Abstract
Neoscytalidium dimidiatum is the main causal agent of pitaya canker. Most studies of virulence and pathogenicity genes have measured expression levels using real-time quantitative polymerase chain reaction (RT-qPCR). Suitable reference genes are essential for ensuring that estimates of gene expression levels by RT-qPCR are accurate. However, no reference genes can be robustly applied across all contexts and species. No studies to date have evaluated the most effective reference genes for normalizing gene expression levels estimated by RT-qPCR in N. dimidiatum. In this study, RT-qPCR data for individual candidate reference genes were analyzed using four different methods: the delta Ct method and the geNorm, NormFinder, and BestKeeper algorithms. We evaluated the utility of eight candidate reference genes (18S rRNA, Actin (1), Actin (2), Actin, GAPDH (1), GAPDH (2), UBQ, and Tubulin) for normalizing expression levels estimated by RT-qPCR in N. dimidiatum at different developmental stages, at different temperatures, and during interaction with pitaya. All candidate reference genes were suitable for gene expression analysis except for Actin (2). Tubulin and Actin (1) were the most stably expressed reference genes under different temperatures. Actin (1) and Actin were the most stably expressed reference genes in N. dimidiatum at different developmental stages. Tubulin and UBQ were the most stably expressed reference genes during interaction with pitaya. Actin and 18s rRNA were the most stably expressed across all experimental conditions. Subsequently, Tubulin and UBQ were further investigated in analyses of pectinase expression during the pitaya–N. dimidiatum interaction. Our results provide insights that will aid future RT-qPCR studies of gene expression in N. dimidiatum.
Similar content being viewed by others
Introduction
Neoscytalidium dimidiatum is the main causal agent of pitaya canker in southern China1. Pitaya canker has spread rapidly worldwide and has resulted in major economic losses2. Pitaya canker has become one of the most serious diseases affecting pitaya production. Previously, we have shown that pitaya canker cannot be completely eliminated by chemical treatment with traditional fungicides because the pathogen penetrates the epicuticular wax of the host; consequently, once a plantation is infected with N. dimidiatum, ulcers can reappear when climate conditions are suitable3. The mechanisms underlying pitaya–N. dimidiatum interactions and pathogen infection remain unknown.
The plant cell wall, which represents the first protective barrier against pathogens, is a composite material made of cellulose microfibrils embedded in a matrix of hemicellulose and pectin4. Cell wall-degrading enzymes secreted by the fungus might play an important role during the infection process. For example, lytic polysaccharide monooxygenases can recognize and degrade pectin, and silencing of the genes encoding these enzymes in Phytophthora infestans inhibits infection of potato5. Clarifying the role of pectinases in N. dimidiatum infection requires gene expression analysis.
Real-time quantitative PCR (RT-qPCR) is the most used method for detecting gene expression patterns6,7,8. The selection of appropriate reference genes is important for ensuring that estimates of gene expression are accurate9, as the use of inappropriate reference genes can result in inaccurate results10. Quantitative evaluations of gene expression in N. dimidiatum during infection can be used to identify the genes involved in virulence or pathogenesis. The use of one or more stable reference genes is the most common method for normalizing RT-qPCR data. In RT-qPCR analysis, normalization of the data requires internal control gene(s) that display uniform expression under various biological conditions11. Therefore, a suitable reference gene(s) needs to be selected based on specific sets of experimental conditions relevant to the study objective. Validations of internal reference gene stability have been performed in some fungi, including Valsa mali var. mali12, Magnaporthe oryzae13, Xanthomonas oryzae pv. oryzae14, and Ustilaginoidea virens15. However, no studies to date have systematically screened for stably expressed reference genes for RT-qPCR analysis in N. dimidiatum.
In this study, eight candidate reference genes (18S rRNA, Actin (1), Actin (2), Actin, GAPDH (1), GAPDH (2), Tubulin, and UBQ) were screened in N. dimidiatum. As Actin (2) exhibited non-specific amplification, we evaluated the utility of 18S rRNA, Actin (1), Actin, GAPDH (1), GAPDH (2), Tubulin, and UBQ as reference genes for RT-qPCR in N. dimidiatum at different stages of development, under different temperatures, and during interaction with pitaya. Three Excel add-ins (i.e., BestKeeper, geNorm, and NormFinder) were used to evaluate the stability of the expression of these genes16,17,18,19. The suitable reference genes identified will facilitate future studies of gene function in N. dimidiatum.
Results
Selection of candidate reference genes
Eight candidate reference genes from the N. dimidiatum genome were selected based on non-redundant functional annotations. The length of the primers was approximately 20 bp, the size of the amplified products ranged from 106 bp for GAPDH (1) to 185 bp for Actin (Table 1).
Expression profiles of candidate reference genes
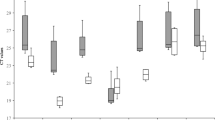
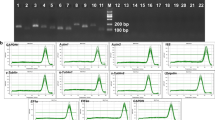
To test the specificity of the RT-qPCR primers, PCR was used to amplify the sequences of the eight candidate genes. All primers except Actin (2) amplified a single PCR product of the expected size according to 1.5% gel electrophoresis and melting curve analysis (Figs. S1, S2). Ct values for the seven candidate genes showing specific amplification (i.e., excluding Actin (2)) ranged from 19.80 to 31.62. The expression of GAPDH (2) was the highest, and the expression of 18S rRNA was the lowest among all candidate genes. Variation in expression was the lowest for GAPDH (2) among all candidate genes (mean Ct value ± SD = 26.77 ± 1.39). Variation in expression was the highest for 18S rRNA and Actin (1) among the seven candidate genes (23.97 ± 2.61 and 22.97 ± 2.67, respectively) (Fig. 1, Table 2).
Ranges of expression of the seven selected reference genes in all analyzed samples. The whisker box plots represent the cycle threshold numbers (Ct value) of the seven reference genes. The horizontal lines inside the boxes are the median values, and the whiskers of the boxes indicate the minimum and maximum.
Expression of candidate genes in N. dimidiatum at different temperatures
The expression levels of the seven candidate genes in N. dimidiatum at different temperatures and developmental stages were first determined by RT-qPCR, and their expression stabilities were assessed using three statistical algorithms (Fig. 2). The average expression stability (M-value) of all genes was calculated by geNorm. Genes with the lowest M-value are considered the most stable. In our analysis, all genes had M-values below the geNorm threshold of 1.5. The expression of Tubulin and Actin (1) were the most stable under different temperature treatments. Analysis of RT-qPCR data using the BestKeeper algorithm indicated that the expression of 18S rRNA was the most stable at different temperatures. The NormFinder algorithms indicated that the expression of Tubulin and Actin (1) was the most stable at different temperatures (Fig. 2A–C).
Rankings of reference gene expression stabilities determined by RT-qPCR followed by geNorm, Bestkeeper, and Normfinder algorithm. (A–C) correspond to different temperatures; (D–F) N. dimidiatum at different developmental stages; (G–I) correspond to pitaya–N. dimidiatum interaction; (J–L) correspond to all conditions (at different developmental stages, at different temperatures, and during the pitaya–N. dimidiatum interaction.
Expression of the candidate genes at different developmental stages
To determine the expression of the seven candidate genes at different developmental stages, we analyzed their expression at 3 days, 5 days, and 10 days. Analyses of the RT-qPCR data using the geNorm algorithm revealed that the expression of Actin (1) and Acin were the most stable at different developmental stages. Analysis of RT-qPCR data using the BestKeeper algorithm revealed that the expression of GAPDH (1) was the most stable at different developmental stages. Based on the NormFinder algorithm, Actin (1) and Acin were the most stably expressed gene at different developmental stages (Fig. 2D–F).
Expression of selected genes during interaction with pitaya
Analysis of RT-qPCR data using geNorm or NormFinder revealed that the expression of Tubulin and UBQ were most stable during interaction with pitaya. However, the expression of GAPDH (1) was determined to be the most stable when the BestKeeper algorithm was used to analyze the RT-qPCR data (Fig. 2G–I).
Suitability of the candidate genes
In addition to identifying the optimal reference genes for each treatment, we used three different algorithms to estimate the overall suitability of these reference genes to be used across all experimental contexts (i.e., at different temperatures, different developmental stages, and during interaction with pitaya). Analysis with the geNorm and NormFinder algorithm revealed that the optimal reference gene was Actin, and analysis with the BestKeeper algorithm revealed that GAPDH (1) was the most suitable reference gene (Fig. 2J–L).
Determination of the optimal number of reference genes needed for the normalization of RT-qPCR data
A single reference gene is often insufficient for accurately quantifying levels of gene expression. It is frequently necessary to use two or more than two reference genes to ensure the accurate normalization of gene expression levels9. To determine whether two or more N. dimidiatum reference genes are needed to normalize the RT-qPCR data, pairwise variations were determined using the geNorm algorithm. The optimal number of reference genes can be determined by using the pairwise variation value (Vn/Vn + 1, V-value) as the normalization factor for reference genes. Additional (n + 1) reference genes are necessary to normalize the genes if the V-value is higher than the threshold of 0.15. The ratio of V2/V3 was lower than 0.15, indicating that the optimal number of reference genes was two. The results in Fig. 3 showed that all V-values were less than 0.15, indicating that the addition of a third gene for correction was unnecessary. That is, two reference genes are needed as internal controls for normalizing gene expression data in N. dimidiatum at different temperatures (Tubulin/Actin (1)), at different developmental stages (Actin/Actin (1)), during interaction with pitaya (Tubulin/UBQ), and at all conditions (18s rRNA/Actin) (Fig. 3).
Reference gene validation
Pectin forms an acidic polysaccharide network and makes up the scaffold of the middle lamella that binds plant cells to one another. Therefore, a large amount of pectinase is often secreted in the early stage of infection to dissolve the cell wall of plants. Once the plant cell wall is breached, the pathogen secretes intracellular effector proteins that manipulate the host and its defense mechanisms20. To determine the reliability of the two optimal reference genes identified (UBQ and Tubulin) and the least optimal reference gene (18S rRNA) during interaction with pitaya, we used RT-qPCR to detect the expression of the pectinase gene (ND3060) during the pitaya–N. dimidiatum interaction (Fig. 4, Table S1). The total RNA was extracted from pitaya infected with N. dimidiatum at 3 days, 4 days, and 5 days, and used to synthesize the first strand of cDNA, which will be used as the template of RT-PCR, and relative expression levels were calculated using the 2−△△Ct method. When Tubulin and UBQ were used as internal controls, the expression of pectinase gradually increased on 3 days, 4 days, and 5 days. This finding was consistent with the pattern observed under N. dimidiatum infection of pitaya. When pitaya was infected by N. dimidiatum from 5 to 7 days, the expression of disease resistance-related genes of pitaya was highest3. However, 18S rRNA overestimated the expression of pectinase on 4 d.
Discussion
Pitaya canker is an emerging global disease that poses a significant threat to pitaya production21. Currently, little is known about the virulence functions of N. dimidiatum3. Most functional analyses of virulence or pathogenicity genes that involve gene expression analysis are performed using RT-qPCR12,22,23. A suitable internal reference gene is essential for correcting for errors in RNA quantity, reverse transcription efficiency, and the stability of different reaction channels of the PCR instrument and standard operating mode and determining the expression level of the target gene15,24. Housekeeping genes such as Tubulin, Actin, and 18S rRNA are often used as reference genes. However, these reference genes show expression instability in various species, at different developmental stages, or in different tissues8,9,14,18. Therefore, the reference genes most appropriate for specific sample types and experimental conditions need to be selected. However, no studies to date have systematically evaluated the set of reference genes most suitable for use in N. dimidiatum.
In this study, we evaluated the stability of eight commonly used housekeeping genes for specific amplification analysis and found that Actin (2) exhibited non-specific amplification. Therefore, we used seven housekeeping genes for specific amplification analysis. A series of experiments were performed as an internal control for the analysis of differential gene expression in N. dimidiatum. We found that GAPDH (2) was the most abundant gene compared with the other six candidate genes tested based on Ct values. However, this gene was not stably expressed in different tissues of N. dimidiatum. In addition, its expression during interaction with pitaya was only moderately stable at different developmental stages of N. dimidiatum and under different temperatures. These findings suggest that GAPDH (2) is not the most suitable reference gene for RT-qPCR.
Actin is an important cytoskeletal protein in cells. It is highly conserved across different species, and it is also the most common reference gene used for RT-qPCR25. We comprehensively analyzed the interaction of the Actin gene with pitaya at different temperatures and different developmental stages. The expression of Actin (1) was the most stable at different developmental stages and temperatures, but its expression was not stable under other sets of experimental conditions. Thus, this gene could be used as a reference gene for N. dimidiatum under specific experimental conditions.
Tubulin is the basic building block of microtubules and plays a role in many fundamental cellular processes, such as the maintenance of cytoskeletal structure26. We found that the expression of Tubulin was stable during interaction with pitaya and under different temperatures; the expression stability of Tubulin was intermediate among all candidate genes analyzed. Therefore, this gene is most suitable as a reference gene for RT-qPCR during interaction with pitaya (Fig. 2).
UBQ (Polyubiquitin) is a ubiquitin gene27. geNorm was used to analyze the expression of genes during the pitaya–N. dimidiatum interaction at different periods; two reference genes are needed to ensure the accuracy of expression levels calculated during the pitaya–N. dimidiatum interaction. During the interaction period, the expression of Tubulin was the most stable, followed by UBQ. Therefore, the use of Tubulin and UBQ to normalize RT-qPCR taxa can maximize the accuracy of gene expression estimates.
Pectinase can destroy the cell wall of plants20. When N. dimidiatum infects pitaya, it secretes a large amount of pectinase to destroy the cell wall of pitaya in the early stage of infection, which allows it to obtain nutrients. Therefore, the expression of the pectinase gene ND3060 was observed in the early stage of N. dimidiatum infection. When a single gene or combination of genes (Tubulin and UBQ) was used as an internal control, the expression of pectinase gradually increased on 3 days, 4 days, and 5 days of N. dimidiatum infection of pitaya, which was consistent with the expression patterns of pitaya during infection. These results indicate that Tubulin is the most stably expressed gene during the pitaya–N. dimidiatum interaction, and the use of both Tubulin and UBQ can increase the accuracy of estimates of the expression of genes related to pitaya–N. dimidiatum interaction.
Conclusion
This study represents the first attempt to select a set of commonly used candidate reference genes in N. dimidiatum at different developmental stages, at different temperatures, and during interaction with pitaya for the normalization of gene expression data using RT-qPCR. Tubulin and Actin (1) were the most stably expressed reference genes under different temperatures. Actin (1) and Actin were the most stably expressed reference genes in N. dimidiatum at different developmental stages. Tubulin and UBQ were the most stably expressed reference genes during interaction with pitaya. 18s rRNA and Actin were the most stably expressed across all experimental conditions.
Materials and methods
Plant material, growth conditions, and infection
The pitaya was sampled from the pitaya germplasm nursery of Hainan University located at Banqiao Town, Dongfang City, Hainan Province, China (longitude: 108°68′ E, latitude: 18°79′ N). The pitaya germplasm nursery can continue to provide us with experimental materials for further experiments. The voucher specimen of pitaya in our study was collected and identified by Professor Hua Tang, it has been deposited in pitaya germplasm nursery as number ‘HD-A38-10’ and with herbarium name ‘Jindu NO. 1. It is a publicly available herbarium by some procedure. The ‘Jindu NO. 1’ is an important and major pitaya variety in pitaya production, it was bred and named by Mr. Jindu Wang. The use of plants in the present study complies with international, national and/or institutional guidelines. N. dimidiatum was isolates from diseased pitaya stem tissues and stored at -80 °C at Hainan Key Laboratory for Sustainable Utilization of Tropical Bioresources, Hainan University, Haikou, China2 (Fig. 5). The hyphae were collected from N. dimidiatum by culture on PDA medium at 15 °C, 28 °C, and 37 °C for 5 days; N. dimidiatum was cultured on PDA medium at 28 °C, and the hyphae were collected at 3 days, 5 days, and 10 days.
Pitaya twigs with the same growth state were collected from the pitaya field; their surfaces were sterilized with 75% ethanol in the laboratory of the School of Tropical Crops, Hainan University, rinsed with tap water, and then dried. N. dimidiatum that had been cultured in PDA liquid medium for a week was collected, filtered through three lens cleaning tissue, and diluted with sterile water to a concentration of 1 × 105 spores. The spore suspension was evenly sprayed on the surface of pitaya, and the twigs were kept moist at room temperature. Notably, all the above treatments had three biological replicates. The remaining pitaya twigs were sprayed with sterile water as a control. Lastly, collect the obviously diseased parts of pitaya infected by N. dimidiatum at the 3 days, 4 days and 5 days, frozen in liquid nitrogen, and stored at − 80 °C.
RNA extraction and cDNA synthesis
Total RNA from pitaya–N. dimidiatum interaction samples was extracted following the protocol of Xu et al.2. Harvested mycelia were ground to a fine powder in liquid nitrogen using a pre-cooled mortar and pestle. Total RNA of N. dimidiatum from the samples was extracted using TRI Reagent RNA Isolation Reagent (Sigma, Shanghai, CHN) per the manufacturer’s instructions and then treated with RNase-free DNase I at 37 °C for 30 min. Purification with TRI Reagent RNA Isolation Reagent was repeated to remove DNase I. RNA quality was analyzed by 2.0% agarose gel electrophoresis, and RNA integrity was assessed using a NanoPhotometer (Implen, GER). Only RNA samples with an A260/A280 ratio of 1.8–2.2 and an A260/A230 ratio > 1.8 were used for further analysis. cDNA was synthesized from 1.0 μg of total RNA samples using a HiScript III cDNA Synthesis Kit per the manufacturer’s instructions (Vazyme Biotech Co., Ltd., Nanjing, China, Cat. No. R333).
Candidate gene selection and primer design
The expression of eight candidate reference genes (18S rRNA, Actin (1), Actin (2), Actin, Tubulin, GAPDH (1), GAPDH (2), and UBQ) was analyzed. The specific primers (Table 1) were designed using Primer Premier 5.0 with the following parameters: melting temperature (Tm), 48–61 °C; GC percent, 40–60%; primer lengths, 18–22 bp; and product lengths, 100–200 bp.
To confirm that the correct region was amplified by the selected primers, primer pairs were used to prime amplification in 100 ng of the first-strand cDNA in an end-point PCR reaction. For each genotype, one sample of synthesized first-strand cDNA was selected randomly and used for all primer tests. The master mix for this reaction was prepared using the 2 × Rapid Taq Master Mix (Vazyme Biotech Co., Ltd., Nanjing, China, Cat. No. R222) per the manufacturer’s instructions. The amplification was carried out using the following thermal cycling parameters: initial denaturation at 95 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing for 30 s at the primer pair’s average annealing temperature, and an extension of 45 s at 72 °C. After completion of the last cycle, a final extension was carried out at 72 °C for 5 min19. To confirm that a single product was amplified, the PCR products were loaded into a 1.5% agarose-TAE gel stained with ethidium bromide and run at 120 V for 15 min at room temperature. Gel images were obtained using Gel Doc XR+ (Bio-Rad, USA).
RT‑qPCR analyses
RT-qPCR assays were performed in a 20 μL reaction system using the ChamQ Universal SYBR qPCR Master Mix kit (Vazyme Biotech Co., Ltd., Nanjing, China, Cat. No. Q711–02/03) per the manufacturer’s protocol. Candidate genes were selected for RT-qPCR assays using an Applied Biosystems 7500 RT-qPCR System (Life Tech, 81 Wyman Street, Waltham, MA, 02454, USA). The thermal cycling conditions were as follows: 50 °C for 2 min; 95 °C for 10 min; 40 cycles at 95 °C for 15 s; 56 °C for 30 s; and 72 °C for 40 s. The melting curve was generated by heating the amplicon from 60 to 95 °C to confirm primer specificity. Three biological replicates were carried out for each PCR reaction. Relative fold changes in gene expression were calculated using the comparative 2−ΔΔCT method28.
Analysis of gene expression stability
NormFinder, geNorm, and BestKeeper were used to evaluate the expression stability of the reference genes in N. dimidiatum at various development stages, at different temperatures, and during interaction with pitaya.
The geNorm program determines the stability and optimal number of genes required to calculate the M-value and pairwise variation Vn/Vn + 1 between two sequential normalization factors29. The optimal candidate reference genes were determined based on the average expression stability between samples and the results of variance analysis. The BestKeeper program was used to identify stable reference genes, which calculates pairwise correlations based on values of standard deviation (SD) and percentage covariance (Cov)30.
Validation of reference genes
To verify the reliability of the selected reference genes. Pectate lyase31, an enzyme involved in the digestion of the cell wall of pitaya, was analyzed using RT-qPCR in the early stages of N. dimidiatum infection of pitaya. The specific primers used are shown in Table 1.
Data availability
All data generated and analyzed during the study is included in the published article and its Supplementary Information files.
References
Pan, L., Zhang, R., Lu, F., Liu, C. & Fu, J. Comparative proteomic analysis of the fungal pathogen Neoscytalidium dimidiatum infection in the pitaya. Hortic. Environ. Biotechnol. 62, 649–659. https://doi.org/10.1007/s13580-021-00341-2 (2021).
Min, X. et al. Transcriptomic de novo analysis of pitaya (Hylocereus polyrhizus) canker disease caused by Neoscytalidium dimidiatum. BMC Genom. 20, 10. https://doi.org/10.1186/s12864-018-5343-0 (2019).
Min, X. et al. Molecular characterization and expression analysis of pitaya (Hylocereus polyrhizus) HpLRR genes in response to Neoscytalidium dimidiatum infection. BMC Plant Biol. 20, 160. https://doi.org/10.1186/s12870-020-02368-6 (2020).
Burton, R. A., Gidley, M. J. & Fincher, G. Heterogeneity in the chemistry, structure and function of plant cell walls. Nature Chem. Biol. 6, 724–732. https://doi.org/10.1038/nchembio.439 (2010).
Federico, S. et al. Secreted pectin monooxygenases drive plant infection by pathogenic oomycetes. Sci. (N. Y., N. Y.) 373, 774–779. https://doi.org/10.1126/science.abj1342 (2021).
Ying, W. et al. Establishment of real-time quantitative reverse transcription polymerase chain reaction assay for transcriptional analysis of duck enteritis virus UL55 gene. Virol. J. 8, 266. https://doi.org/10.1186/1743-422X-8-266 (2011).
Keisuke, Y. et al. Evaluation of consistency in quantification of gene copy number by real-time reverse transcription quantitative polymerase chain reaction and virus titer by plaque-forming assay for human respiratory syncytial virus. Microbiol. Immunol. 62, 90–98. https://doi.org/10.1111/1348-0421.12563 (2018).
Wu, W. et al. Determination of suitable RT-qPCR reference genes for studies of gene functions in Laodelphax striatellus (Fallén). Genes (Basel) 10, 887. https://doi.org/10.3390/genes10110887 (2019).
Chen, C. et al. Identification of reliable reference genes for quantitative real-time PCR normalization in pitaya. Plant Methods 15, 70. https://doi.org/10.1186/s13007-019-0455-3 (2019).
Jo, V. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. https://doi.org/10.1186/gb-2002-3-7-research0034 (2002).
Xun, T., Ning, Z., Huaijun, S. & Alejandro, C.-U. Selection and validation of reference genes for RT-qPCR analysis in potato under abiotic stress. Plant Methods 13, 85. https://doi.org/10.1186/s13007-017-0238-7 (2017).
Zhiyuan, Y. et al. Validation of reference genes for gene expression analysis in Valsa mali var. mali using real-time quantitative PCR. World J. Microbiol. Biotechnol. 29, 1563–1571. https://doi.org/10.1007/s11274-013-1320-6 (2013).
Sarena, C. O., Bentley, M. A., Giulia, M., Preston, G. M. & Gurr, S. J. Validation of reference genes for robust qRT-PCR gene expression analysis in the rice blast fungus Magnaporthe oryzae. PLoS ONE 11, e0160637. https://doi.org/10.1371/journal.pone.0160637 (2016).
Chen, X., Laborda, P., Dong, Y. & Liu, F. Evaluation of suitable reference genes for normalization of quantitative real-time PCR analysis in rice plants under Xanthomonas oryzae pv. oryzae–infection and melatonin supplementation. Food Prod. Process. Nutr. 2, 21. https://doi.org/10.1186/s43014-020-00035-9 (2020).
Dawei, Z., Yi, W., Yu, H., Jin-Rong, X. & Chenfang, W. UvHOG1 is important for hyphal growth and stress responses in the rice false smut fungus Ustilaginoidea virens. Sci. Rep. 6, 24824. https://doi.org/10.1038/srep24824 (2016).
Andersen, C. L., Jensen, J. L. & Ørntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. https://doi.org/10.1158/0008-5472.Can-04-0496 (2004).
Pfaffl, W. M., Ales, T., Christian, P. & Neuvians, P. T. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. https://doi.org/10.1023/b:bile.0000019559.84305.47 (2004).
Qi, W. et al. Stability of endogenous reference genes in postmortem human brains for normalization of quantitative real-time PCR data: Comprehensive evaluation using geNorm, NormFinder, and BestKeeper. Int. J. Legal Med. 126, 943–952. https://doi.org/10.1007/s00414-012-0774-7 (2012).
Derveaux, S., Vandesompele, J. & Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods 50, 227–230. https://doi.org/10.1016/j.ymeth.2009.11.001 (2009).
Florian, P., Jérôme, P. & Catherine, R. Plant pectin acetylesterase structure and function: New insights from bioinformatic analysis. BMC Genom. 18, 456. https://doi.org/10.1186/s12864-017-3833-0 (2017).
Min, X. et al. Identification of Neoscytalidium dimidiatum causing canker disease of pitaya in Hainan, China. Austral. Plant Pathol. 47, 547–553. https://doi.org/10.1007/s13313-018-0588-2 (2018).
Nan, Z. et al. The essential effector SCRE1 in Ustilaginoidea virens suppresses rice immunity via a small peptide region. Mol. Plant Pathol. 21, 445–459. https://doi.org/10.1111/mpp.12894 (2020).
Tianqiao, S. et al. The N-terminus of an Ustilaginoidea virens Ser-Thr-rich glycosylphosphatidylinositol-anchored protein elicits plant immunity as a MAMP. Nat. Commun. 12, 2451. https://doi.org/10.1038/s41467-021-22660-9 (2021).
Zhang, S., Chen, C., Xie, T. & Ye, S. Identification and validation of reference genes for qRT-PCR studies of the obligate aphid pathogenic fungus Pandora neoaphidis during different developmental stages. PLoS ONE 12, e0179930. https://doi.org/10.1371/journal.pone.0179930 (2017).
Carlsson, A. E. Membrane bending by actin polymerization. Curr. Opin. Cell Biol. 50, 1–7. https://doi.org/10.1016/j.ceb.2017.11.007 (2018).
Carsten, J. & Magiera, M. M. The tubulin code and its role in controlling microtubule properties and functions. Nature Rev. Mol. Cell Biol. 21, 307–326. https://doi.org/10.1038/s41580-020-0214-3 (2020).
Liliana, M., Andreia, M., Ricardo, C. P. & Célia, M. Reference gene selection for quantitative real-time PCR normalization in Quercus suber. PLoS ONE 7, e35113. https://doi.org/10.1371/journal.pone.0035113 (2012).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. https://doi.org/10.1006/meth.2001.1262 (2001).
Paolinelli-Alfonso, M., Galindo-Sánchez, C. E. & Hernandez-Martinez, R. Quantitative real-time PCR normalization for gene expression studies in the plant pathogenic fungi Lasiodiplodia theobromae. J. Microbiol. Methods 127, 82–88. https://doi.org/10.1016/j.mimet.2016.05.021 (2016).
Rego, E. C. S. et al. Stable reference genes for RT-qPCR analysis of gene expression in the Musa acuminata-Pseudocercospora musae interaction. Sci. Rep. 9, 14592. https://doi.org/10.1038/s41598-019-51040-z (2019).
Alex, W. et al. Gene deletion and constitutive expression of the pectate lyase gene 1 (MoPL1) lead to diminished virulence of Magnaporthe oryzae. J. Microbiol. 60, 526. https://doi.org/10.1007/s12275-022-1074-7 (2021).
Acknowledgements
This work was supported by Hainan Provincial Natural Science Foundation high-level Talents Project (320RC487), Hainan Major Science and Technology project (ZDKJ2021014), Natural Science Fund of Hainan Province (311025), Hainan Province Science and Technology Special Fund (ZDYF2022XDNY199).
Author information
Authors and Affiliations
Contributions
Conceptualization, M.W.; data curation, M.W.; funding acquisition, H.T.; investigation, J.H., D.L.; methodology, S.W., J.X. and W.H.; project administration, H.T.; resources, H.L.; software, Z.W.; writing—original draft, M.W. All authors consent on the submission of this paper for publication in scientific reports.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, M., Wang, Z., Wei, S. et al. Identification of RT-qPCR reference genes suitable for gene function studies in the pitaya canker disease pathogen Neoscytalidium dimidiatum. Sci Rep 12, 22357 (2022). https://doi.org/10.1038/s41598-022-27041-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-27041-w
- Springer Nature Limited