Abstract
Exogenous melatonin (MT) was found to be an interesting tool for enhancing the resistance of rice to Xanthomonasoryzaepv. oryzae (Xoo)-caused bacterial blight (BB). However, the accurate comparison of the expression levels across samples was a challenging task. In this work, the stability of 10 common used housekeeping genes under Xoo-infection and MT supplementation in rice was analyzed using quantitative real-time PCR (qRT-PCR), and algorithms geNorm, NormFinder and BestKeeper. Our results indicated that most reference genes remained stable in Xoo-infected rice plants, while a number of reference genes were affected by MT supplementation. Among all studied genes, the transcript levels of 18S(18S ribosomal RNA) and UBC (Ubiquitin-conjugating enzyme E2) remained unaltered by Xoo infection, while UBC and UBQ5(Ubiquitin 5) were the most stable genes when examining simultaneous Xoo-infection and MT supplementation, demonstrating that UBC is a suitable reference gene for qRT-PCR data normalization in rice under Xoo-infection and MT supplementation.
Similar content being viewed by others
Introduction
Rice is one of the most important crops worldwide, providing essential food supply to most of the world's population. However, rice is highly susceptible to a wide range of pathogens that limit its production and quality. Among rice pathogens, Xanthomonas oryzae pv. oryzae (Xoo), the causal agent of bacterial blight (BB), is a member of γ-proteobacteria, and is able to invade plant through wounds and hydathodes to colonize the xylem (White and Yang 2009). This pathogen can rapidly spread through the vascular system, leading to systemic infection (Yuan et al. 2010). Its pathogenicity is partially dependent on a type III protein secretion system (TTSS) (Pfeilmeier et al. 2016). During the early infection stages, the expression levels of 541 genes were altered in response to Xoo in susceptible rice JG30 (Tariq et al. 2019), while the expression levels of 115 genes were altered in resistant hybrid rice Y73 in response to Xoo infection, which corresponds to 0.22% of the rice genome (Wang et al. 2012). Many of these differential expressed genes (DEGs) were related to signaling, transcription and metabolic processes.
Melatonin (N–acetyl–5–methoxytryptamine) consists of an indole structure with low molecular weight, and has been found in a wide range of plant species (Wang et al. 2020). MT regulates gene expression in many cellular and physiological aspects (Li et al. 2019b). It is reported that 457 differentially expressed genes were identified in response to salt stress under MT supplementation (Liang et al. 2015). Those DEGs were associated to defense responses, protein phosphorylation, hormone-mediated signaling pathways and metabolic processes. Interestingly, Xoo-infection increased the mRNA expression level of OsAMT1, one of the key genes in MT biosynthesis (Wei et al. 2016). Recently, our research group demonstrated that melatonin can enhance rice resistance against Xoo by increasing the expression of pathogensis-related genes (PRs), and showed antibacterial activity against Xanthomonas spp. (Chen et al. 2018, 2019, 2020).
qRT-PCR is a remarkably robust technique used for the quantification of gene expression in different samples (Bustin and Nolan 2017). However, the accuracy of quantitative analysis maybe influenced by several biasing factors, such as sample type, sample integrity and experimental conditions (Shen et al. 2010; Yang et al. 2018). Therefore, normalization of gene expression using stable internal standards, also called housekeeping genes or reference genes, is critical for the accurate comparison of gene expression across samples (Robledo et al. 2014). Theoretically, reference genes should remain stable under different experimental conditions and may show the same mRNA level in all type of cells and tissues. However, there is no universal internal standard gene that fulfills completely this criterium (Sundaram et al. 2019). Hence, the validation of the expression stabilities of reference genes is necessary for the accurate acquisition of qRT-PCR data. In this work, we have investigated for the first time the expression stability of 10 candidate reference genes, including 18S ribosomal RNA (18S), 25S ribosomal RNA (25S), Ubiquitin 5 (UBQ5), Ubiquitin 10 (UBQ10), Actin (ACT), β-Tubulin (β-TUB), Eukaryotic elongation factor 1-alpha (eEF-1α), Eukaryotic initiation factor 4-alpha (eIF-4α), UBC and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), under Xoo-infection and MT supplementation (Jain et al. 2006; Kim et al. 2003; Li et al. 2010). The final outcome of this feasible study will benefit further quantifications of gene expression by qRT-PCR and RNA-seq-based transcriptomic studies in rice.
Materials and methods
Reagents
MT was purchased from Sigma-Aldrich (St. Louis, USA). Analytical grade methanol was purchased from Sinopharm Chemical Reagent (Shanghai, China). TRIzol was obtained from Invitrogen (Carlsbad, USA). SYBR Green PCR Master Mix and cDNA Synthesis kits were purchased from Takara Bio (Shiga, Japan). Ultra pure water was purified in a Milli-Q water purification system (Millipore,Burlington, USA).
Plant and bacterial strains
Rice seeds (O. sativa cv. Nipponbare) were grown in a growth chamber with a photon flux density of 200 μmol/m2s1 under alternating 30 °C/12 h dark and 28 °C/12 h light cycles. The seedlings were cultured in 1 L of IRRI nutrient solution (28.6 mg/L NH4NO3, 40.5 mg/L MgSO4·7H2O,36.7 mg/L CaCl2·2H2O, 89.3 mg/L K2SO4, 50.3 mg/L Na2HPO4·2H2O, 1.2 mg/L H3BO3, 11.6 mg/L Na2EDTA·2H2O, 8.7 mg/L FeSO4·7H2O, 9.25 mg/L (NH4)6MoO24·4H2O, 43.8 mg/L ZnSO4·7H2O, 38.8 mg/L CuSO4·5H2O and 1.8 mg/L MnCl2·4H2O) for 5 weeks. Xoo strain PXO99 was grown in liquid nutrient broth (NB) medium (5 g/L polypeptone, 3 g/L beef extract, 10 g/L sucrose and 1 g/L yeast extract, pH 7.0–7.2) or on nutrient agar (NA) medium (NB with 15 g/L agar).
MT treatment and bacterial pathogen inoculation
Five-week-old rice plants were used for the experiments. The plant roots were submerged in water containing MT (0 and 20 μg/mL), and the plant leaves were inoculated with water or Xoo (OD600 = 1.0). Xoo was cultured in NB medium for 24 h with shaking at 180 rpm at 28 °C. The bacterial cells were centrifuged for 10 min at 6000 rpm and 4 °C, and suspended in water to OD600 = 1.0. Rice leaves were inoculated with PXO99 using the leaf clipping method (Kauffman et al. 1973), following the same conditions previously reported by Laborda et al. (2020). Four parallel treatment conditions were studied: 1. rice leaves were treated with water, while the plant roots were cultivated in water in the absence of MT (water group); 2. rice leaves were inoculated with Xoo, and the plant roots were submerged in water in the absence of MT (Xoo group); 3. rice leaves were inoculated with water, and the plant roots were submerged in an aqueous solution containing 20 μg/mL melatonin (MT-water group); and 4. rice leaves were inoculated with Xoo, and the plant roots were submerged in an aqueous solution containing 20 μg/mL melatonin (MT-Xoo group). Twelve hours after inoculation of the pathogen, the infected leaves were collected for RNA extraction.
RNA isolation and cDNA synthesis
Specific primersfor candidate reference genes ACT, eEF-1α, eIF-4α, GAPDH, UBC, UBQ5, UBQ10, β-TUB, 18S and 25S were designed using Primer 5 (version 5.0). The gene sequences were obtained from the rice genome annotation project (RGAP; http://rice.plantbiology.msu.edu/) database (Table 1). The total RNA from rice leaves was extracted using the TRizol reagent method (Invitrogen, Germany). Total RNA extraction and quality test were determined by UV spectrophotometric analysis and 2% agarose gel electrophoresis, and pure RNA should yield an A260/A230 ratio of around 2 or slightly above. To eliminate the genomic DNA, 2 μg of total RNA was treated with 1 μLg DNA Eraser (Takara, Japan). Finally, cDNA was synthetized using a Primerscript™ RT reagent kit (Takara, Japan). qRT-PCR was performed with diluted first strand cDNA, specific gene primers, and SYBR Green PCR Master MIX (TakaRa, Japan). All samples were diluted 10 times and were run on a LightCycler® 480 Instrument II Real Time PCR system (Roche, Switzerland). The PCR amplification program consisted of 1 cycle of 95 °C for 30 s followed by 45 cycles of 95 °C for 5 s, and 60 °C for 30s. Individual well fluorescence data were generated at the end of PCR cycles, with 95 °C for 5 s followed by60 °C for60s. The experiments were repeated at least three times using different biological samples.
qRT-PCR assay and expression stability analysis
Total RNA concentration of each sample was determined with a NanoDrop spectrophotometer by using the RNA application (Eppendorf, BiophotometerPlus Spectrophotometer, Germany). Efficiency of amplification curves (E value), cycle threshold (Ct) and correlation coefficient (R2) were determined using LinReg PCR software (http://LinRegPCR.nl/). In LinReg analysis, efficiency of amplification value was set to 2.0. The expression stability of the candidate reference genes was analyzed using the BestKeeper version (http://www.gene-quantification.de/bestkeeper.html), NormFinder_0953 (http://moma.dk/normfinder-software) and geNorm (http://medgen.ugent.be/jvdesomp/genorm/) following the developer’s instructions. For BestKeeper analysis, the average Ct value from each sample were input directly. The relative expression of each target gene was calculated using the 2-ΔΔCt method. Before inputting into the software program of Norm Finder and geNorm, the raw Ct values of each gene were converted into relative quantities and the highest relative expression was set to 1.0 for each reference gene. Through the geNorm analysis, the ranking of tested reference genes was based on their expression stability value (M) as the average pairwise variation(V) between a particular gene to all other candidate genes.
Results and discussion
Total RNA extraction and RNA quality assessment
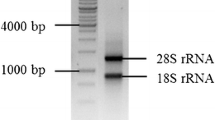
In plant leaves, the isolation of high-purity RNA with sufficient quantity is essential to study the stability of reference genes. To achieve this goal, total RNA from four groups (water, Xoo, MT-water and MT-Xoo) were extracted and isolated by using the TRizol reagent method (Chomczynski and Mackey 1995). The concentration and purity of RNA from different samples were determined using Eppendorf Biophotometer plus and agarose gel electrophoresis. In the eppendorf Biophotometer plus analysis, the values of A260/A280 of the isolated RNA were close to 2.0. The overall quality of total RNA was further assessed by agarose gel electrophoresis assay. As shown in Fig. 1, the two ribosomal components (25S and 18S) were readily identified in the agarose gel without obvious degradation (Srivastava et al. 2012). The isolated RNA showed five bands (Fig. 1). The results suggested that the total RNA had good integrity and purity, and could be used for further studies.
Optimization of qRT-PCR amplification conditions for candidate reference genes
After synthesis of the first-strand cDNA from the four treatment groups (water, Xoo, MT-water and MT-Xoo), the specificity and PCR amplification conditions for the 10 candidate reference genes were tested by PCR. The cDNA with length from 167 to 238 base pairs containing normal bases were amplified. As shown in the agarose gel of Fig. 2, all 10 pairs of primers successfully produced a unique amplicon with the target product lengths. Then, specificity and amplification efficiency of the primers for the 10 candidate reference genes was tested by using qRT-PCR (Table 2). A single peak for each primer set was observed in the amplification plot, indicating that all candidate primers produced unique amplicon without any primer dimers or other nonspecific amplification products (Fig. 2). The obtained results indicated that all primers had good specificity and could be used for the gene stability assay.
It is well known that amplification efficiency plays a key role in the accuracy and reliability of reference genes, while the challenge of determining amplification efficiency is known to be a predominant aspect of implementing qRT-PCR (Rutledge and Stewart 2008). Here, the amplification efficiency (E) and the correlation coefficient (R2) values were calculated using the LinRegPCR software. The E values of the candidate reference genes varied from 0.97 for ACT to 0.9998 for UBQ10, and the R2 values of candidate reference genes ranged from 1.8358 to 1.9935 (Table 2). The reactions provided accurate linear relationships, with R2 > 0.99 and E value close to 2.0. The overall variability of candidate reference genes under different treatment conditions was assessed by the average values of Ct (cycle threshold) and SD (standard deviation) (Table 2). LinRegPCR analysis determined the average Ct values of the 10 candidate reference genes, and the minimum Ct value was found for the 18S gene (7.50 ± 0.05), indicating that this gene showed the highest transcript abundance, whereas the lowest transcript abundance was found for the β-TUB gene (28.44 ± 0.18). The Ct values of eEF-1a and UBQ5 genes were relatively constant (17.36–18.01 and 17.94–18.63, respectively), withlow SD values throughout the infection-groups (Xoo and MT-Xoo) and mock inoculated-groups (water and MT-water). For the rest eight candidate genes, the mean Ct values ranged from 7.51 to 28.43 in the water group, from 7.54 to 28.56 in the Xoo group, from 7.36 to 28.36 in the MT-water group, and from 7.25 to 28.94 in the MT-Xoo group. The mean Ct values of 18S changed over a narrow range from 7.25 ± 0.06 to 7.59 ± 0.057 in mock inoculated (water and MT-water) and infection (Xoo and MT-Xoo) groups (Table 2).
The gene of UBC has been commonly used as reference gene in rice under pathogen infection (Bi et al. 2019; Li et al. 2019). Previous studies indicated that the mean Ct values for UBC were 22.53, 22.55 and 22.43 in virus-free rice plants, Rice Black-Streaked Dwarf Virus (RBSDV)-infected rice plants and Rice Strip Virus (RSV)-infected rice plants, respectively (Shen et al. 2014). In agreement with that work, the mean Ct values of UBC were 21.94, 22.16, 22.21 and 23.38 in the water, Xoo, MT-water, and MT-Xoo groups, respectively.
Evaluation of expression stability of reference genes for qRT-PCR
It was reported that some reference genes, such as TUB and ACT, were associated with low stability in rice plants (Jain et al. 2006; Zhao et al. 2019). Using unstable reference genes may lead to large errors in normalization of reference genes, resulting in incorrect interpretations (Sheshadri et al. 2018). In order to identified the most stable reference genes under MT supplementation in presence and absence of Xoo infection in rice plants, three commonly used algorithms for qRT-PCR normalization, including geNorm, NormFinder and BestKeeper, were employed (Vandesompele et al. 2002; Andersen et al. 2004; Pfaffl et al. 2004). The geNorm algorithm provides the stability value (M) for each candidate gene based on the pairwise standard deviation of cycle quantification (Cq) value. In the geNorm algorithm, the lower is the M value, the higher is the expression stability of the studied gene. The NormFinder evaluates the stability value of all tested candidate genes based on the variations between intra-group and inter-group. BestKeeper determines the stability value of the candidate reference genes based on the standard deviation (SD) and coefficient of variance (CV).
In our work, the default value was fixed at M = 0.5 in the geNorm program (Silveira et al. 2009). Thus, the candidate reference genes with M value lower than 0.5 were considered as genes with stable expression, whereas the genes with M value higher than 1.5 were excluded directly. In the water treatment group, the expression stability of the 10 candidate genes ranked as follows: UBC = 18S > β-TUB > ACT > GAPDH > eIF-4α > UBQ5 > eEF-1α > UBQ10 > 25S. Our study identified that UBC and 18S genes had the lowest M values (M = 0.07), while 25S had the highest M value (M = 0.21), indicating that UBC and 18S were the most stably expressed genes for this treatment group (Fig. 4a). In the Xoo group, the expression stability of the 10 candidate genes ranked as follows: UBQ10 = UBQ5 > eEF-1α > eIF-4α > 18S > β-TUB > ACT > 25S > GAPDH > UBC. UBQ10 and UBQ5 genes showed the lowest M value (M = 0.04) (Fig. 4b), while 25S ranked as the least stable (M = 0.19). In the MT-water group, the expression stability of the 10 candidate genes ranked as follows:eEF-1α = UBC > UBQ10 > UBQ5 > 25S > eIF-4α > GAPDH > ACT > 18S > β-TUB. The β-TUB gene showed the highest M value (M = 0.13), while eEF-1α and UBC were found the most stable genes (M = 0.06) for this treatment group (Fig. 4c). In the MT-Xoo group, the expression stability of 10 candidate genes ranked as follows: eEF-1α = eIF-4α > UBQ5 > UBC > β-TUB > UBQ10 > ACT > GAPDH > 18S > 25S, demonstrating that genes eEF-1α and eIF-4α (M = 0.05) were the most stable (Fig. 3d). In contrast, the 25S gene was ranked as least stable, with the highest M-value (M = 0.13). The obtained results suggested that all the 10 candidate genes were acceptable as reference genes (M < 0.5).
Expression stability of the candidate reference genes assessed by geNorm analysis. a-d Expression stability of candidate reference genes in water-inoculated rice plants, in Xoo-infected rice plants, in MT-water infected rice plants and in MT-Xoo infected rice plants; e-f The pairwise variation value of reference genes in water and Xoo rice plants, in MT-water and MT-Xoo rice plants
The optimal number of candidate reference genes required for normalization factors (NF) was also determined by geNorm, by calculating the pairwise variation (Vn/n + 1) between two sequential normalization factors. The V-value cut-off was set at 0.15 in this case. The results showed that all V values of the selected genes were less than 0.15 (Fig. 4e and f), in agreement with the stability results.
In the NormFinder analysis the candidate reference gene with the lowest stability value (SV) have the highest stable expression. As shown in Fig. 4a, the expression stability of the 10 candidate genes in the water group ranked as follows: UBC > 18S > eIF-4α > β-TUB > ACT > eEF-1α > GAPDH > UBQ10 > UBQ5 > 25S, whereas, in the Xoo group, the stability of the 10 candidate genes was as follows: UBC > 18S > β-TUB > eIF-4a > GAPDH > UBQ10 > UBQ5 > ACT > eEF-1a > 25S. Thus, in both cases, the most stable genes were UBC and 18S. In contrast, the gene 25S showed the lowest stability in the water and Xoo groups. In the MT-water group, the expression stability of the 10 candidate genes ranked as follows: UBQ5 > UBC > UBQ10 > eEF-1α > eIF-4α > GAPDH > 18S > β-TUB > ACT > 25S, whereas, in the MT-Xoo group, the stability of the 10 candidate genes ranked as follows: UBQ5 > UBC > eEF-1α > eIF-4α > ACT > UBQ10 > 18S > GAPDH > β-TUB > 25S. Thus, UBQ5 and UBC were the most stably expressed genes under melatonin treatment. In general, UBC was the most stably expressed gene across the four groups, and the 25S was found to be the most unstably expressed gene.
In the Bestkeeper analysis, genes with the highest correlation coefficient (r) and the lowest SD and CV values were considered as the most stably expressed. Genes with SD value greater than 1 were considered to be unacceptable reference genes. As shown in Table 3, in the water group, the stability of the 10 candidate genes ranked as follows: UBC > 18S > β-TUB > ACT > GAPDH > eIF-4α > UBQ5 > eEF-1α > UBQ10 > 25S;and, in the Xoo group, the stability of the 10 candidate genes ranked from UBC > 18S > GAPDH > β-TUB > eIF-4α > eEF-1α > ACT > UBQ5 > UBQ10 > 25S. In both cases, UBC and 18S, were the most stable. In the MT-water group, the stability of the 10 candidate genes ranked from 18S > UBQ5 > 25S > UBC > UBQ10 > ACT > eEF-1α > GAPDH > eIF-4α > β-TUB; and, in the MT-Xoo group, the following order was detected: 18S > ACT > UBC > UBQ5 > 25S > UBQ10 > eEF-1α > GAPDH > eIF-4α > β-TUB. Thus, the most stably expressed genes were 18S and ACT in the MT-Xoo group, whereas 18S and UBQ5 were the most stable in the MT-water group. In this case, the results suggested that 18S was the most stably expressed gene across the four groups (Tables 4 and 5).
The expression stability analysis using the three most common software algorithms, including geNorm, NormFinder and BestKeeper, gives a ranking of the candidate reference genes, but none of them is currently considered to be the best one (Robledo et al. 2014). The geNorm method ranks candidate reference genes mainly by their correlations, assuming that none of the above genes are co-regulated (Manjarin et al. 2011). If there is co-regulation between two genes, then geNormmay spoil the analysis. Since SD is a direct measure of variation, the BestKeeper method is considered to be of “common sense” to measure stability (Robledo et al. 2014). However, genes with a lower overall intergroup an intragroup variation still cannot be recognized as a good reference gene if this variation can not reflect the errors produced during the sample preparation steps. While if none of the candidate reference genes are co-regulated, then this above problem may be circumvented. Compared to geNorm and BestKeeper, the software algorithm of NormFinder relies on the intragroup and intergroup variation by using a different mathematical model. This helps NormFinder to avoid the drawback of co-regulated genes. In many cases, the application of geNorm and NormFinder yields very similar results. However, the candidate reference genes can’t be recognized as good reference genes if there is a lower overall intergroup an intragroup variation. Hence, the advantages and disadvantages of these three methods should be taken into consideration when evaluating candidate reference genes.
Previous studies revealed that 18S was the most stable reference gene for qRT-PCR in rice under various growth stages and times after UV-irradiation treatment (Kim et al. 2003). In agreement, 18S was recognized as the most stable reference gene in rice under Xoo-infection and water treatment in this work. It was reported that UBC was identified as the most unaltered reference gene under RBSDV and RSV treatments (Shen et al. 2014). Here, UBC and UBQ were the most reliable genes across all rice samples under Xoo infection and MT treatment, and the application of UBC and UBQ would provide more accurate comparison across samples by qRT-PCR.
Conclusions
To the best of our knowledge, this is the first report on evaluation of suitable candidate reference genes for normalization of gene expression of qRT-PCR in rice under MT supplementation. In this work, we tested 10 common used candidate reference genes. We found that some reference genes were unstably expressed under the studied conditions. However, we observed that 18S + UBC and UBQ5 + UBC were the most reliable reference genes in Xoo infection and simultaneous MT supplementation with Xoo infection, respectively. These fundamental but vital outcomes will facilitate the gene expression studies of related biological processes, and will help to better understand MT-induced signal pathways in rice plants.
Availability of data and materials
Please contact authors for data request.
Abbreviations
- MT:
-
Melatonin
- Xoo :
-
Xanthomonasoryzaepv. oryzae
- 18S :
-
18S ribosomal RNA
- 25S :
-
25S ribosomal RNA
- UBQ5 :
-
Ubiquitin 5
- UBQ10 :
-
Ubiquitin 10
- ACT :
-
Actin
- β-TUB :
-
β-Tubulin
- eEF-1α :
-
Eukaryotic elongation factor 1-alpha
- eIF-4α:
-
Eukaryotic initiation factor 4-alpha
- UBC :
-
Ubiquitin-conjugating enzyme E2
- GAPDH :
-
Glyceraldehyde-3-phosphate dehydrogenase
- qRT-PCR:
-
Quantitative real-time PCR
- TTSS:
-
Type III protein secretion system; DEGs, differential expressed genes
- PRs:
-
Pathogenesis-related proteins; NB, nutrient broth
- NA:
-
Nutrient broth with agar
- UV:
-
Ultraviolet
- E value:
-
Efficiency of amplification curves
- Ct:
-
Cycle threshold
- R2 :
-
Correlation coefficient
- SD:
-
Standard deviation
- CV:
-
Coefficient of variance
- SV:
-
Stability value
- Cq:
-
Cycle quantification value
- RBSDV :
-
Rice Black-Streaked Dwarf Virus
- RSV :
-
Rice Strip Virus
References
Andersen, C. L., Jensen, J. L., & Orntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research, 64, 5245–5250.
Bi, J. A., Yang, Y., Chen, B., Zhao, J., Chen, Z., Song, B., et al. (2019). Retardation of the Calvin cycle contributes to the reduced CO2 assimilation ability of rice stripe virus-infected N. benthamiana and suppresses viral infection. Front Microbiology, 10, 568.
Bustin, S., & Nolan, T. (2017). Talking the talk, but not walking the walk: RT-qPCR as a paradigm for the lack of reproducibility in molecular research. European Journal of Clinical Investigation, 47, 756–774.
Chen, X., Laborda, P., & Liu, F. Q. (2020). Exogenous melatonin enhances rice plant resistance against Xanthomonas oryzae pv. Oryzae. Plant Disease, 104, 1701–1708.
Chen, X., Sun, C., Laborda, P., He, Y., Zhao, Y., Li, Z., & Liu, F. (2019). Melatonin treatments reduce the pathogenicity and inhibit the growth of Xanthomonas oryzae pv. oryzicola. Plant Pathology, 68, 288–296.
Chen, X., Sun, C., Laborda, P., Zhao, Y., Palmer, I., Fu, Z., et al. (2018). Melatonin treatment inhibits the growth of Xanthomonas oryzae pv. oryzae. Frontiers in Microbiology, 9, 2280.
Chomczynski, P., & Mackey, K. (1995). Short technical reports. Modification of the TRI reagent procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. Biotechniques, 19, 942–945.
Jain, M., Nijhawan, A., Tyagi, A. K., & Khurana, J. P. (2006). Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochemical and Biophysical Research Communications, 345, 646–651.
Kauffman, H., Reddy, A., Hsieh, S., & Merca, S. (1973). Improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Disease Reporter, 57, 537–541.
Kim, B. R., Nam, H. Y., Kim, S. U., Kim, S. I., & Chang, Y. J. (2003). Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnology Letters, 25, 1869–1872.
Laborda, P., Chen, X., Wu, G., Wang, S., Lu, X., Ling, J., et al. (2020). Lysobacter gummosus OH17 induces disease resistance systemic resistance in Oryza sativa ´Nipponbare´. Plant Pathology, 69, 838–848.
Li, Q., Lu, J., Zhou, Y., Wu, F., Tong, H., Wang, J., et al. (2019). Abscisic acid represses rice lamina joint inclination by antagonizing brassinosteroid biosynthesis and signaling. International Journal of Molecular Sciences, 20, 4908.
Li, Q., Sun, S., Yuan, D., Yu, H., Gu, M., & Liu, Q. (2010). Validation of candidate reference genes for the accurate normalization of real-time quantitative RT-PCR data in rice during seed development. Plant Molecular Biology Reporter, 28, 49.
Li, T., Wu, Q., Zhu, H., Zhou, Y., Jiang, Y., Gao, H., & Yun, Z. (2019b). Comparative transcriptomic and metabolic analysis reveals the effect of melatonin on delaying anthracnose incidence upon postharvest banana fruit peel. BMC Plant Biology, 19, 289.
Liang, C., Zheng, G., Li, W., Wang, Y., Hu, B., Wang, H., et al. (2015). Melatonin delays leaf senescence and enhances salt stress tolerance in rice. Journal of Pineal Research, 59, 91–101.
Manjarin, R., Trottier, N. L., Weber, P. S., Liesman, J. S., Taylor, N. P., & Steibel, J. P. (2011). A simple analytical and experimental procedure for selection of reference genes for reverse-transcription quantitative PCR normalization data. Journal of Diary Science, 94, 4950–4961.
Pfaffl, M. W., Tichopad, A., Prgomet, C., & Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--excel-based tool using pair-wise correlations. Biotechnology Letters, 26, 509–515.
Pfeilmeier, S., Caly, D. L., & Malone, J. G. (2016). Bacterial pathogenesis of plants: Future challenges from a microbial perspective: Challenges in bacterial molecular plant pathology. Molecular Plant Pathology, 17, 1298–1313.
Robledo, D., Hernandez-Urcera, J., Cal, R. M., Pardo, B. G., Sanchez, L., Martinez, P., & Vinas, A. (2014). Analysis of qPCR reference gene stability determination methods and a practical approach for efficiency calculation on a turbot (Scophthalmus maximus) gonad dataset. BMC Genomics, 15, 648.
Rutledge, R. G., & Stewart, D. (2008). Critical evaluation of methods used to determine amplification efficiency refutes the exponential character of real-time PCR. BMC Molecular Biology, 9, 96.
Shen, G., Jiang, H., Wang, X., & Wang, J. (2010). Evaluation of endogenous references for gene expression profiling in different tissues of the oriental fruit fly Bactrocera dorsalis (Diptera: Tephritidae). BMC Molecular Biology, 11, 76–76.
Shen, J., Li, J., Sun, L., & Chen, J. (2014). Reference gene selection for real-time fluorescence quantitative PCR analysis in rice plants infected by Rice black-streaked dwarf virus or Rice stripe virus. Acta Phytopathologica Sinica, 44, 276–286.
Sheshadri, S. A., Nishanth, M. J., Yamine, V., & Simon, B. (2018). Effect of melatonin on the stability and expression of reference genes in Catharanthus roseus. Scientific Reports, 8, 2222.
Silveira, É. D., Alves-Ferreira, M., Guimarães, L. A., da Silva, F. R., & Carneiro, V. T. D. C. (2009). Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biology, 9, 84.
Srivastava, N., Chaudhary, S., Kumar, V., Katudia, K., Vaidya, K., Vyas, M. K., & Chikara, S. K. (2012). Evaluation of the yield, quality and integrity of total RNA extracted by four different extraction methods in rice (Oryza sativa). Journal of Crop Science and Technology, 1, 1–9.
Sundaram, V. K., Sampathkumar, N. K., Massaad, C., & Grenier, J. (2019). Optimal use of statistical methods to validate reference gene stability in longitudinal studies. PLoS One, 14, e0219440.
Tariq, R., Ji, Z., Wang, C., Tang, Y., Zou, L., Sun, H., et al. (2019). RNA-Seq analysis of gene expression changes triggered by Xanthomonas oryzae pv. oryzae in a susceptible rice genotype. Rice, 12, 44.
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., & Speleman, F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3, 34.
Wang, S., Shi, X., & Laborda, P. (2020). Indole-based melatonin analogues: Synthetic approaches and biological activity. European Journal of Medicinal Chemistry, 185, 11847.
Wang, X., Zhou, J., Yang, Y., Yu, F., Chen, J., Yu, C., et al. (2012). Transcriptome analysis of a progeny of somatic hybrids of cultivated rice (Oryza sativa L.) and wild rice (Oryza meyeriana L.) with high resistance to bacterial blight. Journal of Phytopathology, 161, 324–334.
Wei, Y., Zeng, H., Hu, W., Chen, L., He, C., & Shi, H. (2016). Comparative transcriptional profiling of melatonin synthesis and catabolic genes indicates the possible role of melatonin in developmental and stress responses in rice. Frontiers in Plant Science, 7, 676.
White, F. F., & Yang, B. (2009). Host and pathogen factors controlling the rice-Xanthomonas oryzae interaction. Plant Physiology, 150, 1677–1686.
Yang, X., Pan, H., Yuan, L., & Zhou, X. (2018). Reference gene selection for RT-qPCR analysis in Harmonia axyridis, a global invasive lady beetle. Scientific Reports, 8, 2689.
Yuan, M., Chu, Z., Li, X., Xu, C., & Wang, S. (2010). The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell, 22, 3164–3176.
Zhao, Z., Zhang, Z., Ding, Z., Meng, H., Shen, R., Tang, H., et al. (2019). Public-transcriptome-database-assisted selection and validation of reliable reference genes for qRT-PCR in rice. Science China Life Sciences, 63, 92–101.
Acknowledgements
We want to thank the Natural Science Foundation of Jiangsu Province of China, Agro-scientific Research in the Public Interest, the National Natural Science Foundation of China and the National Key R&D Program of China for supporting this project.
Funding
This research was supported by grants from the State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products (2010DS700124-KF2007). the National Natural Science Foundation of China (31571974), the Special Fund for Agro–scientific Research in the Public Interest (201303015), the National Key R&D Program of China (2017YFD0200900) and the Natural Science Foundation of Jiangsu Province of China (BK20170606).
Author information
Authors and Affiliations
Contributions
FL and XC designed the study; XC and YD performed the experiments; XC and YD analyzed the data; XC drafted the manuscript; FL and PL reviewed and edited the manuscript. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, X., Laborda, P., Dong, Y. et al. Evaluation of suitable reference genes for normalization of quantitative real-time PCR analysis in rice plants under Xanthomonas oryzae pv. oryzae--infection and melatonin supplementation. Food Prod Process and Nutr 2, 21 (2020). https://doi.org/10.1186/s43014-020-00035-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43014-020-00035-9