Abstract
The present study aims to investigate the genotype distribution of Human papillomavirus (HPV) and variations of HPV18 and HPV58 infection among 6538 females in Luoyang city during 2019–2021. The overall positive rate of females with HPV infection was 12.34%, with 9.74% were infected with single HPV and 2.60% with multiple HPVs. The prevalent rate of high-risk HPV (HR-HPV) was 9.85% and the top five HR-HPV genotypes were HPV52 (1.94%), HPV16 (1.93%), HPV58 (1.48%), HPV51 (1.02%) and HPVV39 (0.99%). Two peaks of HPV infections rates were observed in females aged ≤ 20 and 61–65 years old. To characterize mutations, 39 HPV18 and 56 HPV58 L1, E6 and E7 genes were sequenced and submitted to GenBank. In the HPV18 E6-E7-L1 sequences, 38 nucleotides changes were observed with 10/38 were non-synonymous mutations (5 in E6 gene, 1 in E7 gene and 4 in L1 gene). In the HPV58 E6-E7-L1 sequences, 53 nucleotides changes were observed with 23/53 were non-synonymous mutations (3 in E6 gene, 5 in E7 gene and 15 in L1 gene). Phylogenetic analysis based on L1 gene showed that 92.3% (36/39) of HPV18 isolates fell into sublineage A1 and 7.7% (3/39) belonged to A5. For HPV58, 75.0% (42/56) isolates belonged to sublineage A1 and 25.0% (14/56) were sublineage A2. There was no association between amino mutation and cervical lesions. The present study provides basic information about the distribution, genotypes and variations of HPV among females population in Luoyang city, which would assist in the formulation of HPV screening and vaccination programs and preventive strategies for HPV-attributable cancer in this region.
Similar content being viewed by others
Introduction
Globally, it was estimated that there were 604,000 cases and 342,000 deaths were caused by cervical cancer in 2020, which ranked the fourth most frequently diagnosed cancers1. Females lived in developing countries contributed about 85% cervical cancer and the death rate was higher than developed countries (12.4 vs 5.2 per 100,000)1,2. In 2022, there would be approximately 111,820 new cases of cervical cancer and 61,579 cancer deaths in China3.
Human papillomavirus (HPV) are identified in most patients with cervical carcinoma and the persistent infection with one or more genotypes of oncogenic HPV are the necessary causes of cervical cancer4,5,6. HPV is a small circular non-enveloped double-stranded DNA virus that belongs to the family Papillomavirus7. There are more than 200 different HPVs genotypes have been identified and classified into high-risk HPV (HR-HPV) and low-risk HPV (LR-HPV) based on their potential to cause cancer8. Globally, the most common HPV genotypes in invasive cervical cancer were 16, 18, 31, 33, 35, 45, 52 and 589. Meanwhile, it was reported that the distribution of HPV genotypes exhibited significant differences among countries, even in different areas of a country. For example, the top prevalent HR-HPV genotype was HPV52 in Beijing city and HPV16 in Shanghai city of China10,11. The preventive effect of HPV vaccines on cervical cancer and other HPV-associated diseases has been confirmed in multiple studies12,13. Currently, commercial 2-valent (HPV16 and HPV18), 4-valent (HPV 6, 11, 16 and 18) and 9-valent (HPV 6, 11, 16, 18, 31, 33, 45, 52 and 58) HPV vaccines have been approved by the National Medical Products Administration of China. Some provinces of China, such as Guangdong, have piloted the HPV vaccination program among the native females. Understanding the distribution of HPV genotypes in a region will provide baseline information for the implementation of vaccine-based HPV prevention strategies. Until now, there is limited information about the genotype distribution of HPV in Henan province, located in central of China.
Epidemiological study had showed that except HPV16, HPV18 and HPV58 were the most prevalence genotypes among females with cervical lesions in Henan province of China14. HPV18 can be divided into three lineages and ten sublineages: (1) A, A1–A6; (2) B, B1, B2 and B3; (3) C15. HPV58 has been classified into four lineages and eight sublineages: (1) A, A1–A3; (2) B, B1 and B2; (3) C; (4) D, D1 and D215. The sublineage of HPV18 and HPV58 had been determined in Hunan and Zhejiang province of China and the predominant sublineage was determined to be sublineage A116,17. To our knowledge, there are no reports on the sublineages and sequence variations of HPV18 and HPV58 in Henan province.
Object of the present study is to investigate the genotypes distribution and variations of HPV18 and HPV58 among females in Luoyang city, Henan province, located in central China. The investigation would assist on the formulation and development of vaccine-based HPV prevention strategies against cervical cancer.
Results
Characteristics of the study participants
A total of 6538 females were included in this study and 807 (12.34%) were infected with HPV. As shown in Table 1, the positive rate of HR-HPV was 9.85%, higher than LR-HPV (3.79%) (χ2 = 188.673, P < 0.01). The top five HR-HPV genotypes were HPV52 (1.94%), HPV16 (1.93%), HPV58 (1.48%), HPV51 (1.02%) and HPV39 (0.99%). The most prevalent LR-HPV genotypes were HPV61 (0.89%), followed by HPV54 (0.72%), HPV81 (0.60%), HPV6 (0.37%) and HPV11 (0.29%). Among the 807 HPV positive females, 637 were infected with single HPV genotype, 170 were infected with more than one HPV genotype. Among the 637 females infected with single HPV genotype, the HR-HPV infection accounted for 76.45% (487/637). The multiple HR-HPV infection rate was 2.40%, higher than the multiple LR-HPV infection (1.50%) (χ2 = 13.922, P < 0.01). The top prevalent of the multiple HPV infection was HPV52 (0.70%), followed by HPV58 (0.55%), HPV16 (0.47%), HPV39 (0.41%) and HPV51 (0.41%).
Prevalence of HPV infection in different age groups
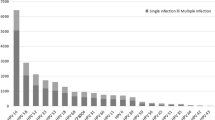
There are significant differences in the positive rates of HPV infection in different age groups (χ2 = 149.128, P < 0.01). The highest rate of any HPV type infection was observed in the ≤ 20 year-old group (29.73%, 11/37), followed by 61–65 year-old group (25.44%, 43/169), while the 31–35 year-old group had the lowest prevalence rate (9.95%, 120/1206). As shown in Fig. 1, the LR-HPV, HR-HPV, single and multiple infection groups showed an identical tendency to the “Any HPV type” infection in different age groups.
Variations of L1 genes on HPV18 and HPV58
Thirty-nine HPV18 and fifty-six HPV58 L1 genes were sequenced successfully. Except for the same sequences, thirteen different HPV18 (18HNL01-18HNL13) and twenty-four HPV58 (58HNL01-58HNL24) sequences were submitted to GenBank (the accession numbers are OP684028-OP684040 and OP684041-OP684064). The HPV18 (AY262282) and HPV58 (D90400) reference sequences were used as the standard for comparison and position of the gene polymorphism sites, separately. The nucleotide sequence mutations of the studied sequences were shown in Tables 2 and 3.
For HPV18 L1 gene, 18HNL02 sequence accounted for 48.7% (19/39) and was the predominant strain. Twenty-one variations were observed in HPV18 L1 gene and four were non-synonymous mutations, including G5503A (R25Q, 39/39), C5920T (A164V, 5/39), A5796G (I123V, 3/39) and C5875A (T149N, 3/39). The most frequency synonymous mutations were A5832C (6/39) and A5924C (6/39) in HPV18 L1 gene.
For HPV58 L1 gene, fifteen nucleotide changes were non-synonymous mutations. The most prevalence synonymous variation was A6560G (27/56), which was found in 26 HPV58 sublineage A1 isolates. The highest rate of non-synonymous mutations was C6688A (T375N, 24/56) and was observed in 24 HPV58 sublineage A1 isolates. The T6434C, A6539G and G6641A variations were only found in sublineage A2. The A6014C synonymous variation was found in 15 HPV58 sequences and 14 were sublineage A2.
Variations of E6–E7 genes on HPV18 and HPV58
A total of thirty-nine HPV18 and fifty-six E6–E7 genes were gained and the nucleotide and amino acid sequences variations are summarized in Tables 4 and 5. For HPV18, thirteen different E6-E7 gene sequences (18HNE01–18HNE13) were submitted to GenBank (the accession numbers are OP684065–OP684077 for HPV18 E6, OP684078–OP684090 for HPV18 E7). The 18HNE01 represented the most predominant strain (27/39), which shared the same sequence with the HPV18 reference strain (AY262282). Five non-synonymous and four synonymous variations were identified on E6 gene. One non-synonymous and seven synonymous were observed on E7 gene. The non-synonymous mutations on E6 gene were E29Q, E40K, R74K and L93R; and on E7 was Q222H.
For HPV58, eleven different E6-E7 sequences (58HNE01–58HNE11) were gained and submitted to GenBank (the accession numbers are OP684091-OP684101 for HPV58 E6, OP684017-OP684027 for HPV58 E7). A total of thirteen gene mutations were observed and five were on E6 and eight on E7. The most frequency synonymous mutation was C307T (11/56) in E6 gene and T744G (42/56) in E7 gene. The most prevalent non-synonymous mutations were A388C (K93N) on E6 gene and T803C (V77A) on E7 gene.
Phylogenetic analysis
Phylogenetic trees based on the full length of HPV18/58 L1 genes were constructed, together with those of reference HPV18/58 L1 sequences that represent individual variant lineages/sublineages. As shown in Fig. 2, 92.3% (36/39) of HPV18 isolates fell into sublineage A1 and 7.7% (3/39) belonged to sublineage A5. Among the fifty-six HPV58 isolates, 75.0% (42/56) belonged to sublineage A1 and 25.0% (14/56) were sublineage A2 (Fig. 3).
Phylogenetic tree generated using nucleotide sequences of the HPV18 L1 gene. Study sequences are labeled in dots, others without dots are reference strain, including: A1 (AY262282, EF202143, MF288710, LC509001, KC470208, GQ180788, MF288706, LC508998), A2 (EF202146, KC470210, KC470211), A3 (EF202147, EF202148, EF202149), A4 (EF202150, EF202151, KC470213), A5 (GQ180787, MF288727), A6 (MF288723, MF288724, MF288725), B1 (EF202153, EF202154, EF202155), B2 (KC470224, KC470225), B3 (EF202152) and C (KC470229, KC470230). Phylogenetic trees were constructed by the Maximum Likelihood by MEGA 6.0 package. Only bootstrap values above 70% are displayed in the branches.
Phylogenetic tree generated using nucleotide sequences of the HPV58 L1 gene. Study sequences are labeled in dots, others without dots are reference strain, including: A1 (D90400, KY225918, KY225919, FJ385262-FJ385268), A2 (KY225926, KY225931, KY225934, HQ537752), A3 (KY225936, KY225937, KY225940, HQ537756, HQ537758), B1 (HQ537761-HQ537763), B2 (HQ537764, HQ537765, KY225956, KY225957), C (KY225961, KY225962, HQ537773, HQ537777), D1 (HQ537766-HQ537768) and D2 (KY225966, KY225967, HQ537768-HQ537770). Phylogenetic trees were constructed by the Maximum Likelihood method by MEGA 6.0 package. Only bootstrap values above 70% are displayed in the branches.
Risk association with cervical lesions
As each non-synonymous mutation in HPV18 E6–E7 gene has only one sample, thus, the risk association of amino mutation with cervical lesions was estimated on HPV58 only. Among the fifty-six females infected only with HPV58, thirty-one were diagnosed with normal cervix by colposcopy examination and twenty-five were diagnosed with CIN2 or worse. It showed that there was no association between amino mutation and cervical lesions (Table 6).
Discussion
Cervical cancer is the leading cause of deaths in China and it was estimated that there were 111,820 new cases and 61,579 deaths in Chinese females in 20223. Persist infection with HR-HPV is known to be the necessary causes of cervical cancer4,5,6. A retrospective study showed that HPV16 and HPV18 were detected in 71% invasive cervical cancer in the world9. In the present study, the distribution of HPV genotypes among 6538 females who underwent gynecological outpatient clinic during 2019–2021 was investigated in Luoyang city. It showed that the overall prevalence of HPV was 12.34%, which was similar to Zhengzhou city of Henan province, but lower than Beijing (21.06%) or Shanghai (18.98%) city10,11,18. The most prevalent HR-HPV genotypes in Luoyang city were HPV52, 16, 58, 51 and 39. The 2-valent, 4-valent and 9-valent HPV vaccines cover 28.6%, 34.0% and 73.1% of HR-HPV positive samples in the present study (data not show). Vaccines contained HPV51 and HPV39 would cover 87.6% of HR-HPV infection in Luoyang city (data not show), which should be taken into consideration in future. In China, the knowledge score and proportion of females who were willing to receive HPV vaccine were relative low19. More efforts should be made by the government to increase the awareness and knowledge of HPV vaccine.
The prevalence of HPV infection in different age population was calculated. It showed that there were two peaks among females with HPV infections, one was ≤ 20, and the other was 61–65 year-old females. The two peaks of HPV infection age group were also observed in other reports18,20,21,22. The first peak of HPV infection may be due to the lack of immunity to HPV in ≤ 20 year-old females23. Thus, the adolescent girls should take priority in the HPV vaccine program. The second peak occurred within the age group 61–65, which was assumed to be caused by the physiologic and immunologic deregulation24. In China, though the government has made enormous investment on cancer screen since 2009, more attention should be paid for females around 60 years old25.
Globally, HPV18 is the second most carcinogenic HPV genotype and has a higher proportion in cervical adenocarcinomas (ADC)26. HPV58 accounted for 6.4% in invasive cervical cancer worldwide, which was especially higher in Eastern Asia27,28. It was reported that HPV18 and HPV58 had a significant association with an increased risk for cervical cancer in China14,29,30,31. Genetic variations and sublineage of HPV may affect the pathogenic potential and host immune responses32,33,34,35. In the present study, the L1, E6 and E7 gene sequences of HPV18 and HPV58 were sequenced. Phylogenetic tree based on the L1 genes showed that the most common HPV18 sublineage was A1, which was similar to other provinces of China16,17. In other countries in Eastern Asia and Pacific, such as Korea and Japan, the predominant HPV18 sublineage was also A136,37. Compared with HPV18 lineage A, the HPV18 B/C tend to cause higher cancer risks38. Four non-synonymous mutations were found in HPV18 L1 gene, including R25Q, I123V, T149N and A164V, which had been detected in Zhejiang province of China17. The R25Q, T149N and A164V mutations were also prevalent in Korea HPV18 sublineage A1 isolates36. In China, it was reported that the distribution of R25Q mutation differed with geographical region and racial characteristic36,39. The A5474G, A5741G, A5796G, C5875A, G6089A, G6143C, A6406G, A7079T and G7130A were only found in HNL03, which represented sublineage A5. Compared with the reference HPV18 sublineage A5 (Accession number: GQ180787), A5468G, A5790G, T5914C, A7073T substitutions were found in L1 sequence. For HPV18 E6 and E7 gene, there were twenty-seven HNE01 isolates that shared the same sequence with the reference. The E29Q, E40K and L93R mutations in E6 protein were also reported in Zhejiang province of China17. Due to the limited numbers of HNE02-HNE13 sequences, associations between amino mutation and cervical lesions were not conducted.
For HPV58 L1 gene, five non-synomous mutations, including N82T, L150F, F318Y, I325M and T375N, had been detected in Liaoning province of China35. The L150F (15/56) was located in the DE loop of L1 protein, which played an important role in the recognition of VLP35. The A6014C, A6416G, T6434C and A6539G in L1 gene variations were present in the whole HPV58 sublineage A2. It was reported that the variations in the fragement of L1 gene (nucleotides 6014–6539) were characterized in HPV58 sublineage A235. In the current research, HPV58 sublineage A1 was the predominant sublineage, which was similar to other provinces of China, such as Zhejiang, Hunan and Liaoning provinces (56.9%)16,34,35. In Japan, most HPV58 isolates belonged to lineage A, with more were sublineage A240. Nevertheless, it was reported that there was no association between HPV58 (sub) lineages and cervical lesions34. For HPV58 E6 gene, the A388C (K93N) variation was the most predominant mutation (55.4%, 31/56), which have also been reported in Hubei province of China41. In our study, no significant difference was observed among HPV58 (K93N) infected females with normal cervix or low-grade lesions. However, it was reported that the K93N can significantly reduce the risk of cervical lesions in Hongkong and Shanghai42,43. The most common synomous mutations observed in HPV58 E6 and E7 gene were C307T and T744G, which had also been reported in the past44,45,46. The C307T and T744G mutations were also common in other countries, such as Mexico, Korean and Italy35,47,48,49. However, the variations included in the present study had been reported to have no association with cervical lesions34,35,49.
Conclusion
In summary, the present study provides basic information about the distribution, genotypes and variations of HPV among females population in Luoyang city, which would assist in the formulation of HPV screening and vaccination programs and preventive strategies for HPV-attributable cancer in this region.
Methods
Study subjects and specimen collection
From April 2019 to April 2021, 6538 females (rang from 18 to 90 years old, mean, 41.14 ± 11.42) who underwent cervical cancer screening in the 989 Hospital of Joint Service Support Force of Chinese PLA (Luoyang city, Henan province, China) were included in this study. The female was considered if she: (a) had no use of vaginal medication or washing in the previous 72 h; (b) had no sexual activity in the previous 24 h; (c) was not presently during menstruation; (d) had no use of acetic or iodine. Before collection, written informed consent from each participant was obtained. The study protocol adhered to the principles of the Declaration of Helsinki and was approved by the institutional ethics committee (Grant No: LLSC20190305).
HPV genotyping
Cervical specimens were collected by a gynecological practitioner using a cytobrush from the ecto- and endocervix of uterus. The samples were stored at – 20 °C until the HPV genotyping. The HPV genotype was proceed by a commercial gene chip (Chaozhou Hybribio Limited Corporation, Chaozhou, China) according to the manufacturer’s instruction. The gene chip contained 37 genotype-specific oligonucleotides designed to detect 18 high-risk human papillomavirus (HR-HPV: 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73 and 82) and 19 low-risk human papillomavirus (LR-HPV: 6, 11, 34, 40, 42, 43, 44, 54, 55, 57, 61, 67, 69, 70, 71, 72, 81, 83 and 84). The final results were determined by colorimetric change on the chip under direct visualization and blue-purple spots were recognized as HPV positive.
HPV sequencing
Single HPV18 and HPV58 positive samples were chosen and used to amplify the full length of L1, E6 and E7 genes. The primers were designed based on published HPV 18 (AY262282) and HPV58 (D90400) sequences in GenBank and further synthesized by Sangon Biotech, Inc. (Shanghai, China) (Table 7). Each 50 μl PCR reaction mixture contained 2 μl of each primer, 25 μl 2 × PrimeSTAR Max Premix (Takara Biotechnology Co., LTD, Dalian, China), 19 μl of ultrapure water and 2 μl of template cDNA. The PCR reaction conditions were as follows: 94 °C for 10 min; 35 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C for 60 s, 72 °C for 10 min. The amplified products were ligated into p-EASY-Blunt cloning vector (TransGen Biotech, China) according to manufacturer’s instruction and then applied for sequencing by Sangon Biotech, Inc. (Shanghai, China).
Variants and phylogenetic analysis of HPV18 and HPV58
To identify the variations on the HPV18 and HPV58 L1, E6 and E7 genes, the reference HPV18 (GeneBank AY262282) and HPV58 (GeneBank D90400) sequences were selected and compared with the studied sequences. The comparison was proceeding by DNAStar (Madison, WI, USA) and positions of variations were numbered based on the reference sequence.
Phylogenetic trees based on the L1 gene of HPV18 and HPV58 were constructed through Maximum Likelihood method with 1000 bootstrapped replicates using the MEGA (version 6.0). Reference sequences that represent each HPV18 and HPV58 lineage were used to construct the distinct phylogenetic branches.
Statistical analysis
SPSS version 19.0 (IBM, Armonk, NY, USA) was used to assess the significance of differences in HPV positivity rates among groups. Females with no lesion at colposcopy biopsy were grouped as normal cervices, with CIN2 or worse were grouped as outcome variable. The oncogenic risk association of HPV18 and HPV58 E6/E7 amino acid substitutions was assessed using Chi-squared test or Fisher’s exact test. P-value < 0.05 was considered to be statistically significant.
Ethics approval and consent to participate
Females were informed and a written consent was received. The study protocol adhered to the principles of the Declaration of Helsinki and was approved by the institutional ethics committee in the 989 Hospital of Joint Service Support Force of Chinese PLA, Military Training Medical Research Institute of the Whole Army (Grant No: LLSC20190305).
Data availability
The datasets generated during the current study are not publicly available yet, due to privacy concerns and ongoing additional research. Data can be made available for peer review on reasonable request through contacting the corresponding author. The sequences described in manuscript were submitted to GenBank and have gained the accession numbers (OP684017-OP684101).
Abbreviations
- HPV:
-
Human papillomavirus
- LR-HPV:
-
Low-risk human papillomavirus
- HR-HPV:
-
High-risk human papillomavirus
References
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. https://doi.org/10.3322/caac.21660 (2021).
Ferlay, J. et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359-386. https://doi.org/10.1002/ijc.29210 (2015).
Xia, C. et al. Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin. Med. J. (Engl.) 135, 584–590. https://doi.org/10.1097/CM9.0000000000002108 (2022).
Bosch, F. X. & de Sanjose, S. Human papillomavirus in cervical cancer. Curr. Oncol. Rep. 4, 175–183. https://doi.org/10.1007/s11912-002-0079-y (2002).
Munoz, N. et al. Role of parity and human papillomavirus in cervical cancer: The IARC multicentric case-control study. Lancet 359, 1093–1101. https://doi.org/10.1016/S0140-6736(02)08151-5 (2002).
Bosch, F. X., Lorincz, A., Munoz, N., Meijer, C. J. & Shah, K. V. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 55, 244–265. https://doi.org/10.1136/jcp.55.4.244 (2002).
Bzhalava, D., Eklund, C. & Dillner, J. International standardization and classification of human papillomavirus types. Virology 476, 341–344. https://doi.org/10.1016/j.virol.2014.12.028 (2015).
Munoz, N. et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348, 518–527. https://doi.org/10.1056/NEJMoa021641 (2003).
de Sanjose, S. et al. Human papillomavirus genotype attribution in invasive cervical cancer: A retrospective cross-sectional worldwide study. Lancet Oncol. 11, 1048–1056. https://doi.org/10.1016/S1470-2045(10)70230-8 (2010).
Ma, L. et al. Characteristics of women infected with human papillomavirus in a tertiary hospital in Beijing China, 2014–2018. BMC Infect. Dis. 19, 670. https://doi.org/10.1186/s12879-019-4313-8 (2019).
Zhang, C. et al. Prevalence and genotype distribution of human papillomavirus among females in the suburb of Shanghai, China. J. Med. Virol. 90, 157–164. https://doi.org/10.1002/jmv.24899 (2018).
Patel, C. et al. The impact of 10 years of human papillomavirus (HPV) vaccination in Australia: What additional disease burden will a nonavalent vaccine prevent?. Euro Surveill. https://doi.org/10.2807/1560-7917.ES.2018.23.41.1700737 (2018).
Kavanagh, K. et al. Changes in the prevalence of human papillomavirus following a national bivalent human papillomavirus vaccination programme in Scotland: A 7-year cross-sectional study. Lancet Infect. Dis. 17, 1293–1302. https://doi.org/10.1016/S1473-3099(17)30468-1 (2017).
Chen, G. et al. Prevalence and genotype distribution of human papillomavirus in women with cervical cancer or cervical intraepithelial neoplasia in Henan province, central China. J. Med. Virol. https://doi.org/10.1002/jmv.25670 (2020).
Burk, R. D., Harari, A. & Chen, Z. Human papillomavirus genome variants. Virology 445, 232–243. https://doi.org/10.1016/j.virol.2013.07.018 (2013).
Zu, Y. et al. Genetic characteristics of human papillomavirus type 16, 18, 52 and 58 in southern China. Genomics 113, 3895–3906. https://doi.org/10.1016/j.ygeno.2021.09.006 (2021).
Xu, H. H. et al. Human papillomavirus (HPV) 18 genetic variants and cervical cancer risk in Taizhou area, China. Gene 647, 192–197. https://doi.org/10.1016/j.gene.2018.01.037 (2018).
Liu, J. et al. Prevalence and genotype distribution of human papillomavirus in Zhengzhou, China, in 2016. Arch. Virol. 165, 731–736. https://doi.org/10.1007/s00705-019-04515-3 (2020).
Hu, S. et al. A nationwide post-marketing survey of knowledge, attitude and practice toward human papillomavirus vaccine in general population: Implications for vaccine roll-out in mainland China. Vaccine 39, 35–44. https://doi.org/10.1016/j.vaccine.2020.11.029 (2021).
Ge, Y. et al. Prevalence of human papillomavirus infection of 65,613 women in East China. BMC Public Health 19, 178. https://doi.org/10.1186/s12889-019-6487-9 (2019).
Zhao, P. et al. Prevalence and genotype distribution of human papillomavirus infection among women in northeastern Guangdong Province of China. BMC Infect. Dis. 18, 204. https://doi.org/10.1186/s12879-018-3105-x (2018).
Liu, X. X. et al. Human papillomavirus prevalence and type-distribution among women in Zhejiang Province, Southeast China: A cross-sectional study. BMC Infect. Dis. 14, 708. https://doi.org/10.1186/s12879-014-0708-8 (2014).
Mai, Q., Yang, X., Cheng, H., Wu, G. & Wu, Z. Prevalence and genotype distribution of human papillomavirus among women with cervical lesions in Shenzhen city, China. Hum. Vaccin. Immunother. https://doi.org/10.1080/21645515.2020.1805993 (2020).
Althoff, K. N. et al. Correlates of cervicovaginal human papillomavirus detection in perimenopausal women. J. Women’s Health 18, 1341–1346 (2009).
Duan, R., Qiao, Y., Clifford, G. & Zhao, F. Cancer burden attributable to human papillomavirus infection by sex, cancer site, age, and geographical area in China. Cancer Med. 9, 374–384. https://doi.org/10.1002/cam4.2697 (2020).
Li, N., Franceschi, S., Howell-Jones, R., Snijders, P. J. & Clifford, G. M. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: Variation by geographical region, histological type and year of publication. Int. J. Cancer 128, 927–935. https://doi.org/10.1002/ijc.25396 (2011).
Chan, P. K. et al. Geographical distribution and oncogenic risk association of human papillomavirus type 58 E6 and E7 sequence variations. Int. J. Cancer 132, 2528–2536. https://doi.org/10.1002/ijc.27932 (2013).
Chan, P. K. et al. Meta-analysis on prevalence and attribution of human papillomavirus types 52 and 58 in cervical neoplasia worldwide. PLoS ONE 9, e107573. https://doi.org/10.1371/journal.pone.0107573 (2014).
Tao, X. et al. Prevalence and carcinogenic risk of high-risk human papillomavirus subtypes in different cervical cytology: A study of 124,251 cases from the largest academic center in China. J. Am. Soc. Cytopathol. 10, 391–398. https://doi.org/10.1016/j.jasc.2021.03.006 (2021).
Serrano, B. et al. Human papillomavirus genotype attribution for HPVs 6, 11, 16, 18, 31, 33, 45, 52 and 58 in female anogenital lesions. Eur. J. Cancer 51, 1732–1741. https://doi.org/10.1016/j.ejca.2015.06.001 (2015).
Xu, H. et al. Diagnostic accuracy of high-risk HPV genotyping in women with high-grade cervical lesions: evidence for improving the cervical cancer screening strategy in China. Oncotarget 7, 83775–83783. https://doi.org/10.18632/oncotarget.11959 (2016).
Yang, R. et al. Papillomavirus capsid mutation to escape dendritic cell-dependent innate immunity in cervical cancer. J. Virol. 79, 6741–6750. https://doi.org/10.1128/JVI.79.11.6741-6750.2005 (2005).
Xi, L. F. et al. Risk for high-grade cervical intraepithelial neoplasia associated with variants of human papillomavirus types 16 and 18. Cancer Epidemiol. Biomark. Prev. 16, 4–10. https://doi.org/10.1158/1055-9965.EPI-06-0670 (2007).
Yu, J. H. et al. Genetic variability and oncogenic risk association of human papillomavirus type 58 E6 and E7 genes in Taizhou area, China. Gene 686, 171–176. https://doi.org/10.1016/j.gene.2018.11.066 (2019).
Liu, J. H. et al. Variations of human papillomavirus type 58 E6, E7, L1 genes and long control region in strains from women with cervical lesions in Liaoning province, China. Infect. Genet. Evol. 12, 1466–1472. https://doi.org/10.1016/j.meegid.2012.05.004 (2012).
Chen, A. A. et al. Human papillomavirus 18 genetic variation and cervical cancer risk worldwide. J. Virol. 89, 10680–10687. https://doi.org/10.1128/JVI.01747-15 (2015).
Yamaguchi-Naka, M. et al. Molecular epidemiology of human papillomavirus 18 infections in Japanese Women. Infect. Genet. Evol. 83, 104345. https://doi.org/10.1016/j.meegid.2020.104345 (2020).
Sichero, L. et al. High grade cervical lesions are caused preferentially by non-European variants of HPVs 16 and 18. Int. J. Cancer 120, 1763–1768. https://doi.org/10.1002/ijc.22481 (2007).
Liu, H. et al. Cervical human papillomavirus among 19,753 women attending gynecological department of a major comprehensive hospital in north Anhui China 2013–2016: Implication for cervical cancer screening and prevention. J. Med. Virol. 91, 698–706. https://doi.org/10.1002/jmv.25365 (2019).
Tenjimbayashi, Y. et al. Whole-genome analysis of human papillomavirus genotypes 52 and 58 isolated from Japanese women with cervical intraepithelial neoplasia and invasive cervical cancer. Infect. Agent Cancer 12, 44. https://doi.org/10.1186/s13027-017-0155-4 (2017).
Yang, Z. et al. Genetic variability of E6 and E7 genes of human papillomavirus type 58 in Jingzhou, Hubei Province of central China. Virol. J. 19, 71. https://doi.org/10.1186/s12985-022-01801-6 (2022).
Zhao, J. et al. Phylogeny and polymorphism in the E6 and E7 of human papillomavirus: alpha-9 (HPV16, 31, 33, 52, 58), alpha-5 (HPV51), alpha-6 (HPV53, 66), alpha-7 (HPV18, 39, 59, 68) and alpha-10 (HPV6, 44) in women from Shanghai. Infect. Agent. Cancer 14, 38. https://doi.org/10.1186/s13027-019-0250-9 (2019).
Chan, P. K. et al. Association of human papillomavirus type 58 variant with the risk of cervical cancer. J. Natl. Cancer Inst. 94, 1249–1253. https://doi.org/10.1093/jnci/94.16.1249 (2002).
Liu, Y., Pan, Y., Gao, W., Ke, Y. & Lu, Z. Whole-genome analysis of human papillomavirus types 16, 18, and 58 isolated from cervical precancer and cancer samples in Chinese women. Sci. Rep. 7, 263. https://doi.org/10.1038/s41598-017-00364-9 (2017).
Chen, Z. et al. E6 and E7 gene polymorphisms in human papillomavirus types-58 and 33 identified in Southwest China. PLoS ONE 12, e0171140. https://doi.org/10.1371/journal.pone.0171140 (2017).
Yang, L. et al. Genetic variability of HPV-58 E6 and E7 genes in Southwest China. Infect. Genet. Evol. 21, 395–400. https://doi.org/10.1016/j.meegid.2013.12.009 (2014).
Conde-Ferraez, L. et al. Genetic variability in E6 and E7 oncogenes from human papillomavirus type 58 in Mexican women. Intervirology 60, 235–246. https://doi.org/10.1159/000489306 (2017).
Bae, J. H. et al. Distribution of human papillomavirus type 58 variants in progression of cervical dysplasia in Korean women. J. Microbiol. Biotechnol. 19, 1051–1054. https://doi.org/10.4014/jmb.0812.679 (2009).
Cento, V., Rahmatalla, N., Ciccozzi, M., Perno, C. F. & Ciotti, M. Intratype variations of HPV 31 and 58 in Italian women with abnormal cervical cytology. J. Med. Virol. 83, 1752–1761. https://doi.org/10.1002/jmv.22201 (2011).
Acknowledgements
The authors are grateful to the staff at the Department of Gynecological Outpatient Clinic, the 989th hospital of the People's Liberation Army (PLA), Luoyang, Henan, China.
Author information
Authors and Affiliations
Contributions
Y.W. conceptualized the study, took the test, analyzed the data and wrote the manuscript; S.Z.H. designed the study, took the test, analyzed the data and wrote the manuscript; X.C.W. helped to recruit patients and conducted the clinical performance; S.Y.S. reviewed the manuscript; X.L.W. helped to recruit patients and conducted the clinical performance; X.C.W. and X.L.W. had full access to all of the data in the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, Y., Han, S., Wang, X. et al. Characteristics of human papillomavirus infection among females and the genetic variations of HPV18 and HPV58 in Henan province, China. Sci Rep 13, 2252 (2023). https://doi.org/10.1038/s41598-022-24641-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-24641-4
- Springer Nature Limited
This article is cited by
-
The potential use of therapeutics and prophylactic mRNA vaccines in human papillomavirus (HPV)
Virology Journal (2024)
-
Genetic variability of human papillomavirus type 18 based on E6, E7 and L1 genes in central China
Virology Journal (2024)