Abstract
Background
Spinal muscular atrophy (SMA) is a fatal autosomal recessive disorder for which several treatment options, including a gene therapy, have become available. SMA incidence has not been well-characterized in most Arab countries where rates of consanguinity are high. Understanding SMA disease epidemiology has important implications for screening, prevention, and treatment in those populations.
Methods
We perform SMA diagnostic testing in a clinical multi-national patient cohort (N = 171) referred for hypotonia and/or muscle weakness. In addition, we carry out genetic newborn screening for SMA on 1502 healthy Emirati newborns to estimate the carrier frequency and incidence of the disease in the United Arab Emirates.
Results
Patients referred for SMA genetic testing are mostly Arabs (82%) representing 18 countries. The overall diagnostic yield is 33.9%, which is higher (>50%) for certain nationalities. Most patients (71%) has two SMN2 copies and earlier disease onset. For the first time, we estimate SMA carrier frequency (1.3%) and incidence of the disease (1 in 7122 live births) in the United Arab Emirates. Using birth and marriage rates in two Arab populations (United Arab Emirates and Saudi Arabia), as well as disease incidence in both countries, we show that, besides preventing new cases, premarital genetic screening could potentially result in around $8 to $324 million annual cost savings, respectively, relative to postnatal treatment.
Conclusions
The SMA carrier frequency and incidence we document suggests high potential benefit for universal implementation of premarital genomic screening for a wide range of recessive disorders in Arab populations.
Plain language summary
The occurrence of spinal muscular atrophy, a fatal genetic nerve and muscle disease, has been poorly studied in most Arab countries. Individuals who carry a single mutated gene copy (carriers) may be more likely to marry other carriers in regions where marriage rates amongst relatives, who share similar genetics, are high. Here we report the results of a newborn testing program for this disease in 1502 Emiratis and calculate the presence of carriers (1/79) and occurrence of disease (1/7122) in this population. Using this new information along with the annual birth and marriage rates in the United Arab Emirates and Saudi Arabia, we make the case that premarital genomic screening (carrier testing) is the best way to prevent this and other similarly inherited disorders in the Arab population.
Similar content being viewed by others
Introduction
Spinal muscular atrophy (SMA) is a rare genetic neuromuscular disorder characterized by irreversible loss of alpha motor neurons in the spinal cord and lower brainstem resulting in progressive muscle atrophy and weakness1. Its clinical manifestation is highly variable, ranging in onset from intrauterine to adulthood, and in severity from minor muscle weakness to severe paralysis and death. Accordingly, SMA has been classified into four primary subtypes: type I, type II, type III and type IV. Type I, known as infantile SMA, is the most common and severe form of the disease, seen in ~50%2 of affected patients and has an onset before 6 months of age. If untreated, death generally occurs within the first 2 years of life due to respiratory failure3,4.
SMA is an autosomal recessive disorder caused by biallelic pathogenic variants in the telomeric survival motor neuron 1 (SMN1) gene located on chromosome 5q135. Most affected patients (92%) harbor a homozygous deletion of SMN1 exon 7, while a small subset (4%) are compound heterozygotes for exon 7 deletion on one allele coupled with a small intragenic variant on the other allele6. In rare occasions (4%), SMA can be due to over 30 genes identified, other than SMN13,6,7. Though an almost identical homolog to SMN1, SMN2 can only partially compensate for SMN1 loss. Increase in SMN2 copy number has been associated with milder forms of SMA and, therefore, has prognostic value. Of patients with 4 copies of SMN2, 1% of patients had SMA type I, 11% had SMA type II and 88% had SMA type III or type IV4,7.
SMA is globally the second most common fatal autosomal recessive disorder. Based on available data in populations of European ancestry, SMA highest incidence and carrier frequency are around 1 in 8000 and 1 in 45, respectively6,7. These figures are expected to be even higher in regions with high rates of consanguinity, particularly in the Middle East where the incidence has been predicted to range between 10 and 193 in 100,000 live births, almost 40-fold higher than in Western populations8. However, there is a lack of data to accurately estimate SMA epidemiology across Arab populations. Here we estimate the carrier frequency and incidence of SMA in the United Arab Emirates (UAE) and, using this information, we show that premarital genomic screening is the most efficient public health measure for SMA, and other recessive diseases, prevention in Middle Eastern population.
Methods
Study design and participants
Clinical patient cohort
Pediatric patients (age < 18 years old) were referred for diagnostic SMA testing at the Genomics Centre in Al Jalila Childrens’ Specialty Hospital between June 2019 and August 2023. Al Jalila Children’s is a tertiary pediatric center in the UAE and is the main referral center for pediatric patients with rare diseases across the UAE. Peripheral blood samples were obtained from patients suspected with SMA and clinical data was provided by the referring physician either through the electronic medical record, for internal patients, or on requisition forms, for external patients. The de-identified reporting of clinical cohort in this study was approved by the Dubai Healthcare Authority Research Ethics Committee (DSREC-07/203_06), which determined that this study meets the exemption criteria with a waiver of informed consent since all data were de-identified. A subset of this clinical cohort was previously published9, though in addition to increasing cohort size, this current study includes additional information on age ranges, diagnostic yield stratified by nationality, and SMN2 copy number status. Diagnostic yield was calculated by dividing the number of patients positive for the biallelic deletion of the SMN1 gene by the total number of tested patients.
SMA genetic screening initiative in Emirati Newborn
A network of ten local maternity hospitals across the UAE was established to conduct the country’s first large scale representative genetic newborn screening study for SMA. Recruitment and consenting of Emirati newborns’ (age ≤ 3 months) families were carried out by neonatologists/pediatricians at each hospital. Peripheral blood samples were obtained from each participant and clinical data were provided by the recruiting physician through an electronic collection data sheet. Samples were transported to Al Jalila Genomics Centre where SMA testing was performed. The study was approved by the Dubai Health Authority Scientific Research Ethics Committee (DSREC-06/2021_20), Ministry of Health and Prevention Research Ethics Committee (MOHAP/DXB-REC/SOO/No.81/2021) and Abu Dhabi Health Research and Technology Ethics Committee (DOH/CVDC/2022/1626). Parents or legal guardians of all recruited newborns provided written informed consent.
DNA extraction
Genomic DNA was extracted from peripheral whole blood using the QIAsymphony DSP DNA Kit (Qiagen, Hilden, Germany) and QIAsymphony automated nucleic acid extraction instrument, according to the manufacturer’s instructions.
SMA copy number analysis
SMN1 and SMN2 copy numbers were determined by Digital droplet PCR (ddPCR) technology using predesigned proprietary ddPCR assay kits for SMN1 (Catalog No: 186-3500, Bio-Rad) and SMN2 (Catalog No: 186-3503, Bio-Rad). The predesigned Bio-Rad assays include reagents to detect both the target gene (SMN1 or SMN2) and an internal reference gene (RPP30). In addition, experimental controls—0 copy, 1 copy and 2 copy controls for SMN1, and 2 copy, 3 copy and 4 copy controls for SMN2—were included along with a no template control.
Data analysis was performed using QuantaSoft version 1.7.4.0917 (Bio-Rad) to determine the copy number variation (CNV)10,11 (Fig. 1). SMN2 copy number was determined only for SMA-positive cases with a homozygous SMN1 exon 7 deletion to assess disease severity and prognosis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results and discussion
SMA patient distribution and diagnostic yield in a multi-arab population
Between June 2019 and August 2023, 171 pediatric patients (51% females; mostly infants) (Fig. 2A) presenting with hypotonia and/or muscle weakness were referred to Al Jalila Genomics Centre for diagnostic SMA testing. Patients, representing 18 countries, were mostly Arabs (82%) with the majority from Saudi Arabia (29%), UAE (25%) and Iraq (10%) (Fig. 2B).
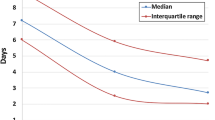
Testing was positive in 58 cases, making up an overall diagnostic yield of 33.9% (Fig. 2C), which varied by nationality (Fig. 2B); yield was significantly higher among patients from Iraq (53%) relative to the UAE (19%) (P = 0.012; Fisher Exact Test). Determination of SMN2 copy number in SMA positive cases showed that the majority (71%) had 2 SMN2 copies, consistent with an earlier onset (Fig. 2D) and a more likely severe phenotype (SMA type I)7,12. Average test turnaround time was 5 working days, with more than half (56%) receiving results in ≤4 working days (Fig. 2E).
SMA epidemiology in the United Arab Emirates
We attempted to estimate the incidence of SMA in the UAE using the number of Emirati SMA patients born in a given year, as diagnosed by our laboratory during this period, divided by total annual live births in this population (which was 31,064 in 2021)13. However, since not all SMA diagnostic testing in the UAE is referred to our laboratory and due to the lack of national SMA registry, it is likely that this clinical-based incidence estimation to be inaccurate.
To gain better insight into SMA epidemiology in the UAE, we genetically screened a total of 1502 newborns (51% males, 49% females) recruited from 10 local maternity hospitals across the country (Fig. 3A). Newborns aged ≤3 months and were generally healthy with no family history of SMA. Results from genetic screening revealed that 19 newborns (47% females, 53% males) were carriers for SMN1 deletion (Fig. 3B), indicating a carrier frequency of 1 in 79 (1.3%) and an SMA allele frequency (q) of 0.0063 (Table 1). None of the newborns tested had a homozygous SMN1 deletion.
Assuming Hardy–Weinberg equilibrium, disease burden or incidence (q2) can be calculated as 1 in 25,000 individuals. However, considering the non-random mating in this population, a more appropriate estimate for incidence would be q x f14, where F represents the coefficient of inbreeding in the Emirati population (F = 0.0222)15, resulting in an incidence of 1 in 7122 individuals (Table 1).
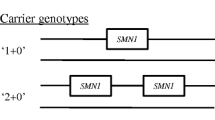
It is important to note that this is likely an underestimate since (1) our testing method only detects deletions that account for 92% of cases, while (2) this method does not distinguish silent carriers, who possess two copies of the SMN1 gene on one allele (0 + 2), from normal noncarriers (1 + 1).
SMA epidemiology in other Arab populations
Genetic newborn screening studies have been limited in most Arab countries. A study from Morocco reported a carrier frequency of 1 in 25 and a prevalence of 1:1900, though this was based on a small sample of 150 Moroccan newborns16. While this work was in preparation, a study was published whereby screening 4189 normal Saudi volunteers determined a carrier frequency of 1 in 3817. Using this SMA allele frequency, and the coefficient of inbreeding in Saudi Arabia18, we estimated SMA incidence in this country to be 1 in 3192 (Table 1), which is consistent with that estimated by authors in this study (32 in 100,000 or 1 in 3125 births)17. Those estimates suggest the SMA carrier frequency and incidence in Saudi Arabia are both higher than those estimated in the UAE; findings which might be consistent with the observed higher SMA diagnostic trend in Saudi patients compared to those in the UAE (Fig. 2B).
The case for premarital genomic screening
The tragic loss of a baby to SMA in Australia has fueled a large-scale reproductive carrier screening program for 750 genetic conditions (“Mackenzie’s Mission”) in Australia19. The estimated SMA incidence in both Arab countries studied here is higher than that in other populations worldwide7, a finding which is consistent with the expectedly higher incidence of recessive disorders in Arab populations, suggesting that premarital genetic screening is likely to be an effective preventive measure in those populations.
Based on the aforementioned estimate of SMA incidence in the UAE (Table 1) and the average annual Emirati birth rate, we conservatively anticipate that 4 births per year will be affected with SMA, necessitating gene therapy at a total cost of $8.4 million ($2.1 million per patient; other long term treatment options, such as Spinraza, have a higher cost). However, if SMA premarital screening is implemented, the projected total cost of screening Emirati marriages would be $0.49 million annually (Table 2). Couples at risk (N = 4) will have the option for pre-implementation genetic testing (PGT) and in vitro fertilization (IVF) at a cost of up to $30,000 per couple20 (Table 2).
Similarly, average annual birth count among Saudi Nationals (18.8 million) is 511,00021,22. Therefore, it is expected that 160 births will be affected with SMA per year, requiring $336 million for treatment. However, the cost of SMA premarital screening for the annual 141,000 Saudi marriages in KSA22, would be around $7 million, along with a total cost of $4.8 million in PGT/IVF for at risk couples (Table 2).
We estimate the disease burden or incidence of SMA in the UAE. Using data from the UAE (this study) and Saudi Arabia17, we demonstrate that, in addition to potentially preventing new SMA cases, premarital screening and subsequent PGT/IVF for at-risk couples, can be a highly cost-effective measure from a public health standpoint, leading to an expected 14 to 28-fold cost reduction compared to postnatal disease treatment. Interestingly, the cost savings due to premarital screening for this one disease (~$8 million in UAE and $324 million in KSA) is sufficient to fund more comprehensive premarital genomic screening programs encompassing hundreds of recessive disorders in both countries. We strongly advocate for the implementation of comprehensive genomic premarital screening in Arab populations with similarly high consanguinity rates.
Data availability
All data is included in the manuscript. Source data used to generate figures can be found in Supplementary Data 1.
References
Lunn, M. & Wang, C. Spinal muscular atrophy. Lancet 371, 2120–2133 (2008).
D’Amico, A. et al. Spinal muscular atrophy. Orphanet. J. Rare Dis. 6, 71 (2011).
Farrar, M. & Kiernan, M. The genetics of spinal muscular atrophy: progress and challenges. Neurotherapeutics 12, 290–302 (2014).
Keinath, M., Prior, D. & Prior, T. Spinal muscular atrophy: mutations, testing, and clinical relevance. Appl. Clin. Genetics 14, 11–25 (2021).
Gene—NCBI. Survival of Motor Neuron 1, Telomeric [Homo Sapiens (Human)]. https://www.ncbi.nlm.nih.gov/gene/6606 (2023).
Verhaart, I. E. C. et al. Prevalence, incidence and carrier frequency of 5q-linked spinal muscular atrophy—a literature review. Orphanet. J. Rare Dis. 4, 124 (2021).
Prior T. W. et al. (eds) Spinal Muscular Atrophy. In GeneReviews® [Internet]. (University of Washington, Seattle (WA), 2020). https://www.ncbi.nlm.nih.gov/sites/books/NBK1352/
Ali, H. G. et al. Gene therapy for spinal muscular atrophy: the Qatari experience. Gene Ther. 28, 676–680 (2021).
El Naofal, M. et al. The genomic landscape of rare disorders in the Middle East. Genome Med. 15, 5 (2023).
Tayoun, A. N. et al. Targeted droplet-digital PCR as a tool for novel deletion discovery at the DFNB1 locus. Hum. Mutat. 37, 119–126 (2015).
Amr, S. et al. Allele-specific droplet digital PCR combined with a next-generation sequencing-based algorithm for diagnostic copy number analysis in genes with high homology: proof of concept using stereocilin. Clin. Chem. 64, 705–714 (2018).
Calucho, M. et al. Correlation between SMA type and SMN2 copy number revisited: an analysis of 625 unrelated Spanish patients and a compilation of 2834 reported cases. Neuromuscul. Disord. 28, 208–215 (2018).
UAE.Stat. UAE.Stat. https://uaestat.fcsc.gov.ae/en (2023).
Abouelhoda, M. et al. Clinical genomics can facilitate countrywide estimation of autosomal recessive disease burden. Genet. Med. 18, 1244–1249 (2016).
al-Gazali, L. I. et al. Consanguineous marriages in the United Arab Emirates. J. Biosoc. Sci. 29, 491–497 (1997).
Lyahyai, J., Sbiti, A., Barkat, A., Ratbi, I. & Sefiani, A. Spinal muscular atrophy carrier frequency and estimated prevalence of the disease in Moroccan newborns. Genetic Testing Mol. Biomarkers. 16, 215–218 (2012).
Al Jumah, M. et al. Spinal muscular atrophy carrier frequency in Saudi Arabia. Mol. Genet. Genomic Med. 10, e2049 (2022).
el-Hazmi, M. A. et al. Consanguinity among the Saudi Arabian population. J. Med. Genet. 32, 623–626 (1995).
Archibald, A. D. et al. The Australian reproductive genetic carrier screening project (Mackenzie’s mission): design and implementation. J. Pers Med. 12, 1781 (2022).
Antero, M. F. et al. Cost-effectiveness of preimplementation genetic testing for aneuploidy for fresh donor oocyte cycles. F S Rep. 2, 36–42 (2021).
Saudi Census. Factors Influencing the Decision Process within Seeking Orthodontic Care among the Saudi Population: A Cross-sectional Survey. https://portal.saudicensus.sa/portal/public/1/15/45?type=DASHBOARD (2023).
GCC-STAT. Accessed December 14, 2023. https://dp.gccstat.org/en/DataAnalysis.
Acknowledgements
The spinal muscular atrophy pilot study is being supported by an unrestricted medical grant from Novartis/AveXis. We would like to thank Haneen Farajallah, Najeeb Muringakodan, Hanan Alraeesi, Saleh Ali, Adel Salem and Fatima AlKaabi from Emirates Health Services for supporting their respective teams in this project.
Author information
Authors and Affiliations
Contributions
A.A.T. conceived study and obtained funding. M.K., N.A., K.A., M.E., S.D., Y.S., A.A., S.A., K.F., J.S., and S.G.A. recruited and consented families for SMA genetic screening. F.R., M.E., I.C. and A.A.T. performed all analysis. A.A.T., F.A. and A.A.K. obtained ethical approvals. F.A., T.L. and A.A.A. helped with epidemiological analysis. F.R. and A.A.T. wrote first manuscript draft. All authors, including H.E., edited and approved the final draft.
Corresponding author
Ethics declarations
Competing interests
Authors declare no competing interests. Dr. Ahmad Abou Tayoun is an Editorial Board Member for Communications Medicine but was not involved in the editorial review or peer review, nor in the decision to publish this article.
Peer review
Peer review information
Communications Medicine thanks Peter Karachunski and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rabea, F., El Naofal, M., Chekroun, I. et al. Spinal muscular atrophy genetic epidemiology and the case for premarital genomic screening in Arab populations. Commun Med 4, 119 (2024). https://doi.org/10.1038/s43856-024-00548-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-024-00548-1
- Springer Nature Limited