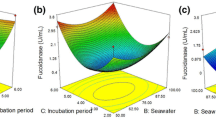
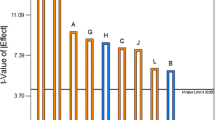
Abstract—
Exopolysaccharides of marine microbial origin are in demand in industry. Seaweeds are marine macroalgae that harbor diverse microorganisms with unique properties. In this perspective, the present study was focused on screening and isolation of marine bacteria from seaweeds for the production of an exopolysaccharide, curdlan gum, which is a linear (1 → 3)-β-d-glucan neutral exopolysaccharide produced by bacteria under conditions of nitrogen starvation. It has unique thermo gelation properties and extensive applications. A total of 78 marine bacteria were screened and isolated from different seaweeds and assessed for their potential for curdlan gum production using aniline blue agar. Curdlan gum was recovered using acidic precipitation. High-yield isolate, screened from red seaweed (Gracilaria foliifera) was designated as “RSW2n” and chosen as a potential producer. The identity of the isolate RSW2n was revealed as Enterobacter cloacae subsp. dissolvens based on its morphology, biochemical characteristics, and 16S rRNA gene sequencing. Optimization of the process parameters such as initial pH, inoculum percentage, production time, and production medium volume by single factor at a time method yielded 4.2 g/L of curdlan gum using Enterobacter cloacae strain RSW2n. Characterization of curdlan gum produced by this marine isolate was carried out using XRD and FT-IR analyses. Further, production of curdlan gum by Enterobacter cloacae RSW2n with different carbon sources (glucose, sucrose, fructose, maltose, glycerol, dates syrup, palm oil and starch) was studied.









Similar content being viewed by others
REFERENCES
Arli, S.D., Trivedi, U.B., and Patel, K.C., Curdlan-like exopolysaccharide production by Cellulomonas flavigena UNP3 during growth on hydrocarbon substrates, World J. Microbiol. Biotechnol., 2011, vol. 27, pp. 1415–1422. https://doi.org/10.1007/s11274-010-0593-2
Arunkumar, K. and Karthik, R., Screening and isolation of bacteria associated with seaweeds occurring along the coast of Thondi (Bay of Bengal, India) for chitinolytic activity, Int. J. Res. Sci. Innov. Appl. Sci., 2013, vol. 1, pp. 14–18.
Baeva, E., Bleha, R., Lavrova, E., Sushytskyi, L., Čopíková, J., Jablonsky, I., and Synytsya, A., Polysaccharides from basidiocarps of cultivating mushroom Pleurotusostreatus: isolation and structural characterization, Molecules, 2019, vol. 24, p. 2740. https://doi.org/10.3390/molecules24152740
Brenner, D.J., and Farmer, III, J., Enterobacteriaceae, in Bergey’s Manual of Systematics of Archaea and Bacteria, 2015, pp. 1–24.
Cai, W.D., Qiu, W.Y., Ding, Z.C., Wu, L.X., and Yan, J.K., Conformational and rheological properties of a quaternary ammonium salt of curdlan, Food Chem., 2019, vol. 280, pp. 130–138. https://doi.org/10.1016/j.foodchem.2018.12.059
Chen, M. and Liang, P., Synthesis and antibacterial activity of quaternized curdlan, Polym. Bull., 2017, vol. 74, pp. 4251–4266.
Evans, S.G., Morrison, D., Kaneko, Y., and Havlik, I., The effect of curdlan sulphate on development in vitro of Plasmodium falciparum, Trans. R. Soc. Trop. Med., 1998, vol. 92, pp. 87–89. https://doi.org/10.1016/s0035-9203(98)90969-5
Eweda, W.E., Sharaf, M.S., and El Sayed, A.M., Production of curdlan by some bacteria isolated from Egyptian soils, MiddleEast J. Appl. Sci., 2015, vol. 1, pp. 102–118.
Fang, J., Kawano, S., Tajima, K., and Kondo, T., In vivo curdlan/cellulose bionanocomposite synthesis by genetically modified Gluconacetobacter xylinus, Biomacromolecules, 2015, vol.16, pp. 3154–3160. https://doi.org/10.1021/acs.biomac.5b01075
FDA, Food additives permitted for direct addition to food for human consumption: curdlan 21 CFR Part 172, Federal Register, 1996, vol. 61, pp. 65941–65942.
Felsenstein, J., Confidence limits on phylogenies: an approach using the bootstrap, Evol., 1985, vol. 39, pp. 783–791. https://doi.org/10.2307/2408678
Godale, C.H. and Injal, A.S., Curdlan Production by Agrobacterium radiobacter and wide array of its applications in medicine, Indian J. Appl. Res., 2016, vol. 6, pp. 574–576.
Harada, T., Fujimori, K., Hirose, S., and Masada, M., Growth and β-glucan 10C3K production by a mutant of Alcaligenes faecalis var. myxogenes in defined medium, Agric. Biol. Chem., 1966, vol. 30, pp. 764–769. https://doi.org/10.1080/00021369.1966.10858682
Iyer, A., Mody, K., and Jha, B., Characterization of an exopolysaccharide produced by a marine Enterobacter cloaca. Indian. J. Exp. Biol., 2005, vol. 43, pp. 467–471.
Jung, D.Y., Cho, Y.S., Chung, C.H., Jung, D.I., Kim, K., and Lee, J.W., Improved production of curdlan with concentrated cells of Agrobacterium sp., Biotechnol. Bioprocess Eng., 2001, vol. 6, pp. 107–111.
Kim, B.S., Jung, I.D., Kim, J.S., Lee, J.H., Lee, I.Y., and Lee, K.B., Curdlan gels as protein drug delivery vehicles, Biotechnol. Lett., 2000, vol. 22, pp. 127–1130. https://doi.org/10.1023/A:1005636205036
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K., MEGA X: molecular evolutionary genetics analysis across computing platforms, Mol. Biol. Evol., 2018, vol.35, pp. 1547–1549. https://doi.org/10.1093/molbev/msy096
Lee, I.Y., Seo, W.T., Kim, G.J., Kim, M.K., Park, C.S., and Park, Y.H., Production of curdlan using sucrose or sugar cane molasses by two-step fed-batch cultivation of Agrobacterium species, J. Ind. Microbiol. Biotechnol., 1997, vol. 18, pp. 255–259. https://doi.org/10.1038/sj.jim.2900378
Mary Josephine, M., Usha, R., and Maria Victorial Rani, S., Current status of seaweed diversity and their seasonal availability at Hare Island, Gulf of Mannar, Sci. Res. Report., 2013, vol. 3, pp. 146–151.
Nei, M. and Kumar, S., Molecular Evolution and Phylogenetics, New York: Oxford Univ. Press, 2000. https://doi.org/10.1111/j.1558-5646.1985. tb00420.x 247
Nicolaus, B., Kambourova, M., and Oner, E.T., Exopolysaccharides from extremophiles: from fundamentals to biotechnology, Environ. Technol., 2010, vol. 31, pp. 1145–1158. https://doi.org/10.1080/09593330903552094
Phillips, K.R., Pik, J., Lawford, H.G., Lavers, B., Kligerman, A., and Lawford, G.R., Production of curdlan-type polysaccharide by Alcaligenes faecalis in batch and continuous culture, Can. J. Microbiol., 1983, vol. 29, pp. 1331–1338. https://doi.org/10.1139/m83-207
Prado, B.M., Kim, S., Özen, B.F and Mauer, L.J., Differentiation of carbohydrate gums and mixtures using Fourier transform infrared spectroscopy and chemometrics, J. Agric. Food Chem., 2005, vol. 53, pp. 2823–2829. https://doi.org/10.1021/jf0485537
Saitou, N. and Nei, M., The neighbor-joining method: a new method for reconstructing phylogenetic trees, Mol. Bio-l. Evol., 1987, vol. 4, pp. 406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Salah, R.B., Jaouadi, B., Bouaziz, A., Chaari, K., Blecker, C., Derrouane, C., and Besbes, S., Fermentation of date palm juice by curdlan gum production from Rhizobium radiobacter ATCC 6466™: purification, rheological and physico-chemical characterization, LWT—Food Sci. Technol, 2011, vol. 44, pp. 1026–1034. https://doi.org/10.1016/j.lwt.2010.11.023
Shivakumar, S. and Vijayendra, S.V.N., Production of exopolysaccharides by Agrobacterium sp. CFR-24 using coconut water–a byproduct of food industry, Lett. Appl. Microbiol., 2006, vol. 42, pp. 477–482. https://doi.org/10.1111/j.1472-765X.2006.01881.x
Stasinopoulos, S.J., Fisher, P.R., Stone, B.A., and Stanisich, V.A., Detection of two loci involved in (1–>3)-beta-glucan (curdlan) biosynthesis by Agrobacterium sp. ATCC31749, and comparative sequence analysis of the putative curdlan synthase gene, Glycobiology, 1999, vol. 9, pp. 31–41. https://doi.org/10.1093/glycob/9.1.31
Synytsya, A. and Novak, M., Structural analysis of glucans, Ann. Transl. Med., 2014, vol. 2, pp. 17–31. https://doi.org/10.3978/j.issn.2305-5839.2014.02.07
Tang, J., Zhen, H., Wang, N., Yan, Q., Jing, H., and Jiang, Z., Curdlan oligosaccharides having higher immunostimulatory activity than curdlan in mice treated with cyclophosphamide, Carbohydr. Polym., 2019, vol. 207, pp. 131–142. https://doi.org/10.1016/j.carbpol.2018.10.120
Zhang, Q., Sun, J., Wang, Z., Hang, H. Zhao, W., Zhuang, Y., and Chu, J., Kinetic analysis of curdlan production by Alcaligenes faecalis with maltose, sucrose, glucose and fructose as carbon sources, Bioresour. Technol., 2018, vol. 259, pp. 319–324. https://doi.org/10.1016/j.biortech.2018.03.059
Zhou, L., Fu, J., Bian, L., Chang, T., and Zhang, C., Preparation of a novel curdlan/bacterial cellulose/cinnamon essential oil blending film for food packaging application, Int. J. Biol. Macromol., 2022, vol. 212, pp. 211–219. https://doi.org/10.1016/j.ijbiomac.2022.05.137
ACKNOWLEDGMENTS
University Research Fellowship provided by management of Kalasalingam Academy of Research and Education to Mrs. Ezhilarasi is gratefully acknowledged. The authors acknowledge the support provided by International Research Centre, Kalasalingam Academy of Research and Management for characterization studies. The authors express their sincere gratitude to Dr. M. Palanisamy, Scientist-E of Botanical Survey of India (BSI), Southern Regional Centre, Coimbatore, for his support in the identification of marine seaweeds.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Rights and permissions
About this article
Cite this article
Ezhilarasi, P., Vanavil, B. Curdlan Gum Production Using Marine Bacteria Isolated from Red Seaweeds: Screening and Optimization Studies. Microbiology 92, 725–733 (2023). https://doi.org/10.1134/S0026261723600131
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261723600131