\) (\(0 < {{{v}}_{m}} < 1\)) of the decision tree is computed by solving the following optimization problem:
5. The trained tree with its weight is added to the ensemble:
6. If the size of the ensemble m is not equal to M, then step 2 is repeated.
7. The resulting model is denoted by \({{F}_{M}}\).
Object classification is carried out by the composition of the responses from the ensemble models: each decision tree \({{h}_{m}}(x)\) returns a real “degree” of class membership for the object, and the result \({{F}_{M}}\) is obtained by applying a threshold rule to the composition.
3.5 Random Forest and Neural Network Ensemble
To enable separate processing of features with different degrees of filling, we aggregate basic predictive models. We use an ensemble of two predictive models: the first model is trained on features with a large number of missings, while the second model is trained on well-filled features and the output of the first model.
The largest number of missings is found in the group of laboratory test features. This group includes all data of complete blood count (CBC) and biochemical analyzes. We propose to train the neural network based on laboratory features. The output of the model is added to the group of the remaining features, based on which the RF algorithm is trained.
The probabilistic prediction of the random forest is used to evaluate the quality of the model. The choice of the neural network for training on partially filled features is due to the best approximation on a large space of numerical features (supplemented with binary features to indicate the presence of missing values) that can correlate with each other. When using tree-based ensembles on features with missing values, some of the features may not be used at all or may have little significance.
3.6 Boosting Ensemble of Decision Trees: Light Gradient Boosting Machine
The light gradient boosting machine (LGBM) [16] is one of the most efficient implementations of the gradient boosting procedure. Being an ensemble of decision trees, LGBM allows one to evaluate the significance of features from a trained model. Generally, the significance indicates the usefulness of each feature for constructing decision trees in the model. The more frequently the feature is used to make key decisions, the higher its relative significance. The significance is evaluated for individual decision trees; then, feature values are averaged over all decision trees in the model.
It should be noted that training in LGBM is carried out only on the data that lead to a larger gradient, which speeds up the algorithm and reduces its computational complexity.
In addition, LGBM allows one to process missings in input features. For numerical features, the missing values are attributed to the branch of the split that minimizes the loss function best of all. For categorial features, the missing values are used as an individual category. In this case, there is no need to use an ensemble of machine learning models that process features with different degrees of filling.
3.7 Ensemble of Regularized Neural Networks
In its pure form, the back-propagation method does not work well [11, 18]. There are problems of slow convergence or divergence and stucking in local minima. To stabilize the training process, we add procedures of initialization and regularization of neural network layers. With the sigmoid being used as the activation function of the neural network, the response of the neural network can be interpreted as a probability.
In addition, different initializations of \(random\_state\) lead to different results of network convergence and different binarization thresholds for network response. To improve the stability of predictions provided by individual neural networks, an approach to construction of neural network ensembles is proposed. For an input observation, a set of responses is computed over all trained neural networks; the responses are summed and normalized. The resulting response is taken as a response of the ensemble.
In addition, to control the training process, a training termination handler is used when the quality begins to deteriorate. The maximum number of epochs without parameter improvement is 10. Moreover, a handler is used to reduce the learning rate when the quality metric deteriorates.
3.8 Calibration of Results and Selection of Thresholds
In the process of classification, it is often required not only to predict the label of a class but also to find the probability of the corresponding label. This probability determines the confidence in the prediction.
The probability distribution can be adjusted for better correspondence to the expected distribution observed in the data. This adjustment is called model calibration [19, 20] and is used to reduce the responses of predictive models to a probabilistic scale. This is important for interpreting predictions, as well as for making decisions about the implementation of models and analyzing their performance.
Suppose that yi is the reference probability of observation xi and \(p({{x}_{i}})\) is the response of the model. The goal of the calibration method is to construct adjusted response \(\hat {p}(x)\).
In this paper, we use Platt calibration [21], which fits the logistic regression model to the estimates provided by the classifier:
Parameters a and b are determined by the maximum likelihood method on a holdout sample. Platt calibration is most efficient when the distortion in the predicted probabilities is sigmoidal.
There is also a problem of attributing observations to classes based on the predicted probability. In fact, not all constructed models have a threshold of 0.5 for output probabilities, and weights of errors can differ when “underestimating” or “overestimating” the result. In this case, the “underestimation” errors are more critical than the “overestimation” errors, and it is required to maximize the recall or sensitivity metrics.
Experts can set the minimum permissible recall based on which thresholds that maximize precision are selected. This approach allows one to use many individual thresholds in various use cases; however, it is not a unified approach to computing thresholds.
4 EXPERIMENTAL RESEARCH
4.1 Description of Performance Metrics
To assess the performance of the predictive models, we use the ROC-AUC metric [22].
The ROC-curve plots the ratio between the percentage of objects from the total number of correctly classified feature carriers and the percentage of objects from the total number of objects misclassified as feature carriers while varying the threshold of the decision rule (type I errors).
The area under the ROC-curve (AUC) takes values from 0 to 1 and is interpreted as a probability that the classifier assigns a larger weight to a randomly selected positive observation rather than to a randomly selected negative observation.
4.2 Description of the Datasets
In this work, we use two different datasets.
The first dataset includes municipal medical data on inpatients and outpatients with COVID-19 (Moscow) for the period from March 2020 to February 2021. These data have six sources. The structure of the dataset and the content of the sources are shown in Table 1. All data sources have a common patient ID. This dataset is sufficiently complete to be used for training and testing the predictive models.
The second dataset obtained from a Moscow city hospital consists of 95 observations: patient ID, anamnesis features (gender, age, and chronic diseases), examination features (severity, saturation, respiratory rate, dyspnea, weakness, congestion, body temperature, presence and type of cough), and laboratory test results (PCR, WBC, PDV, MON, GRA, LYM, PLT, HGB, RBC, MPV, HCT, and RDW).
Due to the small size and obvious bias to the CT-1 degree, this dataset can only be used as a test sample to assess the quality of the models.
4.3 Data Preparation
Before constructing predictive models, it is required to process the source medical data on COVID-19 patients to sort out incorrect instances and normalize the feature values to the same scales, as well as to find and process artifacts, outliers, and inconsistent data.
In this paper, we propose the following algorithm for dataset generation based on a variety of sources.
For each observation from the CT center, the nearest (within a week window) medical tests are found. Each patient is assigned a list of tests based on his or her unique ID. The tests are filtered and sorted with respect to the proximity to the date of the CT procedure. The nearest tests with three features (test value, reference values, and test date) are selected.
Sets of CBC tests and most-filled biochemical tests are considered: ALT, albumin, AST, total bilirubin, direct bilirubin, total potassium, creatinine, lactate dehydrogenase, urea, total sodium, total protein, chlorine, alkaline phosphatase, and relative number of normoblasts.
The source dataset is filtered based on these tests. Thus, a test result (rather than an analysis result) is considered because it can be obtained from several sources. In addition, tests from different sources are combined into a single test feature and the units of measurement are unified.
The most-filled features among similar tests are extracted. For instance, in the case of platelets, there are many features upon combining tests from different sources: total platelet volume (thrombocrit, PCT), platelet count, mean platelet volume, and platelet distribution width. These features correlate with each other; hence, when forming the feature space, the most-filled feature is selected.
The resulting dataset is supplemented with the following tests: PCR, IFA, and saturation (for each patient, the nearest (within a week window) tests are extracted). This dataset is also supplemented with features of chronic diseases. Each patient is assigned a list of chronic diagnoses in the format of ICD-10 codes: coronary heart disease (I11, I20, I24, I25, and I51), arterial hypertension (I10, O10-13, G97, I27, K76, P29, and I15), diabetes mellitus (G63, E10-14, H36, M14, G59, E23, N08, and O24), and obesity (E66).
4.3.1. Unification of feature values. This subsection considers the problem of normalizing the feature values to common scales and dictionaries taking into account the reference values.
When processing the features of the municipal dataset, we composed several rules for unification of feature values. The values that fall outside the permissible limits of a feature are eliminated. For instance, the CT data contained the body temperature values 3.0, 3.6, and 3.8, as well as the respiratory rate values 0, 1, and above 150.
To unify units of measurement for each feature value, we propose an algorithm for reducing these units of measurement to the most frequent (target) one. First, the prefixes and postfixes of the target unit of measurement are extracted. Then, for the other units of measurement, the distance to the target one is calculated. The prefixes and postfixes “m, mk, n, ml, k, and M,” as well as power exponents, are processed. Non-unifiable units of measurement are eliminated.
To unify categorial features, dictionaries of all possible categories were composed.
To unify continuous features, insignificant symbols are eliminated with conversion to real values (in the case of a multiple value, splitting by separators is carried out and the first value is selected).
To unify reference values of features, reference values of two main types are used: interval \(x - y\) and half-intervals < x and >y. All reference values are cast to these types; otherwise, they are considered invalid.
To unify the results of the IFA test, four features are generated: two real values IGG, IGM and two binary indicators IGG > 10, IGM > 2.0.
4.3.2. Processing anomalous values. This section discusses the problem of finding, eliminating, or correcting artifacts, outliers, and inconsistent data.
When processing the features of the municipal dataset, inconsistent data of several types were found. The inconsistencies were found both by analyzing the features and with the help of experts.
First, there is inconsistency between the fields “time of body temperature measurement during CT scan” and “time of CT scan:” there can be several days difference between these dates. In 299 thousand observations, this problem is not observed. However, more than 3800 observations have a non-zero distance between the dates, in particular, one day has 2142 observations, more than two days have 1694 observations, and more than seven days have 763 observations. The maximum time difference is 176 days. In this work, the distance of more than seven days is considered incorrect: these observations are eliminated from the dataset.
Second, there is inconsistency between the fields “CT degree of damage” and “CT result.” In accordance with the description of the source dataset, the CT category and the CT degree of damage are matched as follows: CT-0 is zero damage, CT-1 is mild damage, CT-2 is moderate damage, CT-3 is severe damage, and CT-4 is critical damage. However, in the dataset, this matching is valid only for 167 thousand observations. The complete match between the CT category and the CT degree of damage is presented in Table 2. In addition, 61 thousand observations do not include the degree of damage. In this work, the patient is considered to have a CT-N degree of damage if his or her “CT result” is CT-N and the “CT degree of damage” is not lower than CT-N or is not indicated.
Third, based on expert assessment, an inconsistency due to irreducible units of measurement was found. In the features that represent the absolute number of eosinophils, basophils, monocytes, granulocytes, lymphocytes, and neutrophils, there are two categories of values that indicate their composition in percentage and quantitative terms. These categories cannot be converted to one another by means of general unit conversion. For correct conversion, it is required to multiply the percentage composition by the white blood count (WBC). If WBC is absent, then the composition value is assumed to be a gap.
Finally, based on expert assessment, an inconsistency due to the absence of units of measurement for the RDW, PDW, and D-dimer features was found. To eliminate this problem, we decided to analyze the reference values of the features.
For RDW (and PDW), the following rule is defined: if the right-hand reference boundary is below 20 or the notation contains a comma, then the value is represented as RDW-CV (and PDW-CV) and is measured in %; otherwise, it is represented as RDW-SD (and PDW-SD) and is measured in femtoliters. The units of measurement are matched by formula RDW – CV = \(\frac{{{\text{RDW}} - {\text{SD}}}}{{{\text{MCV}}}}\) × 100, where MCV is the mean cell volume (or by formula PDW – CV = \(\frac{{{\text{PDW}} - {\text{SD}}}}{{{\text{MPV}}}}\) × 100, where MPV is the mean platelet volume).
For the D-dimer feature, the following rule is defined: if the right-hand reference boundary is below one, then the value is measured in µg/ml; otherwise, it is measured in ng/ml. The units of measurement are matched as follows: 1 µg/ml = 1000 ng/ml.
4.3.3. Dataset enrichment. In this subsection, we consider the problem of expanding the feature space by including information about the dates of analyzes and examinations.
As mentioned above, when forming the dataset, observations with CT scans are enriched with clinical analyses, PCR results, IFA results, and saturation, which are obtained over a weekly period from the date of the CT scan.
In practice, analysis data sometimes prove outdated. To control outdated data, two features are added: the CBC test date and the biochemical analysis date. Based on them, the number of days between the analyzes and the CT date is calculated.
Thus, we can filter the input data based on their relevance: if the number of days is less than seven, then the feature is used in the predictive model.
In addition, for each feature N from the dataset, a feature \(none\_N\) is generated to indicate the presence of missings in the original feature (“1” if the feature had a missing value, “0” otherwise).
4.3.4. Imputation of the saturation index. In this subsection, we consider the problem of imputation of the saturation index when assessing the severity of the examination result with recalculation based on the NEWS2 scale.
When predicting the expected CT degree of damage, one of the most important features is the saturation index (blood oxygen level in percentage terms). However, in the generated dataset, the saturation feature is filled only in 103 thousand observations (out of 299 thousand).
To solve this problem, we use the national early warning score (NEWS2) [23], which was proposed in 2020 by the Royal College of Physicians to assess the severity of the coronavirus disease.
Based on NEWS2 , features \(b\_spo2\) and \(bw\_spo2\) are generated. Feature \(b\_spo2 = \frac{{97 - SpO2}}{2}\) determines the filled saturation score, while feature \(bw\_spo2\) determines the expected saturation score based on the weighting scheme with respect to the filled features of respiratory rate, body temperature, and severity level.
Thus, to each observation, one saturation point is assigned if saturation is present or expected saturation points are given if it is absent.
4.3.5. Sample generation.Thus, taking into account the data preparation procedure described above, the dataset with 299 792 observations is formed based on the municipal data sources. The distribution of the target field “CT result” is shown in Fig. 2.
The CBC feature is filled in 176 354 observations (by this, we mean the presence of the value of the hematocrit feature). The biochemical analysis is filled in 176 866 observations (by this, we mean the presence of the “C-reactive protein” value). Both the CBC feature and the biochemical feature are simultaneously filled in 151 532 observations.
Once the initial samples for the CT 01-234 and CT 012-34 classifiers are generated, the missings are imputed by the median value of the feature on the initial samples. In addition, class balancing is carried out: a smaller class of size N is completely included in the sample, while from a larger class, N observations are randomly selected.
Based on the resulting dataset, the training, validation, and test samples are generated.
For the CT 01-234 classifier, the total number of observations is 67648 with the proportion of observations in the training, validation, and test sets being 54118/6765/6765.
For the CT 012-34 classifier, the total number of observations is 12 576 with the proportion of observations in the training, validation, and test sets being 10062/1257/1257.
In addition, we consider two approaches to the generation of test, validation, and training samples. In the case of a randomized partitioning, 80% of the observations from the original sample are randomly included in the training sample, while the remaining observations are equally split between the test and validation samples.
In the case of a time-based partitioning, the first 10% of the observations (for the entire period) with the maximum CT time are included in the test sample, while the next 10% of the observations are attributed to the validation sample. The remaining observations are included in the training sample.
4.4 Setting up the Experiments
First, the dataset is preprocessed, and the feature spaces and target variables are generated for the CT 01-234 and CT 012-34 classification.
Then, the features are post-processed, missings are imputed based on the sample-median values, and class balancing is carried out. The test, validation, and training samples are generated based on the datasets formed.
The following binary classifiers are trained with parameter selection on the validation set: neural network (NN), neural network with random forest (NN + RF), NN ensemble, LGBM with missings, and LGBM without missings (missing values are imputed with the median value over the original classifier sample).
For the constructed models, a prediction on the test sample is computed; then, their prediction quality is estimated using the ROC AUC metric.
4.5 Result Tables
Table 3 contains the values of the ROC AUC metric on the randomized and time-based test sets for the CT 01-234 and CT 012-34 classification problems.
According to the result table, the LGBM model without missings and with Platt calibration shows the best ROC AUC value for these two classification problems on the randomized and time-based test sets.
In addition, the model was tested on the data provided by the Moscow city hospital. For the CT 01-234 classification problem, ROC AUC is 0.8805; for the CT 012-34 classification problem, ROC AUC is 0.9311. The corresponding ROC curves are shown in Fig. 3.
5 IMPLEMENTATION
Based on the results of theoretical and experimental investigations, the web service called CT Calculator was developed. It enables the express assessment of lung tissue damage in COVID-19 patients without using computed tomography of the chest, based on the physical and laboratory analyzes of the patient. This service makes it possible to predict the probability of the mild and severe degrees of lung damage.
5.1 Architecture of the Solution
The software implementation of CT Calculator is based on two independent parts: external environment and internal environment (the whole architecture is shown in Fig. 4).
The external environment is used to fine-tune predictive models. It receives the source dataset as an input, pre-processes the features, and generates the training and test samples. Based on the generated samples, the training procedure is carried out and the best predictive model is selected based on the ROC AUC metric. The test sample is used to generate reports on the predictive models and compute the thresholds. The external environment outputs the file of the best predictive model, as well as contextual information (the dataset-median values and standardization coefficients for the features).
The internal environment is designed to run the web service. It receives the files generated by the external environment as an input, as well as loads and initializes the models. A predictive model of each type generates an environment that includes the model, handler function for observation features, and logger. The user can specify a particular prediction environment.
The internal environment also includes a Waitress-based WSGI server [24] that runs the web application. The web application processes incoming requests and makes predictions based on the previously trained model.
When deploying the service, it is only required to transfer the program code of the internal environment, as well as the files generated by the external environment. This approach uses a small amount of RAM (because the source dataset is processed before deploying the service) and does not require transferring the dataset to the external environment due to information security reasons.
Upon launching the predictive model, the user receives a unique request ID. In the case of an incorrect result, the user can send the request ID to the developer to analyze the log data associated with this request (in particular, perform a consistency check). If the request is correct, then the event is labeled and added to the test set.
Logged events are also used to generate operational statistics for the CT Calculator service. This statistics was used to investigate the source data and was reported in a paper published on the official website of the Mayor of Moscow.Footnote 3
5.2 Interaction Methods
To interact with CT Calculator, the REST architectural style [25] operating over the HTTP protocol is used. Each operation has its own HTTP method: GET obtains data, POST creates new data, PUT updates (modifies) data, and DELETE eliminates data.
A JSON [26] array is sent as a data packet to a specified address of the service. On the side of CT Calculator, a handler function is triggered, and a prediction in a certain format is returned depending on the sent data and the current request.
5.3 Integrating the Service into an External Organization Environment Using Docker
The service allows the user to predict the degree of lung damage based on the course of the disease. For this purpose, a web form or REST API tools can be used.
However, in this case, requests are sent to a single service and request logs are consolidated in a single repository. To support several independent services with local storages, we use Docker software [27, 28].
Docker is designed to automate the deployment and control of applications in environments that support application containerization. It also enables a more efficient use of system resources, rapid deployment of software products, as well as their scaling and porting to other environments while guaranteeing the stability of their operation.
The basic operating principle of Docker is application containerization. This type of virtualization allows software to be packaged in isolated container environments. Each of the environments contains all necessary elements to run the application. This allows a large number of containers to be simultaneously run on the same host.
Docker consists of several components. The first one is the server that initializes the daemon used to control and modify containers, images, and volumes. The daemon controls Docker objects (networks, repositories, images, and containers) and can communicate with other daemons to manage Docker services. The second component is the client that allows the user to interact with the server by sending commands. The third component is the REST API mechanism that enables communication between the Docker client and the Docker daemon.
Docker has a fairly simple syntax and is compatible with all versions of the Linux and Windows operating systems.
Thus, using Docker, CT Calculator can be packaged in a container and deployed in an external organization. Access to this container is also provided through the REST API.
In December 2020, CT Calculator was integrated into the Moscow medical information system, and doctors from all regions of the Russian Federation, as well as ordinary users, received free access to it.
6 CONCLUSIONS
In this paper, we have considered the problem of predicting the degree of lung damage in COVID-19 patients.
Computed tomography (CT) is important for determining adequate treatment strategies. With the mild degree of damage (CT 0-1), the patient does not require hospitalization and can be treated at home; with the severe degree of damage (CT 3-4), the patient must be hospitalized for intensive treatment without an intermediate visit to a CT center or clinic.
The frequent use of CT scans also has certain disadvantages, e.g., the risk of creating artificial epidemic foci, inefficient operation of ambulance services, and high cost of the CT procedure.
As an alternative diagnostic tool, we have proposed the predictive models for assessing the mild (CT 0-1) and severe (CT 3-4) degrees of lung damage in COVID-19 patients based on the examination, clinical, and biochemical features.
It should be noted that, in this work, we have used real-world medical data, which causes several significant problems: the complexity and limitations of the procedure for collecting data from multiple sources, nonuniform filling of certain features, input errors, and data inconsistencies.
As basic machine learning models, we have considered the random forest method, neural networks, gradient boosting, and ensembles of basic models that can process features with different degrees of filling. Platt calibration has been used to reduce the responses of the predictive models to the probabilistic scale.
Based on the results of the experimental investigation, the LGBM method with missing values imputation has showed the best performance in terms of the ROC AUC metric.
The proposed predictive models have been implemented as the CT Calculator web service. In December 2020, this service was integrated into the Moscow medical information system. The service enables quick medical decision making and can reduce the number of examinations for the patient (reduce radiation exposure). Moreover, the service makes it possible to reduce the workload on the medical equipment of CT centers and can be employed in the regions where access to CT scanners is limited or this equipment is not available.
Notes
Russian doctors used CT calculator 10 000 times to diagnose COVID-19. https://www.mos.ru/news/item/86015073/
REFERENCES
García-Cremades, S., Morales-García, J., Hernández-Sanjaime, R., Martínez-España, R., Bueno-Crespo, A., Hernáandez-Orallo, E., López-Espín, J.J., and Cecilia, J.M., Improving prediction of COVID-19 evolution by fusing epidemiological and mobility data, Sci. Rep., 2021, vol. 11, no. 1, pp. 1–16.
Elaziz, M.A., Hosny, K.M., Salah, A., Darwish, M.M., Lu, S., and Sahlol, A.T., New machine learning method for image-based diagnosis of COVID-19, PLOS One, 2020, vol. 15, no. 6.
Levy, T.J., Richardson, S., Coppa, K., Barnaby, D.P., McGinn, T., Becker, L.B., Davidson, K.W., Cohen, S.L., Hirsch, J.S., Zanos, T.P., et al., Development and validation of a survival calculator for hospitalized patients with COVID-19, MedRxiv, 2020.
Jin, J., Agarwala, N., Kundu, P., Harvey, B., Zhang, Y., Wallace, E., and Chatterjee, N., Individual and community-level risk for COVID-19 mortality in the United States, Nat. Med., 2021, vol. 27, no. 2, pp. 264–269.
Yadaw, A.S., Li, Y.-C., Bose, S., Iyengar, R., Bunyavanich, S., and Pandey, G., Clinical features of COVID-19 mortality: Development and validation of a clinical prediction model, Lancet Digital Health, 2020, vol. 2, no. 10, pp. e516–e525.
Shah, S., Majmudar, K., Stein, A., Gupta, N., Suppes, S., Karamanis, M., Capannari, J., Sethi, S., and Patte, C., Novel use of homepulse oximetry monitoring in COVID-19 patients discharged from the emergency department identifies need for hospitalization, Acad. Emerg. Med., 2020, vol. 27, no. 8, pp. 681–692.
Bishop, C.M., Pattern recognition, Mach. Learn., 2006, vol. 128, no. 9.
Ripley, B.D., Pattern Recognition and Neural Networks, Cambridge University Press, 2007.
Breiman, L., Random forests, Mach. Learn., 2001, vol. 45, no. 1, pp. 5–32.
Breiman, L., Friedman, J.H., Olshen, R.A., and Stone, C.J., Classification and Regression Trees, Routledge, 2017.
Rumelhart, D.E., Hinton, G.E., and Williams, R.J., Learning representations by backpropagating errors, Nature, 1986, vol. 323, no. 6088, pp. 533–536.
Minsky, M.L. and Papert, S.A., Perceptrons: Expanded Edition, 1988.
Hunt, K.J., Sbarbaro, D., Zbikowski, R., and Gawthrop, P.J., Neural networks for control systems: A survey, Automatica, 1992, vol. 28, no. 6, pp. 1083–1112.
Breiman, L., Randomizing outputs to increase prediction accuracy, Mach. Learn., 2000, vol. 40, no. 3, pp. 229–242.
Friedman, J.H., Greedy function approximation: A gradient boosting machine, Ann. Stat., 2001, pp. 1189–1232.
Ke, G., Meng, Q., Finley, T., Wang, T., Chen, W., Ma, W., Ye, Q., and Liu, T.-Y., LightGBM: A highly efficient gradient boosting decision tree, Adv. Neur. Inf. Process. Syst., 2017, vol. 30, pp. 3146–3154.
Dreiseitl, S. and Ohno-Machado, L., Logistic regression and artificial neural network classification models: A methodology review, J. Biomed. Inf., 2002, vol. 35, nos. 5–6, pp. 352–359.
Rosen, B.E., Ensemble learning using decorrelated neural networks, Connect. Sci., 1996, vol. 8, nos. 3–4, pp. 373–384.
DeGroot, M.H. and Fienberg, S.E., The comparison and evaluation of forecasters, J. R. Stat. Soc., Ser. D, 1983, vol. 32, nos. 1–2, pp. 12–22.
Niculescu-Mizil, A. and Caruana, R., Obtaining calibrated probabilities from boosting, UAI, 2005, vol. 5, pp. 413–420.
Platt, J., et al., Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods, Adv. Large Margin Classif., 1999, vol. 10, no. 3, pp. 61–74.
Hand, D.J. and Till, R.J., A simple generalisation of the area under the ROC curve for multiple class classification problems, Mach. Learn., 2001, vol. 45, no. 2, pp. 171–186.
Smith, G.B., Redfern, O.C., Pimentel, M.A., Gerry, S., Collins, G.S., Malycha, J., Prytherch, D., Schmidt, P.E., and Watkinson, P.J., The national early warning score 2 (NEWS2), Clin. Med. J. R. Coll. Physicians London, 2019, vol. 19, no. 3.
Gardner, J., The web server gateway interface (WSGI), The Definitive Guide to Pylons, 2009, pp. 369–388.
Ong, S.P., Cholia, S., Jain, A., Brafman, M., Gunter, D., Ceder, G., and Persson, K.A., The materials application programming interface (API): A simple, flexible, and efficient API for materials data based on representational state transfer (REST) principles, Comput. Mater. Sci., 2015, vol. 97, pp. 209–215.
Crockford, D., The application/JSON media type for JavaScript object notation (JSON), RFC 4627, 2006.
Anderson, C., Docker [Software engineering], IEEE Software, 2015, vol. 32, no. 3, p. 102_c3.
Boettiger, C., An introduction to Docker for reproducible research, ACM SIGOPS Oper. Syst. Rev., 2015, vol. 49, no. 1, pp. 71–79.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no conflicts of interest.
Additional information
Translated by Yu. Kornienko
Rights and permissions
About this article
Cite this article
Vasilev, I.A., Petrovskiy, M.I., Mashechkin, I.V. et al. Predicting COVID-19-Induced Lung Damage Based on Machine Learning Methods. Program Comput Soft 48, 243–255 (2022). https://doi.org/10.1134/S0361768822040065
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0361768822040065