Abstract
Acetobacter pasteurianus (Ap) CICC 20001 and CGMCC 1.41 are two acetic acid bacteria strains that, because of their strong abilities to produce and tolerate high concentrations of acetic acid, have been widely used to brew vinegar in China. To globally understand the fermentation characteristics, acid-tolerant mechanisms and genetic stabilities, their genomes were sequenced. Genomic comparisons with 9 other sequenced Ap strains revealed that their chromosomes were evolutionarily conserved, whereas the plasmids were unique compared with other Ap strains. Analysis of the acid-tolerant metabolic pathway at the genomic level indicated that the metabolism of some amino acids and the known mechanisms of acetic acid tolerance, might collaboratively contribute to acetic acid resistance in Ap strains. The balance of instability factors and stability factors in the genomes of Ap CICC 20001 and CGMCC 1.41 strains might be the basis for their genetic stability, consistent with their stable industrial performances. These observations provide important insights into the acid resistance mechanism and the genetic stability of Ap strains and lay a foundation for future genetic manipulation and engineering of these two strains.
Similar content being viewed by others
Introduction
Acetic acid bacteria (AAB) are a group of gram-negative or gram-variable obligate aerobic bacteria within the class alpha-proteobacteria1 and have been isolated from a variety of natural sites, including fermented food2,3,4,5, plant organs6,7, animal organs8 and soil9,10. Since Acetobacter (A.) was recognized in 189811, 17 genera consisting of approximately 88 species in total, have been recorded in the AAB group12,13,14,15. Among them, Acetobacter and Komagataeibacter (K., formerly classified as Gluconacetobacter16) are the main AAB genera that have been widely used to brew vinegar17,18. For example, K. europaeus strains contribute to fruit and alcohol vinegars by liquid-state fermentation in Europe19,20,21, while A. pasteurianus (Ap) strains are mainly used to brew cereal vinegars by liquid-state or solid-state fermentation in Asia, especially in China and Japan17,22,23.
In the vinegar industry, instabilities in the ability of AAB strains to produce and tolerate acetic acid may be the most critical factors leading to instability in acetic acid production. Instabilities of fermentative and genetic characteristics of K. europaeus have been well-recognized24,25. However, only a few papers related to the instabilities of the fermentative and genetic characteristics in Ap have been published. Azuma et al. found that there were a large amount of transposons and some hyper-mutable tandem repeats in the genomes of Ap IFO 3283 substrains, indicating their genetic instability26, while Gullo et al. proved that Ap AB0220 maintained the quondam features, such as ethanol oxidation, acetate assimilation, phenotypic traits and 16S rRNA gene sequences, after 9 years of preservation27. In China, Ap CICC 20001 (once named Huniang 1.01) and CGMCC 1.41 (once named AS1.41), which were isolated from a vinegar factory in Dandong by the Shanghai Institute of Brewing and bred by the Institute of Microbiology Chinese Academy of Sciences, respectively, may be the earliest screened AAB isolates and are still widely used to brew vinegar by solid-state and liquid-state fermentation, displaying high stabilities in acetic acid production28,29,30,31.
In addition to genetic instability, the mechanisms related to acetic acid resistance in AAB have been a hot research topic. Although some mechanisms conferring acetic acid resistance in AAB, such as acetate assimilation, transportation systems, cell membrane composition and stress proteins expression, have been reviewed15,32, the mechanisms of acetic acid resistance in AAB remain unclear. Escherichia (E.) coli is one of the microorganisms whose acid resistance mechanisms have been extensively studied and thoroughly understood. In E. coli, the metabolic responses, the chloride transporters, the oxidative system, the cyclopropane fatty acid, the arginine-dependent system, the glutamate-dependent system and the lysine-dependent system are elegantly regulated systems that permit E. coli to survive when a nurturing environment at pH 7 declines sharply to a harsh pH 2 milieu33,34,35,36,37. Furthermore, other mechanisms, such as arginine deiminase pathway and urease system also have been proved to confer acid resistance in bacteria38,39,40,41. Acid resistance mechanisms in E. coli and other bacteria may provide a reference for investigating acetic acid resistance mechanisms in AAB using comparative genomics.
The combination of whole genome sequencing and subsequent genomic analysis42,43,44,45, is an effective method to investigate gene functions, genetic information and biological characteristics, which may contribute to the dissection of product and genetic stability in Ap. Genomic analysis can be used to construct the metabolic blueprint for Ap, by comparisons with the known mechanisms conferring acid tolerance and the known metabolic pathways in microorganisms. However, as far as we know, no study has been conducted to investigate acid tolerance in microorganisms by analyzing their overall metabolic pathway.
In this study, the fermentation characteristics of Ap CICC 20001 and CGMCC 1.41 were investigated. Then, to globally understand their fermentation characteristics, as well as the mechanisms conferring acetic acid resistance, the complete genomes of Ap CICC 20001 and CGMCC 1.41 were sequenced and analyzed. Comparisons of Ap CICC 20001 and CGMCC 1.41 with other sequenced Ap strains revealed differences among Ap strains. Furthermore, an integral understanding of the molecular mechanisms underlying their acetic acid tolerance has been undertaken by arranging a metabolic blueprint related to acetic acid resistance in Ap strains, which may lead to detailed information for improving their abilities to produce and tolerate acetic acid during vinegar fermentation.
Results
Comparison of fermentation characteristics of Ap CICC 20001 and CGMCC 1.41
Both Ap CICC 20001 and CGMCC 1.41 have been widely used to brew vinegar in China. However, they display different fermentation characteristics in vinegar industry, such as acetic acid resistance and production. To investigate the differences, Ap CICC 20001 and CGMCC1.41 were inoculated into a modified GYP medium containing different concentrations of ethanol and acetic acid.
The effects of initial acetic acid concentration on acetic acid production in Ap CICC 20001 and CGMCC 1.41
To investigate the effects of the initial amount of acetic acid on acetic acid production, Ap CICC 20001 and CGMCC 1.41 were inoculated into modified GYP medium containing 6% ethanol and different concentrations of acetic acid (Fig. 1). The results showed that a low initial concentration of acetic acid (0.5% ~ 1%) could promote acetic acid fermentation in Ap CICC 20001 and CGMCC 1.41. Ap CGMCC 1.41 and CICC 20001 produced the highest concentration of acetic acid at 6% ethanol, with initial concentrations of 0.5% and 1% acetic acid, respectively. However, the high initial acid concentration would also inhibit the fermentation process. The lengths of the lag phase in Ap CICC 20001 and CGMCC 1.41 increased considerably when the initial concentration of acetic acid was greater than 1.5% and 3%, respectively (Fig. 1).
Ap CGMCC 1.41 produced a higher concentration of acetic acid than Ap CICC 20001
Ap CICC 20001 and CGMCC 1.41 were inoculated into modified GYP medium containing a range of concentrations of ethanol and optimized concentrations of acetic acid (1% in Ap CICC 20001 and 0.5% in Ap CGMCC 1.41) (Fig. 2). Ap CICC 20001 yields the maximum acetic acid (6.35%) in the GYP medium with 6% ethanol and 1% initial acetic acid, while Ap CGMCC 1.41 produces the maximum acetic acid (7.15%) in GYP medium with 10% ethanol and 0.5% initial acetic acid. There is a positive relationship between the initial ethanol concentration and the acetic acid produced within a certain range during fermentation by Ap CICC 20001 and CGMCC 1.41. Furthermore, high levels of ethanol also inhibit acetic acid production. The lengths of lag phase in Ap CICC 20001 and CGMCC 1.41 increase considerably, when the initial concentrations of ethanol are greater than 6% and 10%, respectively. Ap CGMCC 1.41 also exhibits an obvious peroxidation in the medium with 2% ethanol and 0.5% acetic acid. However, Ap CICC 20001 has a weak peroxidation ability.
Features of the Ap CICC 20001 and CGMCC 1.41 genomes
To gain insights into the fermentation characteristics of these two Ap strains from genomic analysis, Ap CICC 20001 and CGMCC 1.41 cells were subjected to single molecule real time (SMRT) sequencing using a PacBio RS instrument. Previously, genomic sequencing of 9 Ap strains, including Ap 386B, 7 IFO 3283 substrains and 1 IFO 3283 mutant IFO 3283-01-42C, had been completed and published. Among the Ap strains, 386B was isolated from a spontaneous cocoa bean heap fermentation in Ghana4, while IFO 3283 was isolated from a pellicle formed on the surface during fermentation of traditional Japanese rice vinegar in 195426. Seven substrains of IFO 3283 were isolated from IFO 3283 after it had been stored for more than 30 years and the mutant substrain IFO 3283-01-42C was trained and isolated. In this study, we present genomic sequencing of Ap CICC 20001 and CGMCC 1.41 and the genomic comparison of these strains with 9 other Ap strains. The genome sequences of 11 Ap strains have been published.
Plasmids in Ap CICC 20001 may play a greater role in metabolism than they do in Ap CGMCC 1.41
The chromosome of Ap CICC 20001 contains 2865612 bp, with a 52.94% G + C content, 3006 predicted coding sequences (CDSs), 118 tRNAs, 15 mobile elements and 2 prophages, while that of Ap CGMCC 1.41 contains 2928931 bp, with a 52.95% G + C content, 3012 CDSs, 111 rRNAs, 56 mobile elements and 2 prophages (Table 1). Their chromosomes are analogous, but their plasmids are completely different from each other, not only in encoded genes but also in the number of plasmids. Specific functions are assigned to 67.6% (2450 genes) of the total of 3623 protein-coding genes in Ap CICC 20001 and 69.2% (2248 genes) of the total of 3250 protein-coding genes in Ap CGMCC 1.41 and the remaining genes are hypothetical genes.
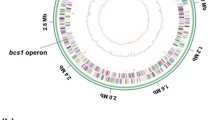
The GC skew [(G − C)/(G + C)] and the cumulative GC skew have proven to be useful as indicators of the DNA leading strand, lagging strand, replication origin and replication terminal46. The putative replication origin (ori) and terminal (ter) of the chromosomes were predicted, based on the GC skew and the cumulative GC skew. The general chromosome features of Ap CICC 20001 and CGMCC 1.41, including ori, ter and G + C content, as well as colony characteristics, are shown in Fig. 3. The ori and ter of Ap CICC 20001 may be located at 1231951 bp and 2862135 bp, respectively, while those of Ap CGMCC 1.41 may be located at 1364449 bp and 2895793 bp, respectively. Colonies of Ap CICC 20001 and CGMCC 1.41 that are cultivated on GYC medium for 5 d have their own features. A colony of Ap CICC 20001 (approximately 0.8 mm) is smaller than one of Ap CGMCC 1.41 (approximately 1.2 mm). Furthermore, both Ap CICC 20001 and CGMCC 1.41 display highly similar orthologous groups (Supplementary Fig. S1).
Chromosomal features of Ap CICC 20001 and CGMCC 1.41.
From outer to inner circles, the first, second, third and fourth circle represent the positive strand ORF, the negative strand ORF, the GC content and the GC skew, respectively. The middle figure shows a colony grown on a GYC medium plate for 5 d.
Chromosomes of Ap CICC 20001 and CGMCC 1.41 display a high homology with other Ap strains
To explore the relationships among the sequenced Ap strains, including Ap CICC 20001, CGMCC 1.41 and the previously sequenced Ap species, their chromosomes were compared using Mauve software. Ap CICC 20001 and CGMCC 1.41 share a high degree of homology with the 9 published Ap strains. However, the chromosome sequence of Ap CGMCC 1.41 is slightly longer than that of Ap CICC 20001 and the IFO 3283 substrains (Fig. 4). Although there are high degrees of similarity among the 11 Ap strains, there are also deletions, amplifications, insertions, inversions and translocations. Compared to Ap IFO 3283 substrains and 386B, some DNA fragments are missing.
The locations of orthologs between chromosomes of 11 Ap strains.
The lines represent (from top to bottom): line 1, chromosome of Ap CGMCC 1.41; line 2, chromosome of Ap CICC 20001; line 3, chromosome of Ap 386B; line 4, chromosome of Ap IFO 3283-01; line 5, chromosome of Ap IFO 3283-01-42C; line 6, chromosome of Ap IFO 3283-03; line 7, chromosome of Ap IFO 3283-07; line 8, chromosome of Ap IFO 3283-12; line 9, chromosome of Ap IFO 3283-22; line 10, chromosome of Ap IFO 3283-26; and line 11, chromosome of Ap IFO 3283-32. Fragments of the same color represent orthologous regions in different Ap strains.
The plasmids of Ap CICC 20001 and CGMCC 1.41 are unique
To compare the plasmids of different Ap strains, their plasmids were integrated severally and aligned using Mauve 2.3.1 (Fig. 5). The results show that plasmids have specific characteristics of Ap strains from different sources. Compared to Ap IFO 3283 substrains and 386B, CICC 20001 and CGMCC 1.41 possess few similar genotypes. The plasmids of Ap CICC 20001, CGMCC 1.41, 386B and IFO 3283 are largely different. Most notably, Ap CICC 20001 has a large plasmid of approximately 474484 bp, which has almost no homology to plasmids from other Ap strains. This plasmid may play a key role in protecting the genomic integrity of Ap CICC 20001 because it contains 3 CRISPR (clustered regularly interspaced short palindromic repeats) elements, as discussed later. Furthermore, it appears that similar to the chromosome, the plasmids display genetic stability after storage for more than 30 years. Plasmids of all IFO 3283 substrains also have very similar genetic sequences. The relative positions of the genes in the plasmids of the substrains are identical, with the exception of two transposases, APA01_40012 in IFO 3283-01-42C and APA42C_40012 in IFO 3283-0126, which were inserted into related plasmids without interrupting the coding sequence of a gene. Among Ap IFO 3283 substrains, IFO 3283-32 possesses the least variation in its chromosome and plasmids; therefore, Ap IFO 3283-32 is used to represent all Ap IFO 3283 substrains in subsequent analyses.
The locations of orthologs between plasmids of 11 Ap strains.
The lines represent (from top to bottom): line 1, plasmids of Ap CGMCC 1.41; line 2, plasmids of Ap CICC 20001; line 3, plasmids of Ap 386B; line 4, plasmids of Ap IFO 3283-01; line 5, plasmids of Ap IFO 3283-01-42C; line 6, plasmids of Ap IFO 3283-03; line 7, plasmids of Ap IFO 3283-07; line 8, plasmids of Ap IFO 3283-12; line 9, plasmids of Ap IFO 3283-22; line 10, plasmids of Ap IFO 3283-26; and line 11, plasmids of Ap IFO 3283-32. The red vertical lines on each line represent the edges of different plasmids.
More than 100 essential genes support the survival of Ap strains
Essential genes are thought to be critical for the survival of organisms. Essential genes in Ap strains were predicted using ZCURVE 3.0 software. Ap IFO 3283 substrains have extremely similar chromosomes with the same essential genes and Ap IFO 3283-32 are used as a representative of Ap IFO 3283 substrains. The distribution of essential genes in chromosomes of Ap IFO 3283-32, 386B, CICC 20001 and CGMCC 1.41 are shown in Supplementary Fig. S2. The essential genes pervade chromosomes. In each chromosome, there is more than one cluster of essential genes and some large regions (longer than 300 kb) without essential genes. These essential genes encode proteins that primarily participate in maintaining basic cellular structure, replicating DNA, translating genes into proteins, mediating transport processes into and out of the cell and maintaining central metabolism.
Amino acid composition among Ap strains is similar
Ap IFO 3283-32 is characterized to be a representative of IFO 3283 substrains, which are highly similar in their gene distributions and coded proteins. Amino acid components from Ap IFO 3283-32, 386B, CICC 20001 and CICC 1.41 were computed. Amino acid components in different Ap species are highly similar, although Ap strains share a high homology in the chromosome and almost no similarity in their plasmids (Fig. 6). All of them have a high concentration of alanine, leucine and glycine. Ap IFO 3283-32 and CICC 20001 have higher levels of arginine and serine, respectively.
Plausible mechanisms conferring acetic acid resistance and related metabolism in Ap
Because Ap strains have highly similar genomes, in the presence of a high concentration of acetic acid, they may also exhibit similar mechanisms conferring acetic acid resistance. Some mechanisms related to acid resistance in AAB and E. coli, such as transportation systems, stress proteins, metabolic responses, chloride transporters, arginine-dependent system, glutamate-dependent system and lysine-dependent system, have been discovered15,33,35,47,48. The comparison of protein-coding genes that contribute to acid resistance can be used to discover mechanisms conferring acetic acid resistance and to construct a metabolic blueprint related to acetic acid tolerance in Ap.
Comparative genomic analysis emphasizes the roles of PQQ-ADH in acetic acid production and resistance in AAB
Pyrroloquinoline quinine dependent alcohol dehydrogenase (PQQ-ADH) and aldehyde dehydrogenase (PQQ-ALDH) are membrane-bound enzymes in AAB, which catalyze the oxidation of alcohol to aldehyde and aldehyde to acetic acid, respectively, producing abundant extracellular acetic acid49. Both PQQ-ADH and PQQ-ALDH play a key role in the respiratory chain in Ap CGMCC 1.41, CICC 20001 and other AAB31,50. Trcek et al. demonstrated that PQQ-ADH is involved in acetic acid resistance by comparing the enzymatic activities of PQQ-ADH from the AAB strains with different acetic acid production abilities21. K. europaeus, K. oboediens and Ap produce a high concentration of acetic acid of up to 18%, 8% and 6%, respectively51,52. In combination with their gradient abilities to produce acetic acid, the gene number of ADH and ALDH may further reveal the relation between ADH, as well as ALDH and acetic acid tolerance.
To emphasize the effects of ADHs and ALDHs on producing a high level of acetic acid, genes coding ADH and ALDH in Ap CICC 20001, Ap CGMCC 1.41, Ap 386B, Ap IFO 3283-32, K. europaeus 5P3 and K. oboediens 174Bp2 were investigated (Table 2). Membrane-bound ADHs (Supplementary Data S1) and ALDHs (Supplementary Data S2), together with their topologies (Supplementary Fig. S3 and Supplementary Fig. S4), have been predicted using three algorithms, as described in the methods. Ap species possess fewer genes coding ADHs and more genes coding ALDHs than K. europaeus 5P3. There are 7, 2, 2, 2, 1 and 1 membrane-bound ADHs, as well as 1, 1, 1, 3, 0 and 0 membrane-bound ALDHs in K. europaeus 5P3, K. oboediens 174Bp2, Ap 386B, Ap IFO 3283-32, Ap CICC 20001 and Ap CGMCC 1.41, respectively. Clearly, compared to the others, K. europaeus 5P3 possesses more than 3 times the genes coding membrane-bound ADHs but a similar number of genes coding membrane-bound ALDHs. Such high levels of ADHs, especially PQQ-ADH, may be a key factor that allows K. europaeus 5P3 to accumulate such a high concentration of acetic acid. Moreover, during vinegar manufacture, a single gene coding membrane-bound ALDH is able to satisfy demand by producing a high level of acetic acid. Interestingly, Ap CICC 20001 and CGMCC 1.41 have one PQQ-ADH and no membrane-bound ALDH, which may explain why they produce a lower concentration of acetic acid than K. europaeus strains. Inserting more copies of genes coding PQQ-ADH to the genome of Ap strains may be an effective method to improve their ability to produce a high level of acetic acid.
The distribution of ADHs and ALDHs are shown in Fig. 7. Genes coding ADHs and ALDHs are distributed throughout the chromosomes of Ap strains at regular intervals. Occasionally, ADH and ALDH are clustered as well. This indicates that ADHs and ALDHs are common enzymes for metabolism in Ap strains, accumulating acetic acid in the full growth process under specific conditions. Therefore, acid resistance and the expression level of ADHs and ALDHs may be key factors for producing a high concentration of acetic acid.
Genes related to producing and tolerating acetic acid in Ap IFO 3283-32, 386B, CGMCC 1.41 and CICC 20001.
From outer to inner, the first, second, third and fourth circles depict chromosomes of Ap IFO 3283-32, CGMCC 1.41, CICC 20001 and 386B. The fifth, sixth, seventh and eighth circles depict genes related to producing and tolerating acetic acid in Ap IFO 3283-32, CGMCC 1.41, CICC 20001 and 386B.
Tactical cooperation may be involved in acetic acid tolerance in Ap strains
The genes coding enzymes that participate in mechanisms conferring acid resistance in microorganisms15,32,33,34,35 were investigated in K. europaeus 5P3, K. oboediens 174Bp2, Ap 386B, Ap IFO 3283-32, Ap CICC 20001 and Ap CGMCC 1.41 (Supplementary Data S3). As indicated in Table 2, there are more genes coding PQQ-ADHs in K. europaeus 5P3 than in Ap CICC 20001 and CGMCC 1.41. A high level of PQQ-ADHs, may contribute not only to producing high concentrations of acetic acid but also to tolerating an extreme acid environment. Acetate kinase (AckA), acetyl-CoA synthetase (Acs), citrate synthase (Cs), aconitate hydratase (AcnA) and phosphate acetyltransferase (Pta) participate in acetic acid assimilation in AAB, maintaining a higher intracellular than extracellular pH value. All of these enzymes are involved in the common metabolism in AAB and are similar to those found in K. europaeus 5P3, K. oboediens 174Bp2, Ap 386B, Ap IFO 3283-32, Ap CICC 20001 and Ap CGMCC 1.41. The ADI pathway plays a key role in acid resistance in Lactobacillus reuteri47. Interestingly, compared to other AAB strains, the ADI pathway is peculiar to K. europaeus 5P3. The ADI pathway may be a key factor that allows K. europaeus 5P3 to accumulate a high level of acetic acid. Genes coding enzymes that contribute to acetic acid resistance cluster in the chromosome of Ap strains. These include molecular chaperones, such as the combination of GroES and GroEL and the combination of DnaK, GrpE and DnaJ and enzymes concerned with the assimilation of acetic acid, including the combination of Pta and AckA and the combination of Cs and Acs (Fig. 7).
Tactical cooperation may be involved in acetic acid tolerance in Ap strains. Because an increasing amount of acetic acid is produced, an extreme acid environment on both sides of the membrane, has been gradually formed. During acetic acid fermentation, a great deal of acetic acid is produced outside and diffuses into the cytoplasm, creating a lower pH environment. To maintain a relatively higher pH value intracellularly, some effective methods are used in Ap CGMCC 1.41 cells. First, PQQ-ADH activities are directly related to acetic acid resistance and thermotolerance in AAB21,53, likely because ADHs act as the main enzyme in energy metabolism of Ap. Second, in the cytoplasm, acetic acid is transformed to acetyl-CoA by acetyl-CoA synthetase (AS. 1613–1614, AS. 2105–2106 and AS. 2938), or by acetate kinase (AS. 74) and phosphotransacetylase (AS. 75). Then, acetyl-CoA is transformed to isocitrate by sequential catalytic action of citrate synthase (AS. 2465) and aconitate hydratase (AS. 1043) in the tricarboxylic acid (TCA) cycle. Third, chaperones, such as GroES (AS. 388), GroEL (AS. 389), DnaK (AS. 1527), DnaJ (AS. 1168, AS. 1449, AS. 1528 and AS. 2523) and GrpE (AS. 1525) confer stress resistance to stressors including temperature shifts, ethanol and acetic acid. Fourth, cyclopropane-fatty-acyl-phospholipid synthase (AS. 2381) catalyzes the formation of cyclopropane fatty acid, which is a major component of the phospholipids of many species of gram-negative bacteria and considered a conditional and post-synthetic modification of bacterial membrane lipid bilayers. The formation of cyclopropane fatty acid may lead to a change in the morphology and permeability to acetic acid involved in acetic acid resistance. Fifth, lysine decarboxylase (AS. 1249) and ornithine decarboxylase (AS. 1047) replace the α-carboxyl groups of their amino acid substrates with a proton from the cytoplasm. CO2 is produced, as well as either cadaverine, which is the end product of the reaction catalyzed by lysine decarboxylase, or putrescine, the end product of that catalyzed by ornithine decarboxylase. These polyamines, consisting of multi-basic group, provide an alkaline environment in the cytoplasm. Then, the putrescine ABC transporter (AS. 2264–2265, AS. 2726–2728 and AS. 2730) removes the superfluous putrescine. Furthermore, in the amino acid decarboxylation-antiporter system, the cognate antiporters expel the decarboxylation products and import new amino acid substrates.
In Ap CICC 20001, similar methods are adopted to maintain a relatively high pH value in the cytoplasm. Acetyl-CoA synthetase (HN. 126, HN. 1654 and HN. 2080), acetate kinase (HN. 787), phosphotransacetylase (HN. 786), citrate synthase (HN. 1277), aconitate hydratase (HN. 2323–2424), GroES (HN. 3179), GroEL (HN. 3196), DnaK (HN. 2159), DnaJ (HN. 1225, HN. 2158, HN. 2234 and HN. 2500), GrpE (HN. 2161), cyclopropane-fatty-acyl-phospholipid (HN. 1360), Ornithine decarboxylase (HN.3419), lysine decarboxylase (HN. 2459) and putrescine ABC transporter (HN. 2264–2265, HN. 2726–2728, HN. 2730) contribute to acetic acid resistance in an extreme acid environment.
The metabolic blueprint reveals the approaches to improve acetic acid production and resistance in Ap
To understand the overall mechanisms of acetic acid resistance in Ap, the main metabolic pathways related to acetic acid tolerance and production in Ap CGMCC 1.41 were arranged in combination with the discovered acid-tolerant mechanisms in E. coli and the published acetic acid resistance in AAB (Fig. 8). These pathways have been shown to be the common pathways in Ap species, including Ap CGMCC 1.41 and CICC 20001. In Ap, there is a special TCA cycle, in which succinate-semialdehyde dehydrogenase (EC 1.2.1.24), rather than succinyl coenzyme A synthetase (EC 6.2.1.4), is the intermediate enzyme in the transformation of 2-oxoglutarate to succinate. This substitution shortens the process of the TCA cycle, gaining a higher metabolic efficiency. In the metabolic blueprint of Ap, pyruvate metabolism is the main process that integrates other metabolic processes, including the special TCA cycle, EMP pathway, pentose phosphate pathway, terpenoid biosynthesis, glycine metabolism and lipopolysaccharide biosynthesis. From these metabolic pathways, Ap can use glucose, fructose, ethanol and acetic acid as ideal carbon sources to grow. Although Ap species have no uniform mechanisms that confer acid tolerance in E. coli, such as the arginine-dependent system, the glutamate-dependent system and the lysine-dependent system, some analogous pathways, as well as published mechanisms, may contribute to acetic acid resistance in Ap. In the presence of ammonia-lyases, NH3 is produced from ornithine, threonine and S-amino-methyl-dihydrolipoyl protein. Moreover, NH3 also can be integrated into L-aspartate and L-glutamate, forming the corresponding amino acids. These opposed reactions may balance the contents of NH3 and control the intracellular pH value in cells. Furthermore, ornithine decarboxylase (EC 4.1.1.17) catalyzes the transformation of ornithine to putrescine, which is a polyamine that greatly modifies intramembrane pH in microorganisms54.
Metabolic pathways related to acetic acid resistance in Ap CGMCC 1.41.
These include the TCA cycle, pyruvate metabolism, the EMP pathway, the pentose phosphate pathway, terpenoid biosynthesis, glycine metabolism, lipopolysaccharide biosynthesis and peptidoglycan biosynthesis. Enzymes in these pathways are included in Supplementary Text 1.
The genetic stabilities of Ap strains may vary with the individual
Similar to other organisms, the genome of Ap strains is remarkably stable from one generation to the next but is plastic on an evolutionary timescale. Bacterial chromosomes are complex and dynamic, thereby maintaining a balance between genome integrity and instability and allowing the survival of organisms and their offspring55. Genomic rearrangements, including deletions, duplications, amplifications, insertions, inversions and translocations, lead to instability of the genome. Some of these mutations are silent, while others bring about phenotypic variation, evolution and speciation. Genomic instability plays two roles in organismal survival. On the one hand, specialized genetic elements, including mobile elements, inteins, introns, retroelements and integrons and recombination methods, homologous or illegitimate, can mediate genome instability, generating phenotypic variation; on the other hand, restriction-modification (RM) systems and the CRISPR-Cas system (comprising CRISPR and CRISPR-associated proteins) use genome instability to protect organisms from invasion by phages and mobile elements56,57. Mobile elements, widely present in organisms, contain insertion sequences (IS), miniature inverted-repeat transposable elements (MITEs), repetitive extragenic palindromic (REP) sequences, bacterial interspersed mosaic elements (BIMEs), transposable elements (TEs), transposable bacteriophages and genomic islands and involve genome instability. The long terminal repeats (LTRs) of Ap were predicted using LTR Finder (Supplementary Data S4). Ap CGMCC 1.41 and all Ap 3283 substrains have an LTR of 1269 bp in the chromosome and 833 bp in the plasmid, respectively. Meanwhile, Ap 386B and Ap CICC 20001 have no LTR in their genomes. Like LTR, all Ap 3283 substrains have a confirmed CRISPR element (Supplementary Data S5) of 1431 bp in almost the same location of the genome and Ap 386B has no CRISPR element in either its chromosome or its plasmids. The chromosomes of Ap CGMCC 1.41 and Ap CICC 20001, have 1 and 2 putative CRISPR elements, respectively. Furthermore, Ap CICC 20001 have a large plasmid containing two confirmed CRISPR elements and four putative CRISPR elements. This plasmid may greatly contribute to the gene stability of Ap CICC 20001 by escaping insertions from phages and mobile elements.
Stability and instability factors direct the balance between genome integrity and instability. As previously mentioned, after 30 years of storage, 7 substrains with different phenotypic characteristics were isolated from Ap IFO 3283. Moreover, when one of these substrains IFO 3283-01 was exposed to a high temperature, a mutant strain IFO 3283-01-42C, which tolerated temperatures as high as 42 °C, was isolated and sequenced. Compared to other substrains, a DNA fragment is missing in Ap 3283-01-42C. Further genomic analysis showed that the genomes of all Ap IFO 3283 substrains contained more than 280 transposons and five genes with hyper-mutable tandem repeats, revealing the genetic instability of Ap26. However, Gullo, et al.27 believed that Ap strains are stable because at different ages of the culture and frequencies of subculture, Ap AB0220 showed a high stability over 9 years of preservation. Instability and stability factors in the genome of Ap strains were summarized (Table 3).
There are approximately 270 transposases and 1 LTR in the genomes of Ap IFO 3283 substrains, while Ap 386B, Ap CICC 20001 and Ap CGMCC 1.41 possess fewer transposases or mobile elements than do the Ap IFO 3283-32 substrains. Importantly, Ap CICC 2001 contains 5 CRISPR elements that contribute to genome integrity and no more than 80 transposases that relate to genetic instability. Therefore, Ap CICC 20001 may have a very stable genome. The stability of Ap strains may vary with the individual. In a given environment, the balance of stability factors and instability factors control the stability of Ap strains.
Discussion
Ap CICC 20001 and CGMCC 1.41 display strong abilities in both producing and tolerating acetic acid, with more than 6%, by liquid-state fermentation. Furthermore, Ap CGMCC 1.41 tolerates higher concentrations of ethanol and acetic acid and produces a high level of acetic acid in the modified GY medium in an Erlenmeyer flask. However, the ability to produce acetic acid in Ap strains may vary by each individual strain. Ap CICIM B7003, which was isolated from industrial vinegar bioreactors in China, yielded 7.00% final acetic acid in a semi-continuous regime by an optimized protocol, which was less than Ap CGMCC 1.4122. However, Ap CICIM B7003-02, an ultraviolet mutant from Ap CICIM B7003, produces a high acidity vinegar with an acetic acid concentration that reached up to 9.33% in the semi-continuous mode in the Frings Pilot-Acetator 9 L58. A bioreactor is a more effective piece of equipment for brewing vinegar than an Erlenmeyer flask. This study indicates that Ap CGMCC 1.41 may be an ideal strain for producing high levels of acetic acid (up to 9%).
Comparative genomic analysis of 11 Ap strains reveals that the chromosomes of Ap CICC 20001 and CGMCC 1.41 are evolutionarily conserved, sharing a high degree of homology with other Ap strains, whereas their plasmids are unique, suggesting a separate evolution between Ap chromosomes and plasmids. All of the Ap strains also share almost identical proportions of amino acid components in their genomes. All IFO 3283 substrains possess an almost identical chromosome, although IFO 3283-01-42C lost a DNA fragment and tolerates a higher temperature. In these substrains, 4 transposon insertions (SecB2, glycosyl transferase, two component kinase and intergenic), 3 SNPs (glycerol kinase, RopA and hypothetical) and 3 hyper-mutable Tandem Repeats (HTRs) were identified as chromosomal variations, while a transposon insertion and a HTR were observed in the largest plasmid26.
Our results comparing the numbers of PQQ-ADH in 3 AABs with a range of abilities to produce acetic acid emphasize that PQQ-ADH contributes to acetic acid tolerance in Ap. Comparison of genes related to acetic acid resistance in AAB reveals that acetate kinase, acetyl-CoA synthetase, citrate synthase, aconitate hydratase and phosphate acetyltransferase jointly participate in acetic acid assimilation in Ap, resisting acetic acid in the presence of a high concentration of acetic acid. Furthermore, the pathway related to acetic acid tolerance shows that in addition to reported mechanisms conferring acetic acid resistance, metabolism of some amino acids, such as degradation of threonine, glycine and ornithine, contribute to acetic acid tolerance by producing a large amount of NH3, which decreases the intracellular pH value, as in E. coli. Ornithine also can be degraded and transformed to putrescine, greatly neutralizing intracellular pH in microorganisms54. Moreover, 3 urease genes were detected in Ap, but no urease gene was found in K. europaeus 5P3 and K. oboediens 174Bp2 (Supplementary Data S3), which indicates that urease system may additionally contribute to acetic acid resistance of Ap. The blueprint constructed in this study benefits the investigation of acetic acid resistance and related regulation.
All Ap strains contain some transposases or mobile elements causing genetic instability and several protection systems, including CRISPR and RM involved in genetic stability by avoiding insertions from phages and mobile elements. A balance between protection systems and transposases directs genetic stability and instability. Ap CICC 20001 and CGMCC 1.41 have been utilized to steadily brew vinegar in the vinegar industry in China for more than 50 years. Ap CICC 20001 and CGMCC1.41 may be more stable than IFO 3283 substrains because genomes of Ap CICC 20001 and CGMCC1.41 contain one-third of the transposases and a greater number of protective systems than the genome of IFO 3283 substrains. The genetic stability of Ap strains may vary by individual.
In summary, we uncovered global insights into acetic acid resistance mechanisms and genetic stability of Ap strains using comparative genomics. These observations provide important insights into the evolution, acid resistant mechanism and genetic stability of these two economically important AAB strains and lay a foundation for future genetic manipulation and engineering of these strains. However, the acid-tolerant mechanisms have only been predicted by comparing the genes related to acid resistance in other microorganisms, without experimental verification. Verifying the mechanisms conferring acetic acid resistance and related regulation systems should be hot topics in the study of AAB.
Methods
Strains and growth conditions
The AAB strains used in this study are Ap CICC 20001 and CGMCC1.41 from the China Center of Industrial Culture Collection (CICC) and China General Microbiological Culture Collection Center (CGMCC), respectively. Ap CICC 20001 and CGMCC 1.41 were grown in glucose yeast medium (10% glucose and 1% yeast extract) and bean sprout glucose ethanol medium (20% bean sprout extract, 1% glucose and 2% ethanol) and then cultivated on an incubator shaker for 24 h at 30 °C. The cells were harvested by centrifugation at 9000 × g for 10 min. Ap CICC 20001 and CGMCC 1.41 were inoculated on GYC medium (5% glucose, 1% yeast extract and 2% CaCO3) using the streak method.
Fermentation characteristics of Ap CICC 20001 and CGMCC 1.41 strains
Ap CICC 20001 and CGMCC1.41 were grown in a modified GYP medium (0.1% glucose, 0.2% peptone, 0.5% yeast extract) containing different concentrations of ethanol (0, 2%, 4%, 6%, 8%, 10% and 12%) or different concentrations of acetic acid (0, 0.5%, 1%, 1.5%, 2%, 2.5%, 3%, 3.5% and 4%). An inoculum of 250 μl (approximately A600 = 0.5) was inoculated in a 250 ml Erlenmeyer flask containing 50 ml GYP medium with 3% ethanol. When the exponential growth phase was reached, 5 ml of this culture was used as an inoculum for acetic acid fermentation in a 250 ml Erlenmeyer flask containing 50 ml GYP medium with a defined concentration of acetic acid and ethanol. The inoculated flasks were incubated on the rotary shaker at 170 rpm and 30 °C. The acidity of the medium was measured with 0.1 M NaOH using phenolphthalein as an indicator.
Genomic sequencing
Ap CICC 20001 and CGMCC 1.41 cells were subjected to SMRT sequencing at the Institute of Medicinal Plant Development (Beijing, China) using a PacBio RS DNA sequencer (Pacific Biosciences, Menlo Park, CA, USA; http://www.pacificbiosciences.com/)59. SMRT bell template libraries with DNA fragments of 2 kb were prepared60. Then, sequencing was performed by utilizing one SMRT cell (http://www.pacificbiosciences.com/products/consumables/SMRT-cells/) and obtaining a zero-mode waveguide61. SMRT reads were mapped to the Ap genome reference sequence using BLASR software (http://github.com/PacificBiosciences/blasr)62 according to standard mapping protocols. Interpulse durations were measured as described for all of the pulses aligned to each position in the Ap genome sequence. The modified bases were identified applying the SMRT Analysis Server v.1.4.0 (Pacific Biosciences). Genomic sequences were assembled using SMRT analysis RS_HGAP_Assembly.2 (https://github.com/PacificBiosciences/SMRT-Analysis/wiki/SMRT-Pipe-Reference-Guide-v2.2.0#PRO_HGAP2). Automatic gene prediction and annotation of the assembled genome sequences were performed using RAST (http://rast.nmpdr.org/)63. The annotated genes were classified using the Clusters of Orthologous Groups of proteins (COG) database (http://www.ncbi.nlm.nih.gov/COG/). The pathways that genes participate in were analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/). The generated data are available for download at the website http://fbfs.hzau.edu.cn/AAB/mulu/genome.asp.
Visualization of data
CGView (http://stothard.afns.ualberta.ca/cgview_server/)64 was utilized to exhibit graphical layouts of the chromosome, as well as the corresponding GC content and GC skew. Circos 0.66 (http://circos.ca/) was used to highlight the distribution of the genes contributing to the production and tolerance of acetic acid in Ap genomes. Metabolic pathways in Ap CGMCC 1.41 were mapped using the KEGG PATHWAY database and expressed using Edrawmax-cn_7.2.
Prediction of special genes or structures
Essential genes were predicted using ZCURVE 3.0 software65 (http://cefg.uestc.edu.cn/zcurve/). To identify membrane-bound ADH and ALDH, Kyte-doolittle (http://gcat.davidson.edu/DGPB/kd/kyte-doolittle.htm), Tmpred (http://www.ch.embnet.org/software/TMPRED_form.html) and TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/), which are based on hydrophobicity, a transmembrane proteins database and Hidden Markov Models, respectively, were utilized to predict the transmembrane helices of ADHs and ALDHs of Ap CICC 20001, Ap CGMCC 1.41, Ap IFO 3283-32, Ap 386B, K. europaeus 5P3 and K. oboediens 174Bp2. Tandem Repeats Finder software (http://tandem.bu.edu/trf/trf.basic.submit.html), LTR Finder software (http://tlife.fudan.edu.cn/ltr_finder/index.php) and CRISPR finder software (http://crispr.u-psud.fr/Server) were applied to predict tandem repeats (TRs), long terminal repeat (LTR) retrotransposons and clustered regularly interspaced short palindromic repeats (CRISPR) in genomes of Ap strains, respectively.
Orthology analysis
The previously published genome sequences of Ap IFO 3283-01, IFO 3283-01-42C, IFO 3283-03, IFO 3283-07, IFO 3283-12, IFO 3283-22, IFO 3283-26 and IFO 3283-32 were downloaded from NCBI. Mauve 2.3.1 (http://darlinglab.org/mauve/mauve.html) was used to compare genome orthologies of Ap CICC 20001 and CGMCC 1.41 with these Ap species. To compare orthologous plasmids, plasmids in all of these Ap species were integrated severally.
Additional Information
How to cite this article: Wang, B. et al. Global insights into acetic acid resistance mechanisms and genetic stability of Acetobacter pasteurianus strains by comparative genomics. Sci. Rep. 5, 18330; doi: 10.1038/srep18330 (2015).
References
Kersters, K., Lisdiyanti, P., Komagata, K. & Swings, J. The family Acetobacteraceae: the genera Acetobacter, Acidomonas, Asaia, Gluconacetobacter, Gluconobacter and Kozakia. The Prokaryotes 5, 163–200, doi: 10.1007/0-387-30745-1_9 (2006).
Dutta, D. & Gachhui, R. Nitrogen-fixing and cellulose-producing Gluconacetobacter kombuchae sp. nov., isolated from Kombucha tea. Int. J. Syst. Evol. Micr. 57, 353–357, doi: 10.1099/ijs.0.64638-0 (2007).
Iino, T. et al. Gluconacetobacter kakiaceti sp. nov., an acetic acid bacterium isolated from a traditional Japanese fruit vinegar. Int. J. Syst. Evol. Micr. 62, 1465–1469, doi: 10.1099/ijs.0.031773-0 (2012).
Illeghems, K., De Vuyst, L. & Weckx, S. Complete genome sequence and comparative analysis of Acetobacter pasteurianus 386B, a strain well-adapted to the cocoa bean fermentation ecosystem. BMC Genomics 14, 1–14, doi: 10.1186/1471-2164-14-526 (2013).
Silva, L. R. et al. Acetobacter oeni sp. nov., isolated from spoiled red wine. Int. J. Syst. Evol. Micr. 56, 21–24, doi: 10.1099/ijs.0.46000-0 (2006).
Lisdiyanti, P. et al. Identification of Acetobacter strains isolated from Indonesian sources and proposals of Acetobacter syzygii sp. nov., Acetobacter cibinongensis sp. nov. and Acetobacter orientalis sp. nov. J. Gen. Appl. Microbiol. 47, 119–131 (2001).
Lino, T. et al. Acetobacter okinawensis sp. nov., Acetobacter papayae sp. nov. and Acetobacter persicus sp. nov.; novel acetic acid bacteria isolated from stems of sugarcane, fruits and a flower in Japan. J. Gen. Appl. Microbiol. 58, 235–243, doi: 10.2323/jgam.58.235 (2012).
Greenberg, D. E. et al. Granulibacter bethesdensis gen. nov., sp. nov., a distinctive pathogenic acetic acid bacterium in the family Acetobacteraceae. Int. J. Syst. Evol. Micr. 56, 2609–2616, doi: 10.1099/ijs.0.64412-0 (2006).
Tazato, N. et al. Gluconacetobacter tumulicola sp. nov. and Gluconacetobacter asukensis sp. nov., isolated from the stone chamber interior of the Kitora Tumulus. Int. J. Syst. Evol. Micr. 62, 2032–2038, doi: 10.1099/ijs.0.034595-0 (2012).
Nishijima, M. et al. Gluconacetobacter tumulisoli sp. nov., Gluconacetobacter takamatsuzukensis sp. nov. and Gluconacetobacter aggeris sp. nov., isolated from Takamatsuzuka Tumulus samples before and during the dismantling work in 2007. Int. J. Syst. Evol. Micr. 63, 3981–3988, doi: 10.1099/ijs.0.051292-0 (2013).
Beijerinck, M. Ueber die arten der essigbakterien. Centralbl. Bakt., Parasitenk Infektionskrank. 4, 209–216 (1898).
Li, L. et al. Acetobacter sicerae sp. nov., isolated from cider and kefir and identification of Acetobacter species by dnaK, groEL and rpoB sequence analysis. Int. J. Syst. Evol. Micr. 64, 2407–2415, doi: 10.1099/ijs.0.058354-0 (2014).
Vu, H. T. L. et al. Nguyenibacter vanlangensis gen. nov., sp. nov., an unusual acetic acid bacterium in the α-Proteobacteria. J. Gen. Appl. Microbiol 59, 153–166, doi: 10.2323/jgam.59.153 (2013).
Li, L. et al. Bombella intestini gen. nov., sp. nov., a novel acetic acid bacterium isolated from bumble bee crop. Int. J. Syst. Evol. Micr. 65, 267–273 doi: 10.1099/ijs.0.068049-0 (2014).
Wang, B., Shao, Y. & Chen, F. Overview on mechanisms of acetic acid resistance in acetic acid bacteria. World J. Microb. Biot. 31, 1–9, doi: 10.1007/s11274-015-1799-0 (2015).
Yamada, Y. et al. Description of Komagataeibacter gen. nov., with proposals of new combinations (Acetobacteraceae). J. Gen. Appl. Microbiol. 58, 397–404, doi: 10.2323/jgam.58.397 (2012).
Wu, J. J., Ma, Y. K., Zhang, F. F. & Chen, F. S. Biodiversity of yeasts, lactic acid bacteria and acetic acid bacteria in the fermentation of “Shanxi aged vinegar”, a traditional Chinese vinegar. Food Microbiol. 30, 289–297, doi: 10.1016/j.fm.2011.08.010 (2012).
Kanchanarach, W. et al. Acetic acid fermentation of Acetobacter pasteurianus: relationship between acetic acid resistance and pellicle polysaccharide formation. Biosci. Biotech. Bioch. 74, 1591–1597, doi: 10.1271/bbb.100183 (2010).
Sokollek, S. J., Hertel, C. & Hammes, W. P. Description of Acetobacter oboediens sp. nov. and Acetobacter pomorum sp. nov., two new species isolated from industrial vinegar fermentations. Int. J. Syst. Bacteriol. 48, 935–940, doi: 10.1099/00207713-48-3-935 (1998).
Schüller, G., Hertel, C. & Hammes, W. P. Gluconacetobacter entanii sp. nov., isolated from submerged high-acid industrial vinegar fermentations. Int. J. Syst. Evol. Micr. 50, 2013–2020, doi: 10.1099/00207713-50-6-2013 (2000).
Trcek, J., Toyama, H., Czuba, J., Misiewicz, A. & Matsushita, K. Correlation between acetic acid resistance and characteristics of PQQ-dependent ADH in acetic acid bacteria. Appl. Microbiol. Biot. 70, 366–373, doi: 10.1007/s00253-005-0073-z (2006).
Qi, Z. et al. A protocol for optimization vinegar fermentation according to the ratio of oxygen consumption versus acid yield. J. Food Eng. 116, 304–309, doi: 10.1016/j.jfoodeng.2012.12.029 (2013).
Shimoji, Y. et al. Isolation and identification of DPPH radical scavenging compounds in Kurosu (Japanese unpolished rice vinegar). J. Agr. Food Chem. 50, 6501–6503, doi: 10.1021/jf020458f (2002).
Sokollek, S. J., Hertel, C. & Hammes, W. P. Cultivation and preservation of vinegar bacteria. J. Biotechnol. 60, 195–206 (1998).
Trček, J., Jernejc, K. & Matsushita, K. The highly tolerant acetic acid bacterium Gluconacetobacter europaeus adapts to the presence of acetic acid by changes in lipid composition, morphological properties and PQQ-dependent ADH expression. Extremophiles 11, 627–635, doi: 10.1007/s00792-007-0077-y (2007).
Azuma, Y. et al. Whole-genome analyses reveal genetic instability of Acetobacter pasteurianus. Nucl. Acids Res. 37, 5768–5783, doi: 10.1093/nar/gkp612 (2009).
Gullo, M., Mamlouk, D., De Vero, L. & Giudici, P. Acetobacter pasteurianus strain AB0220: cultivability and phenotypic stability over 9 years of preservation. Curr. Microbiol. 64, 576–580, doi: 10.1007/s00284-012-0112-9 (2012).
Lu, H., Yao, Y., Wu, W. & Shan, Y. Research on screening Acetobacter spp. for citrus vinegar fermentation. China Brewing (China) 6, 10–12 (2004).
An, Y., Ren, T. & Kan, J. Fermentation technology of Chaenomeles sinensis fruit vinegar. China Brewing (China) 3, 171–175 (2010).
Zhou, B. Discussion on production of high concentration vinegar. China Brewing (China) 4, 26–29 (1991).
Zhu, X., Xia, X., Yang, H. & Wang, W. Study on the key enzymes of ethanol oxidation and acetic acid production in Acetobacter pasteurianus HN 1.01. Sci. Tech. Food Industry (China) 34, 167–170 (2013).
Nakano, S. & Fukaya, M. Analysis of proteins responsive to acetic acid in Acetobacter: molecular mechanisms conferring acetic acid resistance in acetic acid bacteria. Int. J. Food Microbiol. 125, 54–59, doi: 10.1016/j.ijfoodmicro.2007.05.015 (2008).
Foster, J. W. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2, 898–907, doi: 10.1038/nrmicro1021 (2004).
Lin, J. et al. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 62, 3094–3100 (1996).
Kanjee, U. & Houry, W. A. Mechanisms of acid resistance in Escherichia coli. Annu. Rev. Microbiol. 67, 65–81, doi: 10.1146/annurev-micro-092412-155708 (2013).
Bearson, S., Bearson, B. & Foster, J. W. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 147, 173–180 (1997).
Chang, Y. Y. & Cronan, J. E. Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Mol. Microbiol. 33, 249–259, doi: 10.1046/j.1365-2958.1999.01456.x (1999).
Marshall, B., Barrett, L., Prakash, C., McCallum, R. & Guerrant, R. Urea protects Helicobacter (Campylobacter) pylori from the bactericidal effect of acid. Gastroenterology 99, 697–702 (1990).
Marquis, R. E., Bender, G. R., Murray, D. R. & Wong, A. Arginine deiminase system and bacterial adaptation to acid environments. Appl. Environ. Microbiol. 53, 198–200 (1987).
Xiong, L. et al. Molecular characterization of arginine deiminase pathway in Laribacter hongkongensis and unique regulation of arginine catabolism and anabolism by multiple environmental stresses. Environ. Microbiol. in press, doi: 10.1111/1462-2920.12897 (2015).
Xiong, L. et al. Arginine deiminase pathway is far more important than urease for acid resistance and intracellular survival in Laribacter hongkongensis: a possible result of arc gene cassette duplication. BMC Microbiol. 14, 42–42, doi: 10.1186/1471-2180-14-42 (2014).
Pope, W. H. et al. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. eLife 4, e06416, doi: 10.7554/eLife.06416 (2015).
Bartha, I. & Fellay, J. Adaptation on a genomic scale. eLife 4, e06193, doi: 10.7554/eLife.06193 (2015).
Shalem, O., Sanjana, N. E. & Zhang, F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 16, 299–311, doi: 10.1038/nrg3899 (2015).
Cao, J., Lai, Q., Yuan, J. & Shao, Z. Genomic and metabolic analysis of fluoranthene degradation pathway in Celeribacter indicus P73T. Sci. Rep. 5, 7741, doi: 10.1038/srep07741 (2015).
Arakawa, K. & Tomita, M. The GC skew index: a measure of genomic compositional asymmetry and the degree of replicational selection. Evol. Bioinformatics Online 3, 159–168 (2007).
Rollan, G., Lorca, G. & Font de Valdez, G. Arginine catabolism and acid tolerance response in Lactobacillus reuteri isolated from sourdough. Food Microbiol. 20, 313–319 (2003).
Matsushita, K., Inoue, T., Adachi, O. & Toyama, H. Acetobacter aceti possesses a proton motive force-dependent efflux system for acetic acid. J. Bacteriol. 187, 4346–4352, doi: 10.1128/JB.187.13.4346-4352.2005 (2005).
Yakushi, T. & Matsushita, K. Alcohol dehydrogenase of acetic acid bacteria: structure, mode of action and applications in biotechnology. Appl. Microbiol. Biot. 86, 1257–1265, doi: 10.1007/s00253-010-2529-z (2010).
Matsushita, K., Toyama, H. & Adachi, O. Respiratory chains in acetic acid bacteria: membranebound periplasmic sugar and alcohol respirations. In Respiration in Archaea and Bacteria (ed. Zannoni, D. ) 81–99 (Springer, 2004).
Fernández-Pérez, R., Torres, C., Sanz, S. & Ruiz-Larrea, F. Strain typing of acetic acid bacteria responsible for vinegar production by the submerged elaboration method. Food Microbiol. 27, 973–978, doi: 10.1016/j.fm.2010.05.020 (2010).
Sievers, M., Sellmer, S. & Teuber, M. Acetobacter europaeus sp. nov., a main component of industrial vinegar fermenters in central Europe. Syst. Appl. Microbiol. 15, 386–392 (1992).
Kanchanarach, W. et al. Characterization of thermotolerant Acetobacter pasteurianus strains and their quinoprotein alcohol dehydrogenases. Appl. Microbiol. Biot. 85, 741–751, doi: 10.1007/s00253-009-2203-5 (2010).
Young, N. D. & Galston, A. W. Putrescine and acid stress: induction of arginine decarboxylase activity and putrescine accumulation by low pH. Plant Physiol. 71, 767–771, doi: 10.1104/pp.71.4.767 (1983).
Saier, M. H. The Bacterial Chromosome. Crit. Rev. Biochem. Mol. Biology 43, 89–134, doi: 10.1080/10409230801921262 (2008).
Lin, S., Staahl, B. T., Alla, R. K. & Doudna, J. A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3, e04766, doi: 10.7554/eLife.04766 (2014).
Caliando, B. J. & Voigt, C. A. Targeted DNA degradation using a CRISPR device stably carried in the host genome. Nat. Commun. 6, 6989, doi: 10.1038/ncomms7989 (2015).
Qi, Z. et al. Achieving high strength vinegar fermentation via regulating cellular growth status and aeration strategy. Process Biochem. 49, 1063–1070, doi: 10.1016/j.procbio.2014.03.018 (2014).
Powers, J. G. et al. Efficient and accurate whole genome assembly and methylome profiling of E. coli. BMC genomics 14, 675, doi: 10.1186/1471-2164-14-675 (2013).
Travers, K. J., Chin, C.-S., Rank, D. R., Eid, J. S. & Turner, S. W. A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids Res. 38, e159, doi: 10.1093/nar/gkq543 (2010).
Levene, M. J. et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science 299, 682–686, doi: 10.1126/science.1079700 (2003).
Chaisson, M. J. & Tesler, G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC bioinformatics 13, 238, doi: 10.1186/1471-2105-13-238 (2012).
Aziz, R. K. et al. The RAST Server: rapid annotations using subsystems technology. BMC genomics 9, 75, doi: 10.1186/1471-2164-9-75 (2008).
Stothard, P. & Wishart, D. S. Circular genome visualization and exploration using CGView. Bioinformatics 21, 537–539, doi: 10.1093/bioinformatics/bti054 (2005).
Hua, Z. G. et al. ZCURVE 3.0: identify prokaryotic genes with higher accuracy as well as automatically and accurately select essential genes. Nucleic Acids Res. 43(W1), W85–90, doi: 10.1093/nar/gkv491 (2015).
Acknowledgements
This study was supported by Programs of International S & T Cooperation, Ministry of Science and Technology, P. R. China (No. 2014DFG32380) and the Fundamental Research Funds for the Central Universities (Nos. 2013PY006, 2014PY034 and 2662015PY167) and the project of Wuhan Science & Technology Bureau (No. 2015030809020368).
Author information
Authors and Affiliations
Contributions
B.W., W.C. and F.C. wrote the main manuscript text. B.W., W.C., Y.S. and T.C. analyzed the data. F.C. supervised the research activities. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wang, B., Shao, Y., Chen, T. et al. Global insights into acetic acid resistance mechanisms and genetic stability of Acetobacter pasteurianus strains by comparative genomics. Sci Rep 5, 18330 (2015). https://doi.org/10.1038/srep18330
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep18330
- Springer Nature Limited
This article is cited by
-
Genomic Plasticity of Acid-Tolerant Phenotypic Evolution in Acetobacter pasteurianus
Applied Biochemistry and Biotechnology (2023)
-
Comparative pangenomic analyses and biotechnological potential of cocoa-related Acetobacter senegalensis strains
Antonie van Leeuwenhoek (2022)
-
Classification of acetic acid bacteria and their acid resistant mechanism
AMB Express (2021)
-
Enhanced alcohol degradation and hepatic protective effects of an Acetobacter Pasteurianus-derived product, CureZyme-ACE, in an acute intoxication rat model
Laboratory Animal Research (2020)
-
Transcriptome response of Acetobacter pasteurianus Ab3 to high acetic acid stress during vinegar production
Applied Microbiology and Biotechnology (2020)