Abstract
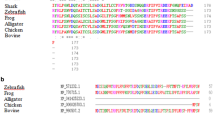
The study was focused on determining transcription levels of γM-crystallin genes in the eye lens of a common carp Cyprinus carpio. The transcription was detected by quantitative RT-PCR, and its relative level was quantified in 5 genes of γM-crystallins and crystallin-like proteins in C. carpio aged 4, 10 and 14 months. In all age groups, the specificity of GCM1, GCM1L, GCM2L, GCM2L3 gene expression in the lenses was found. GCM2 gene expression, apart from the lenses, was also detected in the muscles, liver and brain. Analysis of the role of the amino acid sequence of the identified γM-crystallins in the formation of the refractive properties of the lens was performed based on the assessment of calculated refractive index increments. It is assumed that high values of the refractive index of the lens in C. carpio are due not so much to a relative content of amino acids in γM-crystallins as to their sequences, which ensure the tertiary packing density of these proteins, as well as by γM-crystallin concentrations. To identify conserved domains and evolutionary relationships between γM-crystallins in fish of different taxa, a multiple alignment of amino acid sequences was performed, and a phylogenetic tree was constructed using the neighbor-joining clustering method. The high level of homology, the presence of 26 conserved regions, and the phylogenetic proximity of the compared lens crystallins in the clade Teleostomi may indicate an evolutionary consolidation of the spatial structure of these proteins and their important role in the adaptation of the fish eye’s optical apparatus to vision in water.





Similar content being viewed by others
REFERENCES
Land MF, Nilsson D-E (2012) Animal Eyes. Oxford University Press, Oxford. 288 p. ISBN-13: 9780199581139. https://oxford.universitypressscholarship.com/view/10.1093/acprof:oso/9780199581139.001.0001/acprof-9780199581139
Kröger RHH (2013) Optical plasticity in fish lenses. Prog Retin Eye Res 34: 78–88. https://doi.org/10.1016/j.preteyeres.2012.12.001
Pierscionek BK (2009) Gradient index of refraction (GRIN) profiling of the eye lens. In: Bass M (Ed) The Optical Society of America Handbook of Optics. Volume III: Vision and Vision Optics. McGraw-Hill, New York, 19.1–19.18. ISBN: 9780071498913
Pierscionek BK, Regini JW (2012) The gradient index lens of the eye: an opto-biological synchrony. Prog Retin Eye Res 31(4): 332–349. https://doi.org/10.1016/j.preteyeres.2012.03.001
Lin Y-R, Mok H-K, Wu Y-H, Liang S-S, Hsiao C-C, Huang C-H, Chiou S-H (2013) Comparative proteomics analysis of degenerative eye lenses of nocturnal rice eel and catfish as compared to diurnal zebrafish. Mol Vis 19: 623–637. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3611949/
Posner M, Hawke M, LaCava C, Prince CJ, Bellanco NR, Corbin RW (2008) A proteome map of the zebrafish (Danio rerio) lens reveals similarities between zebrafish and mammalian crystallin expression. Mol Vis 14: 806–814. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2358921/
Greiling TM, Houck SA, Clark JI (2009) The zebrafish lens proteome during development and aging. Mol Vis 15: 2313–2325. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2779061/
Kiss AJ, Cheng CH (2008) Molecular diversity and genomic organisation of the alpha, beta and gamma eye lens crystallins from the Antarctic toothfish Dissostichus mawsoni. Comp Biochem Physiol Part D Genomics and Proteomics 3(2): 155–171. https://doi.org/10.1016/j.cbd.2008.02.002
Chiou SH, Chang WC, Pan FM, Chang T, Lo TB (1987) Physicochemical characterization of lens crystallins from the carp and biochemical comparison with other vertebrate and invertebrate crystallins. J. Biochem. 101(3): 751–759. https://doi.org/10.1093/jb/101.3.751
Zhao H, Chen Y, Rezabkova L, Wu Z, Wistow G, Schuck P (2014) Solution properties of γ-crystallins: hydration of fish and mammal γ-crystallins. Protein Science 23(1): 88–99. https://doi.org/10.1002/pro.2394
Pan FM, Chang WC, Lin CH, Hsu AL, Chiou SH (1995) Characterization of gamma-crystallin from a catfish: structural characterization of one major isoform with high methionine by cDNA sequencing. Biochem Mol Biol Int 35(4): 725–732. https://pubmed.ncbi.nlm.nih.gov/7627123/
Mahler B, Chen Y, Ford J, Thiel C, Wistow G, Wu Z (2013) Structure and dynamics of the fish eye lens protein, γM7-crystallin. Biochemistry 52(20): 3579–3587. https://pubs.acs.org/doi/abs/10.1021/bi400151c
Zhao H, Brown PH, Magone MT, Schuck P (2011) The molecular refractive function of lens γ-crystallins. J Mol Biol 411(3): 680–699. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3146585/
OligoArchitectTM Online. Glossary of Parameters.https://www.sigmaaldrich.com/deepweb/assets/sigmaaldrich/marketing/global/documents/200/845/oligo-architect-glossary-br3011en-mk.pdf. (Date of address: 05.05.2022)
Kolder ICRM, van der Plas-Duivesteijn J, Tan G, Wiegertjes GF, Forlenza M, Guler AT, Travin DY, Nakao M, Moritomo T, Irnazarow I, den Dunnen JT, Anvar SY, Jansen H, Dirks RP, Palmblad M, Lenhard B, Henkel CV, Spaink HP (2016) A full-body transcriptome and proteome resource for the European common carp. BMC Genomics 17: 701. https://doi.org/10.1186/s12864-016-3038-y
Filby AL, Tyler CR (2007) Appropriate ‘housekeeping’ genes for use in expression profiling the effects of environmental estrogens in fish. BMC Mol Biol 8: 10. https://doi.org/10.1186/1471-2199-8-10
Liu W, Yuan X, Yuan S, Dai L, Dong S, Liu J, Peng L, Wang M, Tang Y, Xiao Y (2020) Optimal reference genes for gene expression analysis in polyploid of Cyprinus carpio and Carassius auratus. BMC Genet 21: 107. https://doi.org/10.1186/s12863-020-00915-6
Yuan JS, Reed A, Chen F, Stewart CN (2006) Statistical analysis of real-time PCR data. BMC Bioinformatics 7: 85. https://doi.org/10.1186/1471-2105-7-85
NCBI//URL: https://www.ncbi.nlm.nih.gov/protein (Date of address: 09.02.2022).
Okonechnikov K, Golosova O, Fursov M, UGENE team (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28(8): 1166–1167. https://doi.org/10.1093/bioinformatics/bts091
Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9): 1189–1191. https://doi.org/10.1093/bioinformatics/btp033
Zhao H, Brown PH, Schuck P (2011) On the distribution of protein refractive index increments. Biophys J 100(9): 2309–2317. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3149238/
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM (Ed) The proteomics protocols handbook. Humana Press Inc, Totowa, New Jersey 571–607. https://doi.org/10.1385/1-59259-890-0:571
Gerhard GS, Kauffman EJ, Wang X, Stewart R, Moore JL, Kasales CJ, Demidenko E, Cheng KC (2002) Life spans and senescent phenotypes in two strains of Zebrafish (Danio rerio). Experimental Gerontology 37(8–9): 1055–1068. https://doi.org/10.1016/S0531-5565(02)00088-8
Carey JB, Judge DS (2000) Longevity Records: Monographs on Population Aging. Vol 8: Life Spans of Mammals, Birds, Amphibians, Reptiles, and Fish. University Press of Southern Denmark. 241 p. ISBN-13: 978-8778385390
Brooks CM, Andrews AH, Ashford JR, Ramanna N, Jones CD, Lundstrom CC, Cailliet GM (2011) Age estimation and lead–radium dating of Antarctic toothfish (Dissostichus mawsoni) in the Ross Sea. Polar Biol 34(3): 329–338. https://doi.org/10.1007/s00300-010-0883-z
Wages P, Horwitz J, Ding L, Corbin RW, Posner M (2013) Changes in zebrafish (Danio rerio) lens crystallin content during development. Mol Vis 19: 408–417. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3580975/
Wang K, Vorontsova I, Hoshino M, Uesugi K, Yagi N, Hall JE, Schilling TF, Pierscionek BK (2020) Optical development in the zebrafish eye lens. FASEB J 34(4): 5552–5562. https://doi.org/10.1096/fj.201902607R
Keenan J, Manning G, Elia G, Dunn MJ, Orr DF, Pierscionek BK (2012) Crystallin distribution patterns in Litoria infrafrenata and Phyllomedusa sauvagei lenses. Proteomics 12(11): 1830–1843. https://doi.org/10.1002/pmic.201100393
Axelrod D, Lerner D, Sands PJ (1988) Refractive index within the lens of a goldfish eye determined from the paths of thin laser beams. Vision Res 28(1): 57–65. https://doi.org/10.1016/S0042-6989(88)80006-3
Campbell MC (1984) Measurement of refractive index in an intact crystalline lens. Vision Res 24(5): 409–415. https://doi.org/10.1016/0042-6989(84)90039-7
Van Leen RW, Breuer ML, Lubsen NH, Schoenmakers JG (1987) Developmental expression of crystallin genes: in situ hybridization reveals a differential localization of specific mRNAs. Dev Biol 123(2): 338–345. https://doi.org/10.1016/0012-1606(87)90392-7
Ueda Y, Duncan MK, David L (2002) Lens proteomics: the accumulation of crystallin modifications in the mouse lens with age. Invest Ophthalmol Vis Sci 43(1): 205–215. https://iovs.arvojournals.org/article.aspx?articleid=2123357
Pierscionek B, Bahrami M, Hoshino M, Uesugi K, Regini J, Yagi N (2015) The eye lens: age-related trends and individual variations in refractive index and shape parameters. Oncotarget 6(31): 30532–30544. https://www.oncotarget.com/article/5762/text/
Vendra VP, Khan I, Chandani S, Muniyandi A, Balasubramanian D (2016) Gamma crystallins of the human eye lens. Biochim Biophys Acta 1860(1 Pt B): 333–343. https://doi.org/10.1016/j.bbagen.2015.06.007
Slingsby C, Wistow GJ, Clark AR (2013) Evolution of crystallins for a role in the vertebrate eye lens. Protein Sci 22(4): 367–380. https://doi.org/10.1002/pro.2229
Houston P, Macro N, Kang M, Chen L, Yang J, Wang L, Wu Z, Zhong D (2020) Ultrafast Dynamics of Water-Protein Coupled Motions around the Surface of Eye Crystallin. J Am Chem Soc 142(8): 3997–4007. https://doi.org/10.1021/jacs.9b13506
Kiss AJ, Mirarefi AY, Ramakrishnan S, Zukoski CF, Devries AL, Cheng CH (2004) Cold-stable eye lens crystallins of the Antarctic nototheniid toothfish Dissostichus mawsoni Norman. J Exp Biol 207(Pt 26): 4633–4649. https://doi.org/10.1242/jeb.01312
ACKNOWLEDGMENT
The authors are grateful to P.P. Zhdanov, the director of the Educational and Experimental Fishery of the Kaliningrad State Technical University, for the granted opportunity to collect biological material.
Funding
This work was funded by the Immanuel Kant Baltic Federal University.
Author information
Authors and Affiliations
Contributions
Conceptualization and experimental design (V.V.Zh., I.N.D.), data collection (A.I.K., I.N.D.), data processing (I.N.D., A.I.K.), manuscript writing and editing (A.I.K., I.N.D., V.V.Zh.).
Corresponding author
Ethics declarations
CONFLICT OF INTEREST
The authors declare that they have neither apparent nor potential conflict of interest related to the publication of this article.
Additional information
Translated by A. Polyanovsky
Russian Text © The Author(s), 2022, published in Zhurnal Evolyutsionnoi Biokhimii i Fiziologii, 2022, Vol. 58, No. 4, pp. 333–346https://doi.org/10.31857/S0044452922040052.
Rights and permissions
About this article
Cite this article
Kapitunova, A.I., Dominova, I.N. & Zhukov, V.V. γM Crystallin Genes in the Eye Lens of a Juvenile Common Carp Cyprinus carpio: Transcription Levels and Phylogenetic Aspect. J Evol Biochem Phys 58, 1025–1040 (2022). https://doi.org/10.1134/S0022093022040081
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0022093022040081