Abstract
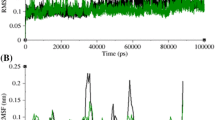
α-crystallin is a major eye lens protein, comprising up to 40% of total lens protein. It is composed of two subunits, αA and αB and even shares a common central domain of about 90 residues with variable N- and C-terminal extensions. For the establishment of an evolutionary inter-relationship, an elucidation of the structure and alignment of protein sequences is crucial. In the present study, a bioinformatics approach was adopted to explore the possible structure, sequence and phylogenetic relationship of α-crystallin (both subunits αA and αB) from ten habitat-specific fish species, (freshwater and saltwater) and compared with a standard sequence of Bos taurus species. The sequence of Bos taurus was predicted to be a close homologue of the fish species. Wet lab experiments such as NMR are not only expensive and time consuming but are suitable for small proteins having less than 150 amino acids, so a preliminary computer-aided approach has been selected for structural analysis of α-crystallin of fish species. Our analysis shows that the secondary structures of bovine α-crystallin revealed no considerable differences as compared to that of the crystallins of the habitat-specific fish and that the presence of β- sheets was predominant in all structures. Though no significant differences in the αA subunits were revealed yet some structural variations were observed for αB subunits which had been confirmed by MSA analysis. The 3D structure of the protein hasn’t been elucidated yet so a computational analysis estimated no major differences in structures of crystallin for either bovine or the fish species except that saltwater fish proteins possess more favourable states and higher reliabilities. At the same time, the RMSF values of α-crystallin computed by CABS Flex 2.0 showed lesser values in the case of freshwater fish species which state the possible favourable structures of freshwater fish species. The stabilities of αB- subunits were revealed from the physiochemical parameters computed as compared to αA subunits of the respective proteins for all species. The Kyle-Doolittle Plots revealed the predominance of hydrophilic amino acids in both subunits of α-crystallin for all species and it is a cytosolic protein that has been determined using the TMHMM server. Considerate differences were revealed in the case of the αB subunit but not for the αA subunit, for all species as deciphered from Clustal omega which may indicate that the differences in α-crystallin occur primarily due to the αB subunit. Homology modeling revealed that bovine α-crystallin showed a greater sequence homology with most fish species, especially zebrafish. From our study, no major differences in α- crystallin structure could be deciphered between fish species in terms of habitat. The structure of human crystallin is a complex model so fish species were chosen as models which would be beneficial for humans in terms of drug designing as well.










Similar content being viewed by others
Data availability
All information collated in this study was gotten from the Google search engine. Data were sourced via relevant references from different journals.
References
Acharya UR, Behera SK, Swain SN, Panda MK, Mistri AR, Sahoo BB (2014) Sequence and structural analysis of β-actin protein of fishes, using bioinformatics tools and techniques. Int J Biosci (IJB) 4(11):249–256. https://doi.org/10.12692/ijb/4.11.249-256
Augusteyn RC (2004) α-crystallin: a review of its structure and function. Clin Exp Optom 87(6):356–366. https://doi.org/10.1111/j.1444-0938.2004.tb03095.x
Baranova EV, Weeks SD, Beelen S, Bukach OV, Gusev NB, Strelkov SV (2011) Three-dimensional structure of α-crystallin domain dimers of human small heat shock proteins HSPB1 and HSPB6. J Mol Biol 411(1):110–122. https://doi.org/10.1016/j.jmb.2011.05.024
Bari KJ, Sharma S (2020) A perspective on biophysical studies of crystallin aggregation and implications for cataract formation. J Phys Chem B 124(49):11041–11054. https://doi.org/10.1021/acs.jpcb.0c07449
Basha E, O’Neill H, Vierling E (2012) Small heat shock proteins and α-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci 37(3):106–117. https://doi.org/10.1002/pro.2229
Biswasa A, Karmakarb S, Banerjee V, Sahac S, Kundud M, Bhattacharyyae J, Das KP (2011) Biophysical studies on the molecular chaperone function, structure and interaction of eye lens protein-crystallin—a review. J Indian Chem Soc 88:1827–1855
Bloemendal H, de Jong WW (1991) Lens proteins and their genes. Prog Nucleic Acid Res Mol Biol 41:259–281. https://doi.org/10.1016/S0079-6603(08)60012-4
Buchan DWA, Jones DT (2019) The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res 47(W1):W402–W407. https://doi.org/10.1093/nar/gkz297
Carver JA, Aquilina JA, Truscott RJ (1993) An investigation into the stability of alpha-crystallin by NMR spectroscopy; evidence for a two-domain structure. Biochem Biophys Acta 1164(1):22–28. https://doi.org/10.1016/0167-4838(93)90107-3
Chang YY, Hsieh MH, Huang YC, Chen CJ, Lee MT (2022) Conformational changes of α-crystallin proteins induced by heat stress. Int J Mol Sci 23(16):9347. https://doi.org/10.3390/ijms23169347
Cheng CHC (2004) Cold-stable eye lens crystallins of the Antarctic nototheniid toothfish Dissostichus mawsoni Norman. J Exp Biol 207(26):4633–4649. https://doi.org/10.1242/jeb.01312
D’Agostino M, Lemma V, Chesi G, Stornaiuolo M, Cannata Serio M, D’Ambrosio C, Bonatti S (2013) The cytosolic chaperone α-crystallin B rescues folding and compartmentalization of misfolded multispan transmembrane proteins. J Cell Sci 126(18):4160–4172. https://doi.org/10.1242/jcs.125443
Dahlman JM, Margot KL, Ding L, Horwitz J, Posner M (2005) Zebrafish alpha-crystallins: protein structure and chaperone-like activity compared to their mammalian orthologs. Mol Vis 11:88–96
Das KP, Surewicz WK (1995) Temperature-induced exposure of hydrophobic surfaces and its effect on the chaperone activity of α-crystallin. FEBS Lett 369(2–3):321–325. https://doi.org/10.1016/0014-5793(95)00775-5
Das KP, Petrash JM, Surewicz WK (1999) Insight into the secondary structure of non-native proteins bound to a molecular chaperone α-crystallin: an isotope-edited infrared spectroscopic study. J Biol Chem 274(47):33209–33212. https://doi.org/10.1074/jbc.274.47.33209
de Jong WW, Leunissen JA, Leenen PJ, Zweers A, Versteeg M (1988) Dogfish alpha-crystallin sequences. Comparison with small heat shock proteins and Schistosoma egg antigen. J Biol Chem 263(11):5141–5149. https://doi.org/10.1016/S0021-9258(18)60691-X
Delaye M, Clark IL, Benedek GB (1982) Identification of the scattering elements responsible for lens opacification in cold cataracts. Biophys J 37(3):647–656. https://doi.org/10.1016/S0006-3495(21)00384-2
Derham BK, Harding JJ (1999) Alpha-crystallin as a molecular chaperone. Prog Retin Eye Res 18(4):463–509. https://doi.org/10.1016/s1350-9462(98)00030-5
Du J, Zhang C, Long Q, Chen W, Guo Z, Liu Q (2022) Bioinformatics analysis of the structure and function of EdeB from Brevibacillus brevis X23. In 2022 10th International Conference on Bioinformatics and Computational Biology (ICBCB) (pp. 1–5). IEEE. doi: https://doi.org/10.1109/ICBCB55259.2022.9802134
Eastman JT, McCune AR (2000) Fishes on the Antarctic continental shelf: evolution of a marine species flock? J Fish Biol 57:84–102. https://doi.org/10.1111/j.1095-8649.2000.tb02246.x
Farnsworth PN, Groth-Vasselli B, Greenfield NJ, Singh K (1997) Effects of temperature and concentration on bovine lens α-crystallin secondary structure: a circular dichroism spectroscopic study. Int J Biol Macromol 20(4):283–291. https://doi.org/10.1016/S0141-8130(97)00028-7
Gawad AEDA, Ibrahim M (2013) Computational studies of the interaction of chitosan nanoparticles and αB-crystallin. BioNanoSci 3:302–311. https://doi.org/10.1007/s12668-013-0096-3
Ghahghaei A, Rekas A, Carver JA, Augusteyn RC (2009) Structure/function studies of dogfish α-crystallin, comparison with bovine α-crystallin. Mol vis 15:2411
Ghosh RK, Kar T, Dutta B et al (2018) Aberration in the structural paradigm of lens protein α crystallin by UV-C irradiation. Appl Biol Chem 61:281–287. https://doi.org/10.1007/s13765-018-0351-y
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis 18(15):2714–2723. https://doi.org/10.1002/elps.1150181505
Guex N, Peitsch MC, Schwede T (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30(S1):S162–S173. https://doi.org/10.1002/elps.200900140
Horwitz J (2003) Alpha-crystallin. Exp Eye Res 76(2):145–153. https://doi.org/10.1016/S0014-4835(02)00278-6
Horwitz J (2009) Alpha crystallin: the quest for a homogeneous quaternary structure. Exp Eye Res 88(2):190–194. https://doi.org/10.1016/j.exer.2008.07.007
Horwitz J, Huang QL, Ding L, Bova MP (1998) Lens α-crystallin: chaperone-like properties. Methods in enzymology, vol 290. Academic Press, pp 365–383. https://doi.org/10.1016/S0076-6879(98)90032-5
Islam S, Do MT, Frank BS, Hom GL, Wheeler S, Fujioka H, Monnier VM (2022) α-crystallin chaperone mimetic drugs inhibit lens γ-crystallin aggregation: potential role for cataract prevention. J Biol Chem. https://doi.org/10.1016/j.jbc.2022.102417
Jamroz M, Kolinski A, Kmiecik S (2014) CABS-flex predictions of protein flexibility compared with NMR ensembles. Bioinformatics (Oxford, England) 30(15):2150–2154. https://doi.org/10.1093/bioinformatics/btu184
Kaiser CJ, Peters C, Schmid PW, Stavropoulou M, Zou J, Dahiya V, Weinkauf S (2019) The structure and oxidation of the eye lens chaperone αA-crystallin. Nat Struct Mol Biol 26(12):1141–1150. https://doi.org/10.1038/s41594-019-0332-9
Khalid S, Idrees S, Khalid H et al (2015) Ab-initio prediction of sequence and structural biology of fish muscle proteins using homology modeling, phylogeny and different computational approaches. MOJ Proteomics Bioinform 2(3):81–91. https://doi.org/10.15406/mojpb.2015.02.00047
Kiss AJ, Cheng CHC (2008) Molecular diversity and genomic organisation of the α, β and γ eye lens crystallins from the Antarctic toothfish Dissostichus mawsoni. Comp Biochem Physiol D 3(2):155–171. https://doi.org/10.1016/j.cbd.2008.02.002
Kiss AJ, Mirarefi AY, Ramakrishnan S, Zukoski CF, DeVries AL, Cheng CHC (2004) Cold-stable eye lens crystallins of the Antarctic nototheniid toothfish Dissostichus mawsoni Norman. J Exp Biol 207(26):4633–4649. https://doi.org/10.1242/jeb.01312
Kopp J, Schwede T (2004) The SWISS-MODEL repository of annotated three-dimensional protein structure homology models. Nucleic Acids Res 32(suppl_1):D230–D234. https://doi.org/10.1093/nar/gkh008
Kumar MS, Kapoor M, Sinha S, Reddy GB (2005) Insights into hydrophobicity and the chaperone-like function of A-and B-crystallins. J Biol Chem 280(23):21726–21730. https://doi.org/10.1093/nar/gky427
Kuriata A, Gierut AM, Oleniecki T, Ciemny MP, Kolinski A, Kurcinski M, Kmiecik S (2018) CABS-flex 2.0: a web server for fast simulations of flexibility of protein structures. Nucleic Acids Res 46(W1):W338–W343. https://doi.org/10.1093/nar/gky356
Lee JS, Satoh T, Shinoda H, Samejima T, Wu SH, Chiou SH (1997) Effect of heat-induced structural perturbation of secondary and tertiary structures on the chaperone activity of α-crystallin. Biochem Biophys Res Commun 237(2):277–282. https://doi.org/10.1006/bbrc.1997.7131
Lusiana E, Irfannuddin I (2022) The biomolecular characteristics of angiotensin II type 1 receptors as parameters in kidney fibrosis: a bioinformatics analysis. Majalah Kedokteran Sriwijaya 54(2):72–84
Mafia K, Gupta R, Kirk M, Wilson L, Srivastava OP, Barnes S (2008) UV-A-induced structural and functional changes in human lens deamidated alphaB-crystallin. Mol vis 14:234–248
Malik A, Khan JM, Alhomida AS, Ola MS (2022) Modulation of the structure and stability of novel camel lens alpha-crystallin by pH and thermal stress. Gels 8(5):273. https://doi.org/10.3390/gels8050273
Moreau KL, King JA (2012) Protein misfolding and aggregation in cataract disease and prospects for prevention. Trends Mol Med 18(5):273–282. https://doi.org/10.1016/j.molmed.2012.03.005
Narberhaus F (2002) α-crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiol Mol Biol Rev 66(1):64–93. https://doi.org/10.1128/MMBR.66.1.64-93.2002
Nielsen H. (2017). Predicting secretory proteins with signalP. In: Clifton NJ (ed) Methods in molecular biology, vol 1611, pp 59–73. https://doi.org/10.1007/978-1-4939-7015-5_6
Pembroke JT (2000) Bio-molecular modelling utilising RasMol and PDB resources: a tutorial with HEW lysozyme. Biochem Mol Biol Educ 28(6):297–300. https://doi.org/10.1111/j.1539-3429.2000.tb00177.x
Persson B (2000) Bioinformatics in protein analysis. EXS 88:215–231. https://doi.org/10.1007/978-3-0348-8458-7_14
Pierscionek BK, Augusteyn RC (1995) The refractive index and protein distribution in the blue eye trevally lens. J Am Optom Assoc 66(12):739–743
Pierscionek BK, Chan DY (1989) Refractive index gradient of human lenses. Optom Vis Sci 66(12):822–829
Posner M, Dahlman J, Margot K, Horwitz J (2003) Comparison of Zebrafish and human-crystallin chaperone activity. Invest Ophthalmol vis Sci 44(13):2368–2368
Posner M, Kiss AJ, Skiba J, Drossman A, Dolinska MB, Hejtmancik JF, Sergeev YV (2012) Functional validation of hydrophobic adaptation to physiological temperature in the small heat shock protein αA-crystallin. PloS one 7(3):e34438. https://doi.org/10.1371/journal.pone.0034438
Posner M, Murray KL, Andrew B, Brdicka S, Roberts A, Franklin K, David LL (2023) Impact of α-crystallin protein loss on zebrafish lens development. Exp Eye Res 227:109358. https://doi.org/10.1016/j.exer.2022.109358
Reddy GB, Kumar PA, Kumar MS (2006) Chaperone-like activity and hydrophobicity of alpha-crystallin. IUBMB Life 58(11):632–641. https://doi.org/10.1080/15216540601010096
Shamsi A, Mohammad T, Anwar S, Nasreen K, Hassan MI, Ahmad F, Islam A (2022) Insight into the binding of PEG-400 with eye protein alpha-crystallin: multi spectroscopic and computational approach: possible therapeutics targeting eye diseases. J Biomol Struct Dyn 40(10):4496–4506. https://doi.org/10.1080/07391102.2020.1858964
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol syst biol 7(1):539. https://doi.org/10.1038/msb.2011.75
Silva FF, Gonçalves D, Lopes D (2020) The use of bioinformatics tools to characterize a hypothetical protein from Penicillium rubens. Genet Mol Res 19(2):1–18. https://doi.org/10.4238/gmr18574
Slingsby C, Wistow GJ, Clark AR (2013) Evolution of crystallins for a role in the vertebrate eye lens. Protein Sci 22(4):367–380. https://doi.org/10.1016/j.tibs.2011.11.005
Sprague-Piercy MA, Rocha MA, Kwok AO, Martin RW (2021) α-Crystallins in the vertebrate eye lens: complex oligomers and molecular chaperones. Annu Rev Phys Chem 72:143–163. https://doi.org/10.1146/annurev-physchem-090419-121428
Surewicz WK, Olesen PR (1995) On the thermal stability of alpha-crystallin: a new insight from infrared spectroscopy. Biochemistry 34(30):9655–9660. https://doi.org/10.1021/bi00030a001
Timsina R, Mainali L (2021) Association of alpha-crystallin with fiber cell plasma membrane of the eye lens accompanied by light scattering and cataract formation. Membranes 11(6):447. https://doi.org/10.3390/membranes11060447
Timsina R, Trossi-Torres G, Thieme J, O’Dell M, Khadka N, Mainali L (2022) α-Crystallin interacts with the model of human eye lens-lipid membrane via hydrophobic interaction. Biophys J 121(3):79a
Upadhyay AK, Mueller NH, Petrash JM, Kompella UB (2022) Nano-assemblies enhance chaperone activity, stability, and delivery of alpha B-crystallin-D3 (αB-D3). J Control Release 352:411–421. https://doi.org/10.1016/j.jconrel.2022.10.026
Valanciute A, Nygaard L, Zschach H, Jepsen MM, Lindorff-Larsen K, Stein A (2023) Accurate protein stability predictions from homology models. Comput Struct Biotechnol J 21:66–73. https://doi.org/10.1016/j.csbj.2022.11.048
van Boekel MA, de Lange F, de Grip WJ, de Jong WW (1999) Eye lens αA-and αB-crystallin: complex stability versus chaperone-like activity. Biochim Biophys Acta (BBA) Protein Struct Mol Enzymol 1434(1):114–123. https://doi.org/10.1016/S0167-4838(99)00178-8
Wang X, Garcia CM, Shui YB, Beebe DC (2004) Expression and regulation of α-, β-, and γ-crystallins in mammalian lens epithelial cells. Invest Ophthalmol Vis Sci 45(10):3608–3619. https://doi.org/10.1167/iovs.04-0423
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1):W296–W303
Woods CN, Ulmer LD, Guttman M, Bush MF, Klevit RE (2023) Disordered region encodes α-crystallin chaperone activity toward lens client γD-crystallin. Proc Natl Acad Sci 120(6):e2213765120. https://doi.org/10.1073/pnas.2213765120
Funding
This research did not receive any particular funding from any funding agencies in the public, commercial, or not- for-profit sectors.
Author information
Authors and Affiliations
Contributions
The study was designed by AC, SG, SS, PD. AC performed the dry lab analyses using various online bioinformatic tools, preparation of figures and preparing the manuscript. SG, SS, PD edited and revised the entire manuscript. All the authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chakraborty, A., Ganguli, S., De, P. et al. An insight into the structural analysis of α-crystallin of habitat-specific fish: a computational approach. J Proteins Proteom 14, 111–127 (2023). https://doi.org/10.1007/s42485-023-00107-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42485-023-00107-7