Abstract
Phage display is a powerful technology that selects specific proteins or peptides to a target. We have used Phage Display to select scFv (single-chain variable fragment) clones from a combinatorial library against total proteins of Strongyloides venezuelensis. After scFv characterization, further analysis demonstrated that this recombinant fragment of antibody was able to bind to an S. venezuelensis antigenic fraction of ~65 kDa, present in the body periphery and digestive system of infective larvae (L3), as demonstrated by immunofluorescence. Mass spectrometry results followed by bioinformatics analysis showed that this antigenic fraction was a heat shock protein 60 (HSP60) of Strongyloides sp. The selected scFv was applied in serodiagnosis by immune complexes detection in serum samples from individuals with strongyloidiasis using a sandwich enzyme-linked immunosorbent assay (ELISA), showing sensitivity of 97.5% (86.84–99.94), specificity of 98.81 (93.54–99.97), positive likelihood ratio of 81.60 and an area under the curve of 0.9993 (0.9973–1.000). Our study provided a novel monoclonal scFv antibody fragment which specifically bound to HSP60 of Strongyloides sp. and was applied in the development of an innovative serodiagnosis method for the human strongyloidiasis.
Similar content being viewed by others
Introduction
Human strongyloidiasis is a neglected condition with worldwide distribution1,2. Immunocompetent individuals usually have a self-limited infection, but the parasite may remain in the body of the individual for up to 10 years without causing infection or being diagnosed. In individuals with immunosuppression, the infection may become life threatening due to hyperinfection and parasite spread to other parts of the body3,4.
The major problem for the serodiagnosis of human strongyloidiasis is caused the difficulty in obtaining larvae of Strongyloides stercoralis for antigen preparation. Because of this difficulty, heterologous antigens from Strongyloides venezuelensis have been used for convenience, due to similarities in transcripts that have key roles in the host-parasite interaction, as well as important molecules for diagnosis such as excretory/secretory proteins. Furthermore, a crude extract is routinely used, which results in cross-reactions with other parasitic infections5.
Phage display is a method which provides the selection of peptides, antibodies or single-chain variable fragment (scFv), expressed on bacteriophages, by standard methods with shorter time of production and important applicability in diagnosis due high specificity of the selected molecule to the target6,7,8,9. An scFv or specific-target antibody fragments represents the smallest functional heavy (VH) and light (VL) chains domain of an antibody10,11.
Here we report a strategy to select scFv clones from a combinatorial phage library against Strongyloides venezuelesis total proteins. The structure of the selected scFv and its binding was characterized by bioinformatics tools and the functionality of this novel scFv was characterized by development of a new serodiagnosis method.
Results
Selection, DNA sequencing, bioinformatic in silico analysis and purification of scFv
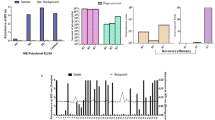
After two cycles of selection against S. venezuelensis total proteins, the selection efficiency was determined. Four out of 96 scFv clones that were expressed have bound to total proteins of S. venezuelensis as demonstrated by ELISA values and clones were named A4, B4, H2 and H3 (Fig. 1A). The nucleotide sequences obtained were subjected to the IgBLAST program to obtain their amino acid sequences and characterize the scFv light and heavy chains, considering both the conserved framework regions (FR1, FR2 and FR3) and the variable complementarity determining regions (CDR1, CDR2 and CDR3). The four clones presented the same amino acid sequence, which was submitted to the Raptor x and PyMOL programs, which are in silico prediction tools, to obtain its 3D structure and identify the CDR regions (Fig. 1B,C). After medium-scale production of the selected clone, the scFv molecules were purified by HPLC over a His-Trap column. Two milliliters of scFv (750 μg/mL) were obtained. The dot blot assay confirmed the expression and efficiency of scFv purification.
Expression and reactivity of scFv clones by ELISA at 492 nm and 3D structure of scFv.
(A) Expression of scFv clones induced with isopropyl β-D-thiogalactopiranoside (IPTG) after Escherichia coli TOP10 transformation and reactivity of scFv clones against Strongyloides venezuelensis total proteins. (B) The 3D structure of the scFv molecule and (C) the predicted antigen-binding site, both analyzed by the PyMOL online tool. (D) Deduced amino acid sequence from CDR regions. Ir: Irrelevant scFv, CDR: complementarity determining region; VH and VL: heavy and light chains, respectively.
Pull-down assay, immunofluorescence antibody test (IFAT) and mass spectrometry
These tests were carried out to identify and characterize the S. venezuelesis antigen that was targeted and bound by the scFv. The 15% SDS-PAGE silver stained, after pull-down assay, showed the profiles of the purified scFv (~29 kDa) and the S. venezuelesis antigenic fraction of ~65 kDa that bound to the scFv (Fig. 2A). Furthermore, the scFv bound to the body periphery and digestive system (arrow), specifically intestine (Fig. 2B) and esophagus (Fig. 2C), from S. venezuelensis infective larvae (L3), as evidenced by IFAT. Taenia solium metacestodes showed no staining by anti-HA-FITC, but only the red color conferred by counterstaining with 2% Evans blue (Fig. 2D). The antigenic fraction was further stained with Coomassie colloidal blue, trypsinized and characterized by mass spectrometry (CID-MS/MS). A BLAST search showed that this antigenic fraction was a heat shock protein 60 (HSP60) of Strongyloides sp. [GenBank:ABY65231.1 and Nematode.net: kq35f01.y1].
Identification and characterization of the antigenic fraction of Strongyloides venezuelensis that bound to the scFv.
(A) Electrophoretic profiles of total proteins detected by silver stained 15% SDS-PAGE after pull-down assay and analyzed by the Image J program. Staining profile of scFv in the body periphery and digestive system (arrow), specifically (B) intestine and (C) esophagus, from S. venezuelensis infective larvae (L3), by immunofluorescence using anti-HA-FITC. (D) Taenia solium metacestode was used as a negative control of staining. MW: molecular weight standard in kilodaltons (kDa); S. v.: Strongyloides venezuelensis; E. coli: Escherichia coli.
Bioinformatic in silico analysis between scFv and HSP60
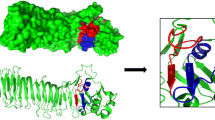
Docking between scFv and HSP60 was performed to determine possible binding regions by in silico prediction tools. The most stable 3D structure of the scFv-HSP60 was submitted and analyzed by the PyMOL online tool and identified the putative CDR regions that are responsible for the antibody binding to the HSP60 epitope with a strong association between them (Fig. 3A, B and C). The binding site analysis also showed that only the CDR2 of the light chain had no binding region to the HSP60 (Fig. 3D,E).
3D structure of scFv binding site on Strongyloides sp. HSP60 analyzed by the PyMOL online tool.
(A) Structures demonstrated the CDR regions of the scFv associated to Strongyloides sp. HPS60 (GenBank:ABY65231.1). (B) Staining of scFv binding site on Strongyloides sp. HSP60 and the amino acids of CDR regions that bound to Strongyloides sp. HSP60 in both (C) frontal and (D) apical views. (E) Amino acids from CRD regions that bound to the HSP60. CDR: complementarity determining region; HSP: heat shock protein.
Sandwich ELISA for immune complexes detection in human sera
The scFv was immobilized in high affinity microtiter plates for the serological detection of immune complexes. The TG-ROC curve obtained the best cut-off (0.7175) (Fig. 4A). The data of reactivity index demonstrated a significant discrimination between sera from S. stercoralis positive individuals (G1) in relation to serum samples from individuals that were positive for other parasitic diseases (G2) and healthy controls (G3) (P < 0.001) (Fig. 4B). Diagnostic parameters of sensitivity and specificity were 97.5% (86.84 - 99.94) and 98.81% (93.54 - 99.97), respectively. The high efficiency of the test was further demonstrated by the significant positive likelihood ratio (LR+ = 81.90) and area under the curve (AUC = 0.9993) (Fig. 4C).
Sandwich ELISA using scFv to detect immune complexes in serum samples.
(A) TG-ROC indicating the optimal cut-off of the method. (B) Reactivity indices of sera from individuals with strongyloidiasis (G1, n = 40), other parasitic diseases (G2, n = 44) and healthy individuals (G3, n = 40). (C) ROC curve showing sensitivity (Se), specificity (Sp), positive likelihood ratio (LR+) and area under the curve (AUC) (P < 0.001). The horizontal lines separating the data represent the median and dashed line indicates the threshold of the reactivity index.
Discussion
The engineering of novel functional combinatorial antibodies expressed on the surface of bacteriophages, such as scFvs, with high specificity for antigens coupled with a phage display strategy is the most promising technology for improved diagnostics and therapeutics of many pathologies10,11. This is due to their small size, great reactivity and specificity and easiness to maintain and express in bacteria, which surpass the hybridoma technology in all aspects. The scFv selected in this study was structurally characterized and applied in the immune complexes detection using human serum samples positive for strongyloidiasis. This diagnostic strategy is an important tool because it is indicative of active infection12,13,14,15.
In this study, we have selected an scFv clone through Phage Display against total proteins of S. venezuelensis as a model for S. stercoralis infection in humans, with a successful application in the serodiagnosis of human strongyloidiasis. Our novel scFv showed strong reactivity to a parasite protein fraction of ~65 kDa from S. venezuelensis, which further identified and characterized to be the heat shock protein 60 (HSP60) from Strongyloides sp., a 64 kDa protein shown in experimental models to have relevance to the parasite-host relationship, with potential significance for the host immune response16. This newly scFv was localized by immunofluorescence staining in the entire parasite body periphery and to regions of the digestive system from the infective larvae. Moreover, the scFv did not bind to T. solium metacestode, indicating specificity.
The scFv functionality was confirmed by a sandwich ELISA using this antibody fragment to detect immune complexes in serum samples from individuals with strongyloidiasis. The TG-ROC curve provided the optimum cut-off value and ROC curve confirmed its high sensitivity and specificity, both values higher than 97%. The test performance indicated by the high LR+ (81.9) and the AUC value close to the maximum (1.00) conferred a high diagnostic value. The assay was able to discriminate all positive serum samples from patients with strongyloidiasis in relation to sera from healthy individuals and with other parasitic diseases, except one false-positive result for an individual with Ascaris lumbricoides, suggesting a possible cross-reaction. However, the very low reactivity indices for the other seven positive sera for A. lumbricoides that was below the threshold suggest a possible co-infection of A. lumbricoides and S. stercoralis, which was not detected by the conventional parasitological method.
To date there have been few studies in the literature employing scFv to diagnose parasitic diseases. The most recent articles have demonstrated its important applicability in the detection of antigenic surface components of Schistosoma mansoni17 and the selection of antigens for diagnosis of neurocysticercosis18. Moreover, there is only one serological assay based on ELISA for the detection of immune complexes in serum samples of individuals with strongyloidiasis19, but with a sensitivity of 93.3% and specificity of 86.1%. Unlike this later study, our present investigation used a phage display technology that enabled the in vitro production of antibody fragments with high affinity for the target molecule and, consequently, did not require animal models for their synthesis, dramatically reducing the time required to obtain the reagent.
Furthermore, this is the first description of the HSP60 as a diagnostic target for the human strongyloidiasis, with additional data on the epitope and paratope binding sites of the scFv-HSP60 by in silico prediction tools. This result confirms previous studies that described HSPs as major immune dominant antigens in several infections, in addition to their original role as molecular chaperones. It has been demonstrated that HSP60 represents a major component of Strongyloides ratti ESPs (excretory/secretory proteins) and presents functions as a target to the humoral and cellular immune response. Interestingly, the vaccination with alum-precipitated HSP60 or a passive immunization with a monoclonal anti-HSP60 IgM antibody, both specifics for Strongyloides ratti, conferred protection to mice against a challenge infection20,21.
This is the first study that has successfully selected and used an scFv biomarker for the diagnosis of human strongyloidiasis, which was applied in the detection of immune complexes in serum samples of patients with great accuracy, aiming the newly found HSP60 as the target antigen. The serological detection of circulating immune complexes of this helminthiasis is indicative of infection, regardless the serum antibody titers and the release of larvae in feces. The development of a better, powerful and innovative diagnostic tool is of public health importance, since strongyloidiasis may be fatal, particularly in immunocompromised individuals.
Methods
Ethics statement
All experimental procedures were performed in accordance with the ethical guidelines of the Brazilian Health Ministry and approved in 2013 (protocol number 307.605) by the Research Ethics Committee from the Federal University of Uberlândia (CEP-UFU), State of Minas Gerais, Brazil. Serum samples from individuals were obtained at the Clinics’ Hospital of the Federal University of Uberlândia (UFU). These samples were based on archived anonymous adult human sera (18–84 years of age) of both genders and were collected in 2009 in the Reference Laboratory for Parasitology Diagnosis at the Federal University of Uberlândia (UFU).
Serum samples
All serum samples coming from endemic areas for strongyloidiasis obtained from individuals undergoing parasitological stool examinations22,23 by two independent technical experts in parasitological analysis. These tests were performed with three samples from each individual for parasite identification. Serum samples from 124 individuals were used and divided into 3 groups: Group 1 (G1) – 40 serum samples from individuals infected only with strongyloidiasis; Group 2 (G2) – 44 serum samples from individuals positive by single infection of other intestinal parasitic diseases including Ascaris lumbricoides (n = 8), hookworm (n = 7), Enterobius vermicularis (n = 5), Trichuris trichiura (n = 5), Schistosoma mansoni (n = 4), Hymenolepis nana (n = 4), Taenia sp. (n = 6) and Giardia lamblia (n = 5); Group 3 (G3) – 40 serum samples from healthy individuals, based on their clinical observation, without contact or previous infection by Strongyloides and three fecal samples tested negative.
Parasites maintenance and preparation
The Strongyloides venezuelensis strain was maintained in Wistar rats (Rattus norvegiccus) at the animal research facility (CBEA - Centro de Bioterismo e Experimentação Animal) of UFU. Animals were subcutaneously inoculated with infective larvae (L3). After 7 days of infection, feces were collected for charcoal culture preparation. After 3 days of culture at 27 °C in BOD (Biochemical Oxygen Demand), the L3 were collected24, washed 3 times in 0.01 M phosphate-buffered saline (PBS) (pH 7.2), centrifuged at 350 × g for 5 min and stored at -20 °C until use.
Preparation of total proteins from Strongyloides venezuelensis
Strongyloides venezuelensis total proteins were isolated using L3 larvae25. Larvae were resuspended in 0.01 M PBS (pH 7.2) containing protease inhibitors and disrupted with five cycles of freezing (1 min, -196 °C) and thawing/sonication (5 min, 40 kHz, 4 °C) (Thornton, Impec Eletrônica, São Paulo, Brazil). After overnight incubation at 4 °C with constant agitation, the suspension was centrifuged at 12,400 × g for 30 min at 4 °C. Protein quantification of the supernatant was performed26 and the extract was stored at -20 °C until use.
Selection of scFvs clones against total proteins of S. venezuelensis
The selection of scFv clones was performed using a human scFv phage library, with approximately 2 × 106 combinatorial sequences with variability for multiple diseases27, against total proteins of S. venezuelensis as target. Two cycles of selection were performed18, being preceded by scFv library reamplification in the competent Escherichia coli XL1-Blue strain and infection by VCSM13 helper phage for assembly and replication of viral proteins28. A microtiter plate well (Nunc MaxiSorpTM) was coated with 50 μL of S. venezuelensis total proteins (1 μg/well) diluted in 0.1 M sodium bicarbonate buffer (pH 8.6) and incubated for 18 h at 4 °C. The well was then blocked with 250 μL PBS/0.05% Tween 20 (PBST) (v/v) and 5% BSA (w/v) for 1 h at 37 °C and washed three times with PBS. Then, 100 μL of the library was added to the well and the plate was incubated for 1 h at 37 °C. Subsequently, the plate was washed 10 times with PBS/0.1% Tween 20 and bound phages to total proteins were eluted with 100 μL of 100 mM glycine-HCl (pH 2.2), followed by neutralization with 16.5 μL of 2 M Tris (pH 9.1). The resulting phages from the first selection were reamplified in E. coli XL1-Blue and a second round of selection was then performed as described above.
Expression of scFv in E. coli TOP 10
Plasmids were extracted from Escherichia coli XL1-Blue bacterial cells infected in the 2nd cycle of selection for further transformation in E. coli TOP 10, a non-suppressor strain28. Aliquots of the transformed cells were plated on LB (Luria-Bertani) agar containing carbenicillin (100 μg/mL). Plates were incubated at 37 °C for 16 h to allow growth of recombinant colonies. Each colony was transferred to a well in a deep well plate containing 1 mL of Super broth (SB) medium containing carbenicillin (100 μg/mL) and 2% 2 M glucose (v/v). After centrifugation, the expression of soluble scFv was induced by incubation with isopropyl β-D-thiogalactopiranoside (IPTG) (Sigma, USA) to a final concentration of 2.5 mM and carbenicillin (100 μg/mL) overnight at 250 × g and 30 °C. The plates were centrifuged at 4000 × g for 15 min at 4 °C and the supernatant containing soluble scFv was transferred to another 96-well plate and stored at 4 °C.28.
Analysis of expression and specificity of scFv clones
For analysis of scFv expression, a microtiter plate was coated with suspensions of 96 scFv clones. For analysis of scFv specificity, total proteins of S. venezuelensis (10 μg/mL) were immobilized in a micotiter plate and the suspension of 96 scFv clones was added. In both cases anti-HA (tag) peroxidase conjugated (1:1000) (Roche, Switzerland) was added. Reactions were revealed by addition of substrate (30% H2O2) and the OPD chromogen (orthophenylenodiamine) diluted in 0.1 M citrate-phosphate (pH 5.0). The optical densities were determined at 492 nm in an ELISA reader (Titertek Plus, Flow Laboratories, USA). An irrelevant scFv clone, from the same library but selected against another target, was used as a negative control.
DNA sequencing and bioinformatics analysis
The sequencing reactions were performed with the DyEnamic ET Dye Terminator Cycle Sequencing Kit (GE Healthcare Life Sciences, USA) and the following primers: MMB4 (5’- GCT TCC GGC TCG TAT GTT GTG T-3’) for the light chain and MMB5 (5’-CGT TTG CCA TCT TTT CAT AAT C-3’) for the heavy chain. The samples were sequenced using an automatic capillary sequencer (MegaBACE 1000 Genetic Analyzer, Amersham Biosciences, USA) and deduced amino acid sequences were then analyzed by IgBlast at NCBI (www.ncbi.nlm.nih.gov/igblast). Furthermore, sequences were submitted to the Raptor-x (http://raptorx.uchicago.edu/) and PyMOL (http://www.pymol.org/) bioinformatics online tools to obtain and analyze the 3D structure of the scFv molecule.
Medium-scale production of soluble scFv and purification of selected scFv by HPLC
The expression of the selected clone was produced in a 1000 mL Erlenmeyer cell culture flask, as described above (expression of scFv section). For medium-scale purification, His6-tagged scFv fragments were purified by immobilized-metal (Nickel) affinity chromatography (HisTrapTM HP, GE Healthcare Life Sciences, USA) according to the manufacturer’s instructions in a HPLC system (AKTATM purifier). The purified scFv was concentrated using a 10 kDa Amicon apparatus (Millipore, USA), the protein concentration was determined26 and a dot-blot assay of the purified scFv was performed18 to confirm the efficiency of the purification process.
Pull-down assay
To characterize the specific protein target that bound to the scFv from the extract of total proteins of S. venezuelensis, 100 μL of mouse anti-His mAb Mag Beads (GenScript Transforming Biology Research, USA) were incubated with 100 μL of purified scFv according to the manufacturer´s instructions. After washing, 100 μL of S. venezuelensis total proteins (1000 μg/mL) were added and, after incubation and additional washings, 20 μL of the SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) sample buffer were added and the tube was incubated at 100 °C for 5 min. Proteins were eluted from beads using a magnetic apparatus and the supernatant was submitted to a 15% SDS-PAGE. Proteins were visualized by silver staining29 and the electrophoretic profile was determined by Image J 1.44 (National Institute of Health, Bethesda, EUA).
Immunofluorescence antibody test (IFAT)
Strongyloides venezuelensis larvae were embedded in Tissue-tek (Tissue Freezing Medium, Durham, NC, USA) and frozen at -70 °C. Sections of 2 μm in thickness were cut using a cryomicrotome and placed on microscope slides. The sections were left at room temperature for 30 min to dry. Twenty microliters of scFv (750 μg/mL) was added to each section and the slides were incubated in a humidified chamber at 37 °C for 1 h. The sections were incubated with fluorescein isothiocyanate conjugated (FITC) mouse anti-HA (1:1000) and counterstained with 2% Evans blue for 40 min at 37 °C. Between each step three washes were performed in PBS. The slides were mounted using glycerol/PBS (pH 9.0) and coverslips. The staining profile of the scFv on the larvae was verified using a LSM 510 confocal microscope (Meta, Carl Zeiss, Germany). A section of Taenia solium metacestode was used as a negative control for staining.
Mass spectrometry (CID-MS/MS) and bioinformatic analyses
For mass spectrometry, the pull-down assay was repeated; however, the gel was staining with Comassie blue G-250 and the band related to the protein of S. venezuelensis that bound to the scFv was trypsinized and submitted to CID-MS/MS30 (Quattro II, Micromass, Manchester, UK). Peptide fragments obtained were submitted and analyzed by BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) in relation to their similarity to proteins of Strongyloides sp. (taxid: 6247). Furthermore, the highest similar protein sequence was submitted to bioinformatics online tools. The Raptor-x (http://raptorx.uchicago.edu/) was used to obtain the 3D structure of the protein. For identification of potential binding regions of the protein and scFv it was performed a docking (http://bioinfo3d.cs.tau.ac.il/PatchDock/). The regions of scFv-protein structure were selected and analyzed by PyMOL (http://www.pymol.org/).
Sandwich ELISA for immune complexes detection in human sera
High affinity microtiter plates (Nunc MaxiSorpTM) were incubated with 50 μL of scFv (10 μg/mL) in carbonate bicarbonate buffer (0.06 M, pH 9.6), overnight at 4 °C. The plates were blocked with PBS/5% BSA for 45 min at 37 °C. Serum samples were diluted 1:50 in PBST, added to the wells and the plates were incubated for 45 min at 37 °C. Subsequently, mouse anti-human I gG peroxidase conjugated diluted in PBST (1:10,000) was added to the wells and the plates were incubated for 45 min at 37 °C. Between each step three washes were performed in PBST. The reaction was revealed by adding orthophenylenediamine (OPD) diluted in 0.1 M citrate-phosphate (pH 5.0) and 30% H2O2. Plates were incubated for 15 min at room temperature and reactions were stopped by adding 2N H2SO4. The optical densities (OD) were determined at 492 nm in an ELISA reader (Titertek Plus, Flow Laboratories, USA). The cut-off value was determined by the TG-ROC curve31, based on the ELISA results. The ELISA reactivity index (RI) was obtained from the ratio: OD of each sample/cut-off value; RI values > 1 were considered positive.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5.0 (GraphPad Software Inc., San Diego, USA). The sensitivity (Se), specificity (Sp) and positive likelihood ratio (LR+) were calculated according to the following formulas: Se (%) = a x 100/(a + c); Sp (%) = d x 100/(b + d); LR + = Se/(1 - Sp) where, a: true positive; b: false positive; c: false negative; d: true negative32. The area under the curve (AUC) was obtained from the ROC curve in order to determine the diagnostic accuracy of the method33. Values were considered statistically significant when p < 0.05 and 95% confidence intervals (CI) were provided for Se, Sp and AUC statistical calculations. The Analysis of Variance (ANOVA) followed by the Tukey post hoc was used to determine differences among groups.
Additional Information
How to cite this article: Levenhagen, M. A. et al. Structural and functional characterization of a novel scFv anti-HSP60 of Strongyloides sp. Sci. Rep. 5, 10447; doi: 10.1038/srep10447 (2015).
Change history
05 August 2015
A correction has been published and is appended to both the HTML and PDF versions of this paper. The error has been fixed in the paper.
References
Olsen, A. et al. Strongyloidiasis - the most neglected of the neglected tropical diseases? Trans R Soc Trop Med Hyg 103, 967–972 (2009).
Paula, F. M. & Costa-Cruz, J. M. Epidemiological aspects of strongyloidiasis in Brazil. Parasitology 138, 1331–1340 (2011).
Fardet, L., Généreau, T., Cabane, J. & Kettaneh, A. Severe strongyloidiasis in corticosteroid-treated patients. Clin Microbiol Infect 12, 945–947 (2006).
Marcos, L. A., Terashima, A., Canales, M. & Gotuzzo, E. Update on strongyloidiasis in the immunocompromised host. Curr Infect Dis Rep 13, 35–46 (2011).
Levenhagen, M. A. & Costa-Cruz, J. M. Update on immunologic and molecular diagnosis of human strongyloidiasis. Acta Trop 135, 33–43 (2014).
Hoogenboom, H. R. Selecting and screening recombinant antibody libraries. Nat Biotechnol 23, 1105–1116 (2005).
Santos, P. S. et al. Epitope-based vaccines with the Anaplasma marginale MSP1a functional motif induce a balanced humoral and cellular immune response in mice. PLoS One 8, e60311 (2013).
Araújo, T. G. P. et al. A novel highly reactive Fab antibody for breast cancer tissue diagnostics and staging also discriminates a subset of good prognostic triple-negative breast cancers. Cancer let 343, 275–285 (2014).
Feliciano, N. D. et al. Bacteriophage-fused peptides for serodiagnosis of human strongyloidiasis. PLoS Negl Trop Dis 5, e2792 (2014).
Ahmad, Z. A. et al. scFv antibody: principles and clinical application. Clin Dev Immunol 2012, 980250 (2012).
Weisser, N. E. & Hall, J. C. Applications of single-chain variable fragment antibodies in therapeutics and diagnostics. Biotechnol Adv 27, 502–520 (2009).
Chaves-Borges, F. A., Souza, M. A., Silva, D. A., Kasper, L. H. & Mineo, J. R. Detection of Toxoplasma gondii soluble antigen, SAG-1 (p30), antibody and immune complex in the cerebrospinal fluid of HIV positive or negative individuals. Rev Inst Med Trop São Paulo 41, 329–338 (1999).
Raja, A., Uma Devi, K. R., Ramalingam, B. & Brennan, P. J. Improved diagnosis of pulmonary tuberculosis by detection of free and immune complex-bound anti-30kDa antibodies. Diagn Microbiol Infect Dis 50, 253–259 (2004).
de Carvalho, C. A. et al. A simple immune complex dissociation ELISA for leishmaniasis: standardization of the assay in experimental models and preliminary results in canine and human samples. Acta Trop 125, 128–136 (2013).
Emmerich, P., Mika, A. & Schmitz H. Detection of serotype-specific antibodies to the four dengue viruses using an immune complex binding (ICB) ELISA. PLoS Negl Trop Dis 7, e2580 (2013).
Tazir, Y. et al. Molecular and functional characterisation of the heat shock protein 10 of Strongyloides ratti. Mol Biochem Parasitol 168, 149–157 (2009).
Sepulveda, J., Tremblay, J. M., DeGnore, J. P., Skelly, P. J. & Shoemaker, C. B. Schistosoma mansoni host-exposed surface antigens characterized by sera and recombinant antibodies from schistosomiasis-resistant rats. Int J Parasitol 40, 1407–1417 (2010).
Ribeiro, V da S. et al. Development of specific scFv antibodies to detect neurocystiscercosis antigens and potential applications in immunodiagnosis. Immunol Lett 156, 59–67 (2013).
Gonçalves, A. L. R., Nunes, D. S., Gonçalves-Pires, M. R. F., Ueta, M. T. & Costa-Cruz, J. M. Use of larval, parasitic female and egg antigens from Strongyloides venezuelensis to detect parasite-specific IgG and immune complexes in immunodiagnosis of human strongyloidiasis. Parasitology 139, 956–961 (2012).
Nouir, N. B. et al. Vaccination with Strongyloides ratti heat shock protein 60 increases susceptibility to challenge infection by induction of Th1 response. Vaccine 30, 862–871 (2012).
Nouir, N. B., Piédavent, M., Osterloh, A. & Breloer, M. Passive immunization with a monoclonal IgM antibody specific for Strongyloides ratti HSP60 protects mice against challenge infection. Vaccine 30, 4971–4976 (2012).
Baermann, G. Eine Einfache Methode Zur Auffindung von Ankylostomum (Nematoden) Larven in Erdproben. Mededeel mit. h. Geneesk, Lab Weltevreden Feestbundel, Batavia, 41–47 (1917).
Lutz, A. Schistosomun mansoni e a esquistosomose segundo observações feitas no Brasil. Mem Inst Oswaldo Cruz 11, 121–155 (1919).
Rugai, E., Mattos, T. & Brisola, A. P. Nova técnica para isolar larvas de nematoides das fezes: modificação do método de Baermann. Rev Inst Adolfo Lutz 14, 5–8 (1954).
Gonzaga, H. T., Ribeiro, V da S., Cunha-Júnior, J. P., Ueta, M. T. & Costa-Cruz, J. M. Usefulness of concanavalin-A non-binding fraction of Strongyloides venezuelensis larvae to detect IgG and IgA in human strongyloidiasis. Diagn Microbiol Infect Dis 70, 78–84 (2011).
Lowry, O. H., Rosebrough, N. J., Farr, A. L. & Randall, R. J. Protein measurement with the Folin phenol reagent. J Biol Chem 29, 265–275 (1951).
Carneiro, A. P. et al. A putative OTU domain-containing protein 1 deubiquitinating enzyme is differentially expressed in thyroid cancer and identifies less-aggressive tumors. Br J Cancer 111, 551–558 (2014).
Barbas III, C. F., Burton, D. R., Scott, J. K. & Silvermam, G. J. Phage display: A laboratory manual. [Barbas III, et al.] [738p.] (Cold Spring Harbor Laboratory Press, New York, 2001).
Friedman, R. D. Comparison of four different silver-staining techniques for salivary protein detection in alkaline polyacrylamide gels. Anal Biochem 126, 346–349 (1982).
Pranchevicius, M. C. S. et al. Characterization and optimization of Artin M lectin expression in Escherichia coli. BMC Biotechnol 12, 44–52 (2012).
Greiner, M., Sohr, D. & Göbel, P. A. A modified ROC analysis for the selection of cut-off values and the definition of intermediate results of serodiagnostic tests. J Immunol Methods 185, 123–132 (1995).
Youden, D. Index for rating diagnostic tests. Cancer 3, 32–35 (1950).
Martinez, E. Z., Louzada-Neto, F. & Pereira, B. B. Analysis of diagnostic tests using ROC curves. Cad Saúde Coletiva 11, 7–31 (2003).
Acknowledgements
This work was supported by the Brazilian funding agencies, Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq (grant N. 302426/2012–4 and 490574/2010–6), CAPES (Rede Nanobiotec/Brasil Project N. 8) and Fundação de Amparo à Pesquisa do Estado de Minas Gerais – FAPEMIG (grant N. CBB-PPM-00396–13 and the Excellence Center Program - PRONEX – APQ 02413–08). We thank Dr. José César Rosa, PhD from Center of Proteins Chemistry, São Paulo University, Ribeirão Preto, for mass spectrometry support.
Author information
Authors and Affiliations
Contributions
M.A.L., F.d.A.A.S., P.T.F., J.M.C.C., L.R.G: Conceived and designed the experiments. M.A.L., F.d.A.A.S., P.T.F: Performed the experiments. M.A.L., F.d.A.A.S., P.T.F., J.M.C.C., L.R.G: Analyzed the data. A.P.C., J.M.C.C., L.R.G: Contributed reagents/materials/analyses tools. M.A.L., J.M.C.C., L.R.G: Wrote the manuscript. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Levenhagen, M., de Almeida Araújo Santos, F., Fujimura, P. et al. Structural and functional characterization of a novel scFv anti-HSP60 of Strongyloides sp.. Sci Rep 5, 10447 (2015). https://doi.org/10.1038/srep10447
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep10447
- Springer Nature Limited
This article is cited by
-
Broad specificity of immune helminth scFv library to identify monoclonal antibodies targeting Strongyloides
Scientific Reports (2021)
-
Direct detection of Strongyloides infection via molecular and antigen detection methods
European Journal of Clinical Microbiology & Infectious Diseases (2021)
-
Upregulation of tropomyosin alpha-4 chain in patients’ saliva with oral squamous cell carcinoma as demonstrated by Phage display
Scientific Reports (2019)
-
eEF1A1 binds and enriches protoporphyrin IX in cancer cells in 5-aminolevulinic acid based photodynamic therapy
Scientific Reports (2016)