Abstract
Background
Persistent high-risk Human papillomavirus (HPV) subtypes infection has been implicated as a causative of cervical cancer. Distribution and genotypes of HPV infection among females and their variations would assist in the formulation of preventive strategy for cervical cancer. The purpose of the present study is to investigate the prevalence of HPV among females in central China.
Methods
The distribution and genotypes of HPV among 9943 females attending the gynecological examinations in central of China during 2015–2021 were investigated. HPV genotypes were detected using a commercial kit. Nucleotides sequences of L1, E6 and E7 genes in HPV16 or HPV52 positive samples collected in 2021 were amplified by polymerase chain reaction (PCR). Variations of L1, E6 and E7 in HPV16 and HPV52 were gained by sequencing and compared with the reference sequence. Sublineages of HPV16 and HPV52 were determined by the construction of phylogenetic tree based on L1 gene.
Results
The overall prevalence of HPV infection was 22.81%, with the infection rate of high-risk human papillomavirus (HR-HPV) was 19.02% and low-risk human papillomavirus (LR-HPV) was 6.40%. The most top five genotypes of HPV infection were HPV16 (7.49%), HPV52 (3.04%), HPV58 (2.36%), HPV18 (1.65%) and HPV51 (1.61%). Plots of the age-infection rate showed that the single HPV, multiple HPV, HR-HPV, LR-HPV infection revealed the same tendency with two peaks of HPV infection were observed among females aged ≤ 20 year-old and 60–65 year-old. The predominant sublineage of HPV16 was A1 and B2 for HPV52. For HPV16, The most prevalent mutations were T266A (27/27) and N181T (7/27) for L1, D32E for E6 and S63F for E7 in HPV16. For HPV52, all of the nucleotide changes were synonymous mutation in L1 (except L5S) and E7 genes. The K93R mutation was observed in most HPV52 E6 protein.
Conclusions
The present study provides basic information about the distribution, genotypes and variations of HPV among females population in Henan province, which would assist in the formulation of preventive strategies and improvements of diagnostic probe and vaccine for HPV in this region.
Similar content being viewed by others
Background
Cervical cancer is the fourth most commonly diagnosed and leading cause of cancer death among females worldwide, with an estimated 570,000 cases and 311,000 deaths in 2018 [1]. Around 85% of women diagnosed and 87% of women who died from cervical cancer live in the developing countries [2]. Human papillomavirus (HPVs) are found in the cervical carcinoma tissues of most patients and the oncogenic HPVs are regarded as the major cause of cervical cancer. In China, it was reported that there are estimated 110,650 new cancer cases and 36,714 cancer deaths are attributable to HPVs infection in 2015, of which cervical cancer accounted for 85.6% and 78.1% [3].
HPVs are small non-enveloped double-stranded DNA viruses that belong to the genus Alpha-Papillomaviridae family [4]. The HPVs genomes are about 7.2–8.0 kb and contain eight open reading frames (ORFs), including: the presumptive early (E1–E2, E4–E7), late (L1 and L2) and Long Control Region (LCR) [5,6,7]. The continued expression of the E6 and E7 genes is related to induce cellular immortalization, transformation, and carcinogenesis [6]. The E6 and E7 proteins would be candidate for the development of therapeutic vaccines [8]. The L1 protein is the primary composition of HPVs and can self-assemble into virus like particles (VLPs) [9]. The first generation commercial HPV vaccines are based on the recombinant expression of L1 protein in system [10, 11]. Human immunized with commercial HPV vaccines can acquire robust immunity against the homology genotype [9]. The polymorphisms of HPV L1 gene affect the generation of neutralization antibody of different binding affinities [12].
More than 200 different HPVs genotypes have been characterized according to the greater than 10% difference within the L1 gene sequence [4, 5, 13]. Based on their association with cervical cancer, HPVs genotypes are classified into high-risk HPV (HR-HPV, including HPV52, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58 and 59) and low-risk HPV (LR-HPV, including HPV6, 11, 40, 42, 43 and 44) [13]. Individual HPV genotypes are referred to as variants or subtypes when less than 10% difference is within the L1 gene sequence [5]. The HPVs variants are grouped into distinct lineages and sublineages based on nucleotides alignments and phylogenetic analyses [5]. Usually, the L1 nucleotides differences among HPV lineages and sublineages are 1.0–10.0% and 0.5–1.0%, respectively [5].
The most common HPV genotypes in invasive cervical cancer were 16, 18, 31, 33, 35, 45, 52 and 58 worldwide [14]. In different regions of China, the most prevalent HPV genotypes are different. For example, in Beijing and Jiangsu province, HPV16 was the predominant type, in Shanghai and Zhejiang province, HPV52 was the most common type [15,16,17,18]. HPV16 has been classified into four major lineages and nine sublineages, including: (1) A, includes A1-3 (European [E]) and A4 (Asian [As]) sublineages; (2)B, includes B1 (African-1 [Afr1a]) and B2 (African-1 [Afr1b]); (3) C (African-2 [Afr2a]); and (4) D, includes D1 (North American 1 [NA]), D2 (Asian-American 2 [AA]) and D3 (Asian-American 1 [AA]) [5, 19]. The lineages and sublineages of HPV16 sequences have geography characteristics [20,21,22]. HPV52 has been classified into four major lineages and seven sublineages, including A (A1 and A2), B (B1 and B2), C (C1 and C2) and D [5]. Sublineages and single nucleotide polymorphism (SNPs) in HPV, especially the E6 and E7, are associated with the disease status of HPV persistent infection and the elevated risk of cervical carcinoma [13, 23,24,25,26,27].
In the present study, the distribution and genotypes of HPV infection among females in Henan province during 2015–2021 were investigated based on commercial HPV test kit. As the L1 gene plays an important role in the classification of HPV sublineage, so L1 gene of HPV16 and HPV52 were sequenced and applied to phylogenetic analysis. The E6 and E7 are the major oncogenes and the variations are correlated with the progression of cervical lesions. The L1, E6 and E7 genes of HPV16 and HPV52 were sequenced and compared with the reference HPVs strain. The distribution and genotypes of HPVs would assist on the formulation of the vaccination program and preventative strategies against cervical cancer. Variations of the HPVs genetic may be useful for the analysis of cervical cancer risk, even provide crucial information for the development of diagnostic tools and vaccine design.
Methodology
Study population and samples
From May 2015 to May 2021, consecutive cervical swabs from females who attended in the gynecological outpatient in the 989 Hospital of Joint Service Support Force of Chinese PLA, Military Training Medical Research Institute of the Whole Army, which is located in Luoyang city, Henan province, central of China were collected. The hospital is open for non-military people in China. The female was eligible to be study if she: (a) had no use of vaginal medication or washing in the previous 72 h; (b) had not had sexual intercourse in the previous 24 h; (c) was not presently during menstruation; (d) had no use of acetic or iodine. Before collection, the females were informed and a written consent was received. The study protocol was approved by the institutional ethics committee in the 989 Hospital of Joint Service Support Force of Chinese PLA, Military Training Medical Research Institute of the Whole Army (Grant No.: LLSC20150305).
HPV genotyping
The DNA of the samples was extracted by kit and then applied to HPV genotypes by flow-through hybridization and gene chip (Chaozhou Hybribio Limited Corporation, Chaozhou, China) according to the manufacturer’s instruction. The PCR reaction volume was 25 μl, which included 1 μl of template DNA, 23.25 μl buffer and 0.75 μl Taq DNA polymerase (Chaozhou Hybribio Limited Corporation, Chaozhou, China). The PCR program was as follows: initial denaturation step at 95°C for 9 min; followed by 40 cycles of 95°C for 20 s, 55°C for 30 s, 72°C for 30 s, and a final 72°C extension for 5 min. The genotyping was performed via hybridization of the PCR products to gene chip containing 37 genotype-specific oligonucleotides and the genotype was analyzed using HybriMax (Chaozhou Hybribio Limited Corporation, Chaozhou, China). The chip can identify 37 genotypes, including 19 LR-HPV (namely 6, 11, 34, 40, 42, 43, 44, 54, 55, 57, 61, 67, 69, 70, 71, 72, 81, 83 and 84) and 18 HR-HPV (16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73 and 82). The final results were detected by colorimetric change on the chip under direct visualization and blue-purple spots were recognized as HPV positive.
HPV sequencing
Samples collected in 2021 that were only positive for HPV16 or HPV52 were chosen and processed for the variant analysis of L1, E6 and E7 genes by sequencing. To amplify the full length of the L1, E6 and E7 genes, primers were designed based on published HPV16 (GeneBank NC 001526) and HPV52 (GeneBank NC 001592) sequences. The primers used for the amplification of L1, E6 and E7 genes were shown in Table1 and synthesized by Sangon Biotech, Inc. (Shanghai, China). The PCR reaction volume was 50 μl, which included 2 μl of template DNA, 25 μl 2 × PrimeSTAR Max Premix(Takara Biotechnology Co., LTD, Dalian, China), 2 μl of each primer and 19 μl of ultrapure water. The PCR program was as follows: initial denaturation step at 94°C for 10 min; followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 30 s, and a final 72°C extension for 10 min. The PCR products were visualized on 1% agarose gels stained with GoldView TM Nucleic Acid Stain. Identified plasmids containing the L1, E6 or E7 genes were used as positive control and the reaction mixture containing no template as negative control. The targets fragments were then purified using TIANgel Midi Purification Kit (TIANGEN BIOTECH, China) and ligated into p-EASY-Blunt cloning vector (TransGen Biotech, China) according to manufacturer’s instruction. The recombinant plasmids were then transformed into Trans1-T1 Phage Resistant Chemically Competent Cells (TransGen Biotech, China) according to manufacturer’s protocols. The positive clones containing the recombinant plasmids were sent to Sangon Biotech, Inc. (Shanghai, China) for sequencing.
Molecular characterization and phylogenetic analysis of HPV16 and HPV52
The variations of the L1, E6 and E7 genes and proteins were gained by the comparison and numbered with the reference strain HPV16 (GeneBank NC 001526) and HPV52 (GeneBank NC 001592) by DNAStar (Madison, WI, USA). Variants between the studied and reference sequence were noted and the frequencies were calculated.
Phylogenetic analyses the L1 gene of HPV16 and HPV52 were constructed using the MEGA (version 6.0). A neighbor-joining algorithm was employed and Kimura 2-parameter distance neighbor-joining trees were built with 1000 bootstrapped replicates. Furthermore, HPV16 and HPV52 reference strains that deposited into NCBI GenBank Database were included to represent each lineage [5]. For HPV16, the reference strains include A1 (K02178), A2 (AF536179), A3 (HQ644236), A4 (AF534061 and LC368960), B1 (AF536180), B2 (HQ644298), C (AF472509), D1 (HQ644257), D2 (AY686579) and D3 (AF402678). For HPV52, the reference strains include A1 (X74481), A2 (HQ537739), B1 (HQ537740), B2 (HQ537743), C1 (HQ537744), C2 (HQ537746) and D (HQ537748).
Statistical analysis
SPSS version 19.0 (IBM, Armonk, NY, USA) was used to assess the significance of differences detected in the frequency of HPV infections among groups. The χ2 test was used to compare the prevalence of HPV infection. A p-value < 0.05 was considered statistically significant.
Results
Characteristics of the study participants
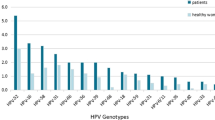
A total of 9943 females (ranging from 18 to 90 years old, the median age is 41.15 ± 11.41) underwent outpatient gynecological examinations met the participation criteria. Overall, 22.81% of females (2268/9943) were found to be HPV positive for any HPV DNA, of whom 19.02% (1891/9943) were found to be HR-HPV infection (including samples are HR-HPV only and both positive for HR-HPV and LR-HPV), substantially higher than the LR-HPV infection (including samples are positive for LR-HPV only and both positive for LR-HPV and HR-HPV) (6.40%, 636/9943) (P < 0.01) (Table 2). The top five commonly identified HPV genotypes were all HR-HPV, including HPV16 (7.49%, 745/9943), HPV52 (3.04%, 302/9943), HPV58 (2.36%, 235/9943), HPV18 (1.65%, 164/9943) and HPV51 (1.61%, 160/9943). The most prevalent LR-HPV subtypes were as follows: HPV81 (CP8304) (1.46%, 145/9943), HPV61 (1.36%, 135/9943), HPV54 (1.24%, 123/9943), HPV6 (0.58%, 58/9943) and HPV40 (0.53%, 53/9943) (Fig. 1).
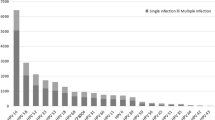
Among the 9943 females, 17.41% (1731/9943) were infected with single HPV subtype, whereas 5.40% (537/9943) were infected with multiple HPV subtypes. In the multiple infection groups, the highest infection rate was double infection 3.71% (369/9943) (Table 2). In the single infection group, HPV16 (7.49%) was the most prevalent subtype, followed by HPV52 (3.04%), HPV58 (2.36%) and HPV18 (1.65%). In the multiple HPV infections group, the most prevalent subtypes were as follows: HPV16 (1.55%), HPV52 (1.29%), HPV58 (1.09%), HPV51 (0.84%), HPV81 (0.82%) (Fig. 1).
Prevalence of HPV infection in different age groups and years
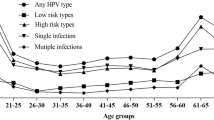
To evaluate the relationship between HPV infection with the age, females were divided into fourteen age groups. There are significant differences in the HPV infection rates among females in different age groups (χ2 = 134.563, P < 0.01). Among the 2268 females with HPV infection, there were two peaks of HPV infection, the first was in the ≤ 20 year-old group (31.48%, 17/54) and the second was in the 61–65 year-old group (38.04%, 151/397). All of the HR-HPV, LR-HPV, single infection and multiple infection groups showed the same tendency with the “Any HPV type” infection in different age groups (Fig. 2).
For all of the groups, the HPV infection rates are significant differences (P < 0.01) during 2015 and 2021. The highest HPV infection rates were observed in 2015, with the any HPV type was 34.98%, the high risk types was 30.26%, the single infection was 27.11% and then declined gradually (Fig. 3).
HPV16 and HPV52 L1 gene nucleotide variations and amino acid mutational analysis
Twenty-seven HPV16 L1 genes were sequenced successfully and twelve different sequences were submitted to GenBank (MZ546238-MZ546249). The twelve HPV16 L1 gene shared 99.6–99.9% identities with the reference sequence (NC 001526). The variation sites and frequencies of HPV16 L1 gene are shown in Table3. Twelve variations (0.8%, 12/1596) were identified in the 1596 bp L1 gene. Specially, four changes were non-synonymous mutations, including: H76Q (1/27), N181T (7/27), E240D (1/27) and T266A (27/27). The A5570G gene variation was found in all of 27 samples and brought the amino acid change from threonine to alanine (T266A). Another high frequent variation was A5316C (25.9%, 7/27), which changed asparagine into threonine (N181T). The most common synonymous mutations in L1 gene were A5803C (66.7%, 18/27) and G6196A (70.4%, 19/27).
Fifteen HPV52 L1 genes were sequenced successfully and five different nucleotide sequences were gained by comparison and then submitted to GenBank (OL589507-OL589511). The five sequences shared 99.0%-99.9% identities with the reference sequence (NC 005192). Compared with the NC 001592, nineteen nucleotide changes were identified among the fifteen sequences (Table 4). All of the changes in HPV52 L1 gene were synonymous mutation except L5S (6.7%, 1/15). Seven synonymous mutations in HPV52 L1 gene, including G6110A, T6701G, T6764C, A6794G and C6824T were found in 93.3% (14/15) samples. The G6218A was detected in all of fifteen sequences.
HPV16 and HPV52 E6-E7 gene nucleotide variations and amino acid mutational analysis
Eighteen HPV16 E6 and E7 genes were sequenced successfully. Thirteen different E6 sequences (MZ546266-MZ546278) and E7 sequences (MZ546295-MZ546307) were obtained and the identity was 99.4–100% for E6 gene and 98.7–100% for E7 gene compared with the HPV16 reference sequence (NC 001526). Compared with the reference sequence, nine nucleotide mutations were observed in the HPV16 E6 genes and eight were non-synonymous mutations (Table 5). The most frequently non-synonymous mutation in HPV E6 genes were T7220G (A) (5/18), which made D32E mutation. Eight nucleotide changes occurred in the HPV16 E7 genes with four were non-synonymous mutations. The most frequently observed non-synonymous in HPV16 E7 genes were C7791T (S63F) (Table 5).
Fifteen HPV52 E6 and E7 genes were sequenced and thirteen sequences were identical (Table 6). The most prevalent non-synonymous mutations in E6 genes was A379G (14/15) and cause the amino acid to change from Lysine to arginine (K93R). For E7 sequences, the high frequent mutations was C751T and A801G, both were synonymous mutations.
Phylogenetic analysis
Phylogenetic analysis based on the full length of HPV16 and HPV52 L1 genes are shown (Figs. 4 and 5). All of the twenty-seven HPV16 L1 sequences, 70.4% (19/27) were A1 sublineage, 22.2% (6/27) were A4 sublineage and 7.4% (2/27) were A3 (Fig. 4). All of the fifteen HPV52 L1 sequences, 93.3% (14/15) were B2 sublineages (Fig. 5).
Neighbor joining phylogenetic tree generated using nucleotide sequences of the HPV16 L1 gene. Legend: Study sequences are labeled in dots, others without dots are reference strain, including: A1 (K02178), A2 (AF536179), A3 (HQ644236), A4 (AF534061 and LC368960), B1 (AF536180), B2 (HQ644298), C (AF472509), D1 (HQ644257), D2 (AY686579), D3 (AF402678). Phylogenetic trees were constructed by the Neighbor-Joining method and the Kimura 2-parameter model by MEGA 6.0 package. Only bootstrap values above 50% are displayed in the branches
Neighbor joining phylogenetic tree generated using nucleotide sequences of the HPV52 L1 gene. Legend: Study sequences are labeled in dots, others without dots are reference strain, including: A1 (X74481.1), A2 (HQ537739), B1 (HQ537740), B2 (HQ537743), C1 (HQ537744), C2 (HQ537746), D (HQ537748). Phylogenetic trees were constructed by the Neighbor-Joining method and the Kimura 2-parameter model by MEGA 6.0 package. Only bootstrap values above 50% are displayed in the branches
Discussion
Globally, cervical cancer is the fourth most common malignancy in females around world and contributes 530 000 new cases per year [1]. Persist infection with HR-HPV has been identified as a major risk factor for cervical cancer. The prevalence of high-risk HPVs infections, such as HPV16, HPV18, HPV52, increased with severity of cervical lesions [28]. In the present study, a retrospective survey of HPV infection among 9943 females who underwent gynecological outpatient clinic during 2015 to 2021 was conducted in a located hospital. Though it is a military hospital, it is open for non-military people. Occupation of females in this study was unclear, which may take deviations in statistical analysis. In this study, the overall prevalence of HPV in Henan province was 22.81%, which was similar to Beijing (21.06%), Zhejiang (22.8%), Sichuan (24.01%) and Jiangxi (22.49%) provinces [29,30,31,32]. However, significant differences on the HPV infection rates were observed in other provinces, such as Shaanxi (30.21%), Xinjiang (9.34%) and Yunnan (7.6%) provinces [33,34,35]. Many factors, including sex, age, ethnicity and socioeconomic status, may contribute to the significant difference on the HPV infection rates in different areas [36].
In the present study, HPV16 was the most prevalent HR-HPV genotype (7.49%), followed by HPV52 (3.04%), HPV58 (2.36%), HPV18 (1.65%) and HPV51 (1.61%). In some regions of China, for example Beijing, the top prevalent HR-HPV genotypes in patients were as follows: HPV16 (27.34%), HPV58 (12.52%), HPV52 (11.89%) and HPV51 (6.33%) [16]. In Shanghai, HPV52 was the most predominant genotype (3.58%), followed by HPV16 (2.85%), HPV58 (2.64%), HPV53 (1.81%) and HPV39 (1.46%) [17]. The top five LR-HPV in the present study were HPV81 (1.46%), HPV61 (1.36%), HPV54 (1.24%), HPV6 (0.58%) and HPV40 (0.53%). Commercial 2-valent (HPV16 and HPV18), 4-valent (HPV 6, 11, 16 and 18) and 9-valent (HPV 6, 11, 16, 18, 31, 33, 45, 52 and 58) HPV vaccine have been approved by the National Medical Products Administration since 2016. However, the proportion of females who were willing to receive HPV vaccine are relative low [37]. The present study may provide valuable data to inform cervical cancer screen and the implementation of HPV vaccination in Henan province. For the development of HPV multiplicity vaccine, vaccines contained HPV51 and HPV81 would be more efficiency for the prevention of HPV infection in China.
The relationship between population age and HPV infection rate was investigated. Two peaks of infection rate were observed in the any type HPV, HR-HPV, LR-HPV, single and multiple HPV infection among ≤ 20 and 61–65 year-old females, which have been observed in other reports [15, 29, 38]. The first peak of HPV infection occurred in the ≤ 20 years group (31.5%, 17/54), with 9.3% (5/54) was LR-HPV and 27.8% (15/54) was HR-HPV infection. The high HPV infection rate was obvious in females aged ≤ 20 years was partly due to the limited sample numbers. On the other hand, high sexual activity and lack of immunity to HPV may contribute to the high HPV infection rate [39]. The second peak was observed in the 61–65 year-old groups (38.0%, 151/397), which consisted of 10.6% (42/397) LR-HPV infection and 33.5% (133/397) HR-HPV infection. It was assumed that viral persistence or reactivation of latent HPV due to the physiologic and immunologic deregulation caused by hormone fluctuations may explain the high HPV infection rate around menopausal women [40]. The present study would assist in the formulation of preventive strategy for cervical cancer and more inspections, including cytology and even colposcopy, should be proceed among women aged > 60 years for the prevention of cervical cancer.
The L1 protein is the major capsid protein and able to induce immune response [12]. Phylogenetic distance and amino variations of the L1 protein have an effect on the immune efficiency of HPV vaccines [11, 41]. The uncontrolled expression of E6 and E7 proteins inactivates the p53 and pRb tumor suppressor proteins and is associated with the HPV persist infection [42]. HPV variants and nucleotide mutations have been suggested to affect the oncogenic potential of HPV persistent infection [22,23,24, 43]. Thus, the L1, E6 and E7 sequences of the most predominant HPV (HPV16 and HPV52) were selected to study lineage phylogeny and the genetic polymorphisms.
Based on the L1 genes, the predominant HPV16 sublineage in Henan province was A1 in 2021. In other areas in China, such as Beijing city, Zhejiang and Yunnan province, the sublineage A4 was the most common genotype [44,45,46,47]. It was reported the sublineage A4 were associated with more severity disease status than A1-3 sublineage in Chinese females and higher risk of cancer [22, 23, 48, 49]. Compared with the reference (NC001526), four non-synonymous mutations were found in HPV16 L1 protein, including H76Q (1/27), N181T (7/27), E240D (1/27) and T266A (27/27). The amino mutations N181T (7/27) and T266A (27/27) were also found in other provinces, such as Shanghai and Sichuan province [47, 50, 51]. Synonymous mutations in L1 gene, including G6196A (19/27, 70.4%), A5803C (18/27, 66.7%) and T5683C (8/27, 29.6%), had also been reported in Sichuan province [51]. In the present study, the most prevalent HPV52 sublineage in Henan province was B2. It was reported that the HPV52 sublineage B2 predominated in Asian, while in Africa, Americas and Europe, lineage A was the most common lineage [52]. Compared with HPV52 lineage A, the B2 sublineage showed a higher risk [52].
For HPV16, the most frequent non-synonymous mutations found in E6 gene was D32E (7/18). Although the D32E mutation in E6 protein did not change the B-cell epitopes, the gene variation altered the other gene profiles [53, 54]. It was suggested that the D32E amino mutation had a significant correlation with the persistent HPV16 infection in females [47]. The S63F was the most prevalent non-synonymous mutations in E7 genes. It was reported that the S63F mutation was more frequent in women with carcinoma cancer [44]. The reason was assumed that the S63F variation had an influence on the E7 epitopes and caused viral persistence and cervical cancer [44]. Compared with the HPV52 reference sequence (NC 001,592), the K93R was the only one non-synonymous mutation. The K93R mutation was also observed in other HPV52 isolates in China [55, 56]. Though the K93R mutation did not increase the cell immortalization ability of HPV52, a higher colony formation and greater cell migration ability was observed when compared to HPV52 prototype [57]. The synonymous mutation C751T and A801G were observed in other report and the roles need be further studied [56].
Conclusion
In summary, the present study provides basic information about the distribution, genotypes and variations of HPV among females population in central China, which would assist in the formulation of preventive strategies and improvements of diagnostic probe and vaccine for HPV in this region.
Availability of data and materials
All data and materials described in manuscript are available.
Abbreviations
- HPV:
-
Human papillomavirus
- LR-HPV:
-
Low-risk human papillomavirus
- HR-HPV:
-
High-risk human papillomavirus
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-386.
Lu Y, Li P, Luo G, Liu D, Zou H. Cancer attributable to human papillomavirus infection in China: Burden and trends. Cancer. 2020;126:3719–32.
Bzhalava D, Eklund C, Dillner J. International standardization and classification of human papillomavirus types. Virology. 2015;476:341–4.
Burk RD, Harari A, Chen Z. Human papillomavirus genome variants. Virology. 2013;445:232–43.
Scheffner M, Romanczuk H, Munger K, Huibregtse JM, Mietz JA, Howley PM. Functions of human papillomavirus proteins. Curr Top Microbiol Immunol. 1994;186:83–99.
Seedorf K, Krammer G, Durst M, Suhai S, Rowekamp WG. Human papillomavirus type 16 DNA sequence. Virology. 1985;145:181–5.
Yao Y, Huang W, Yang X, Sun W, Liu X, Cun W, Ma Y. HPV-16 E6 and E7 protein T cell epitopes prediction analysis based on distributions of HLA-A loci across populations: an in silico approach. Vaccine. 2013;31:2289–94.
Bishop B, Dasgupta J, Klein M, Garcea RL, Christensen ND, Zhao R, Chen XS. Crystal structures of four types of human papillomavirus L1 capsid proteins: understanding the specificity of neutralizing monoclonal antibodies. J Biol Chem. 2007;282:31803–11.
Mo X, Gai Tobe R, Wang L, Liu X, Wu B, Luo H, Nagata C, Mori R, Nakayama T. Cost-effectiveness analysis of different types of human papillomavirus vaccination combined with a cervical cancer screening program in mainland China. BMC Infect Dis. 2017;17:502.
Bogaards JA, van der Weele P, Woestenberg PJ, van Benthem BHB, King AJ. Bivalent human papillomavirus (HPV) vaccine effectiveness correlates with phylogenetic distance from HPV vaccine types 16 and 18. J Infect Dis. 2019;220:1141–6.
Varsani A, Williamson AL, Jaffer MA, Rybicki EP. A deletion and point mutation study of the human papillomavirus type 16 major capsid gene. Virus Res. 2006;122:154–63.
Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ. International agency for research on cancer multicenter cervical cancer study G: epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–27.
de Sanjose S, Quint WG, Alemany L, Geraets DT, Klaustermeier JE, Lloveras B, Tous S, Felix A, Bravo LE, Shin HR, et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11:1048–56.
Ge Y, Zhong S, Ren M, Ge Y, Mao Y, Cao P. Prevalence of human papillomavirus infection of 65,613 women in East China. BMC Public Health. 2019;19:178.
Liu Y, Ang Q, Wu H, Xu J, Chen D, Zhao H, Liu H, Guo X, Gu Y, Qiu H. Prevalence of human papillomavirus genotypes and precancerous cervical lesions in a screening population in Beijing, China: analysis of results from China’s top 3 hospital, 2009–2019. Virol J. 2020;17:104.
Li H, Li P, Huang L, Sun L, Ren H, Li P. Prevalence characteristics of cervical human papillomavirus (HPV) infection in the Zhoupu District, Shanghai City, China. Virol J. 2020;17:84.
Zhu Y, Qian F, Zou W, Wu X, Liu C, Shen G, Lai S, Yang S. Prevalence and genotype distribution of human papillomavirus infection in Huzhou City, eastern China, 2018–2019. Trans R Soc Trop Med Hyg. 2021;115:30–7.
Yamada T, Manos MM, Peto J, Greer CE, Munoz N, Bosch FX, Wheeler CM. Human papillomavirus type 16 sequence variation in cervical cancers: a worldwide perspective. J Virol. 1997;71:2463–72.
Fang L, Lin X, Yang Y, Song Z, Ding X, Tan L, Gao P. Genetic variability, phylogeny and functional implication of the long control region in human papillomavirus type 16, 18 and 58 in Chengdu, China. Virol J. 2020;17:106.
Zhe X, Xin H, Pan Z, Jin F, Zheng W, Li H, Li D, Cao D, Li Y, Zhang C, et al. Genetic variations in E6, E7 and the long control region of human papillomavirus type 16 among patients with cervical lesions in Xinjiang, China. Cancer Cell Int. 2019;19:65.
Sun Z, Lu Z, Liu J, Wang G, Zhou W, Yang L, Liu C, Wang B, Ruan Q. Genetic variations of E6 and long control region of human papillomavirus type 16 from patients with cervical lesion in Liaoning, China. BMC Cancer. 2013;13:459.
Mirabello L, Yeager M, Cullen M, Boland JF, Chen Z, Wentzensen N, Zhang X, Yu K, Yang Q, Mitchell J, et al: HPV16 sublineage associations with histology-specific cancer risk using HPV whole-genome sequences in 3200 women. J Natl Cancer Inst. 2016;108.
van der Weele P, Meijer C, King AJ. Whole-Genome Sequencing and Variant Analysis of Human Papillomavirus 16 Infections. J Virol. 2017;91.
Yu LL, Kang LN, Zhao FH, Lei XQ, Qin Y, Wu ZN, Wang H, Chen W, Qiao YL. Elevated expression of human papillomavirus-16/18 E6 oncoprotein associates with persistence of viral infection: a 3-year prospective study in China. Cancer Epidemiol Biomarkers Prev. 2016;25:1167–74.
Zehbe I, Voglino G, Delius H, Wilander E, Tommasino M. Risk of cervical cancer and geographical variations of human papillomavirus 16 E6 polymorphisms. Lancet. 1998;352:1441–2.
Chan PK, Lam CW, Cheung TH, Li WW, Lo KW, Chan MY, Cheung JL, Xu LY, Cheng AF. Human papillomavirus type 16 intratypic variant infection and risk for cervical neoplasia in southern China. J Infect Dis. 2002;186:696–700.
Chen G, Zheng P, Gao L, Zhao J, Wang Y, Qin W. Prevalence and genotype distribution of human papillomavirus in women with cervical cancer or cervical intraepithelial neoplasia in Henan province, central China. J Med Virol 2020.
Liu XX, Fan XL, Yu YP, Ji L, Yan J, Sun AH. Human papillomavirus prevalence and type-distribution among women in Zhejiang Province, Southeast China: a cross-sectional study. BMC Infect Dis. 2014;14:708.
Ma L, Lei J, Ma L, Cong X, Wang N, Yang H, Liu Q, Yu Y, Cao Y. Characteristics of women infected with human papillomavirus in a tertiary hospital in Beijing China, 2014–2018. BMC Infect Dis. 2019;19:670.
Li B, Wang H, Yang D, Ma J. Prevalence and distribution of cervical human papillomavirus genotypes in women with cytological results from Sichuan province, China. J Med Virol. 2019;91:139–45.
Zhong TY, Zhou JC, Hu R, Fan XN, Xie XY, Liu ZX, Lin M, Chen YG, Hu XM, Wang WH, et al. Prevalence of human papillomavirus infection among 71,435 women in Jiangxi Province, China. J Infect Public Health. 2017;10:783–8.
Yan X, Huang Y, Zhang M, Hu X, Li K, Jing M. Prevalence of human papillomavirus infection and type distribution among Uyghur females in Xinjiang, northwest China. Oncol Lett. 2020;20:25.
Cao D, Zhang S, Zhang Q, Wei X, Zhao M, Ma Q, Li Y, Wang L, Pei M, Yang T, et al. Prevalence of high-risk human papillomavirus infection among women in Shaanxi province of China: a hospital-based investigation. J Med Virol. 2017;89:1281–6.
Su Y, Yuan Z, Xu C, Li Z, Zhu R, Zhang W, Cao R, Yan X, Liu Y. Prevalence and genotype distribution of human papillomavirus infection among women: a population-based study in Dali Bai Autonomous Prefecture, Yunnan Province, China. J Med Virol. 2019;91:1553–61.
Duan R, Qiao Y, Clifford G, Zhao F. Cancer burden attributable to human papillomavirus infection by sex, cancer site, age, and geographical area in China. Cancer Med. 2020;9:374–84.
Hu S, Xu X, Zhang Y, Liu Y, Yang C, Wang Y, Wang Y, Yu Y, Hong Y, Zhang X, et al. A nationwide post-marketing survey of knowledge, attitude and practice toward human papillomavirus vaccine in general population: implications for vaccine roll-out in mainland China. Vaccine. 2021;39:35–44.
Zhao P, Liu S, Zhong Z, Hou J, Lin L, Weng R, Su L, Lei N, Hou T, Yang H. Prevalence and genotype distribution of human papillomavirus infection among women in northeastern Guangdong Province of China. BMC Infect Dis. 2018;18:204.
Mai Q, Yang X, Cheng H, Wu G, Wu Z: Prevalence and genotype distribution of human papillomavirus among women with cervical lesions in Shenzhen city, China. Hum Vaccin Immunother. 2020;1–7.
Althoff KN, Paul P, Burke AE, Viscidi R, Sangaramoorthy M, Gravitt PE. Correlates of cervicovaginal human papillomavirus detection in perimenopausal women. J Womens Health (Larchmt). 2009;18:1341–6.
Godi A, Boampong D, Elegunde B, Panwar K, Fleury M, Li S, Zhao Q, Xia N, Christensen ND, Beddows S. Comprehensive assessment of the antigenic impact of human papillomavirus lineage variation on recognition by neutralizing monoclonal antibodies raised against lineage a major capsid proteins of vaccine-related genotypes. J Virol. 2020;94.
Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, Stanley MA. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(Suppl 5):F55-70.
Xi LF, Schiffman M, Koutsky LA, Hughes JP, Winer RL, Mao C, Hulbert A, Lee SK, Shen Z, Kiviat NB. Lineages of oncogenic human papillomavirus types other than type 16 and 18 and risk for cervical intraepithelial neoplasia. J Natl Cancer Inst. 2014;106.
Zhou Z, Yang H, Yang L, Yao Y, Dai S, Shi L, Li C, Yang L, Yan Z, Yao Y. Human papillomavirus type 16 E6 and E7 gene variations associated with cervical cancer in a Han Chinese population. Infect Genet Evol. 2019;73:13–20.
Ding T, Wang X, Ye F, Cheng X, Lu W, Xie X. Distribution of human papillomavirus 16 E6/E7 variants in cervical cancer and intraepithelial neoplasia in Chinese women. Int J Gynecol Cancer. 2010;20:1391–8.
Hang D, Yin Y, Han J, Jiang J, Ma H, Xie S, Feng X, Zhang K, Hu Z, Shen H, et al. Analysis of human papillomavirus 16 variants and risk for cervical cancer in Chinese population. Virology. 2016;488:156–61.
Yang B, Zhang L, Zhang A, Zhou A, Yuan J, Wang Y, Sun L, Cao H, Zheng W. Variant sublineages of human papillomavirus type 16 predispose women to persistent infection characterized by a sequence analysis of the E6, L1, and LCR regions. Int J Clin Exp Pathol. 2019;12:337–43.
Zhang J, Wang T, Han M, Yang ZH, Liu LX, Chen Y, Zhang L, Hu HZ, Xi MR. Variation of human papillomavirus 16 in cervical and lung cancers in Sichuan, China. Acta Virol. 2010;54:247–53.
Freitas LB, Chen Z, Muqui EF, Boldrini NA, Miranda AE, Spano LC, Burk RD. Human papillomavirus 16 non-European variants are preferentially associated with high-grade cervical lesions. PLoS ONE. 2014;9:e100746.
Yue Y, Yang H, Wu K, Yang L, Chen J, Huang X, Pan Y, Ruan Y, Zhao Y, Shi X, et al. Genetic variability in L1 and L2 genes of HPV-16 and HPV-58 in Southwest China. PLoS ONE. 2013;8:e55204.
Cao M, Chenzhang Y, Ding X, Zhang Y, Jing Y, Chen Z. Genetic variability and lineage phylogeny of human papillomavirus type-16 and -53 based on the E6, E7, and L1 genes in Southwest China. Gene. 2016;592:49–59.
Zhang C, Park JS, Grce M, Hibbitts S, Palefsky JM, Konno R, Smith-McCune KK, Giovannelli L, Chu TY, Picconi MA, et al. Geographical distribution and risk association of human papillomavirus genotype 52-variant lineages. J Infect Dis. 2014;210:1600–4.
Yi JW, Jang M, Kim SJ, Kim SS, Rhee JE. Degradation of p53 by natural variants of the E6 protein of human papillomavirus type 16. Oncol Rep. 2013;29:1617–22.
Jang M, Rhee JE, Jang DH, Kim SS. Gene expression profiles are altered in human papillomavirus-16 E6 D25E-expressing cell lines. Virol J. 2011;8:453.
Li S, Ye M, Chen Y, Gong Q, Mei B. Genetic variation of E6 and E7 genes of human papillomavirus 52 from Central China. J Med Virol. 2020.
Zhang Y, Cao M, Wang M, Ding X, Jing Y, Chen Z, Ma T, Chen H. Genetic variability in E6, E7, and L1 genes of human papillomavirus genotype 52 from Southwest China. Gene. 2016;585:110–8.
Lai TO, Boon SS, Law PT, Chen Z, Thomas M, Banks L, Chan PK. Oncogenicitiy comparison of human papillomavirus type 52 E6 variants. J Gen Virol. 2019;100:484–96.
Acknowledgements
The authors are grateful to the staff at the Department of Gynecological Outpatient Clinic, the 989th hospital of the People's Liberation Army (PLA), Luoyang, Henan, China.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
LM and SZH conceptualized the study. XCW curated the data. SZH analyzed the data and wrote the manuscript. XLW, XCW and SH took the test. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was reviewed and approved by the ethics committees of the No.989 Hospital of the Joint Logistic Support Force of Chinese PLA (Grant No.: LLSC20150305).
Consent for publication
Not applicable.
Competing interests
The authors declare that there have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, X., Han, S., Li, X. et al. Prevalence and distribution of human papillomavirus (HPV) in Luoyang city of Henan province during 2015–2021 and the genetic variability of HPV16 and 52. Virol J 19, 37 (2022). https://doi.org/10.1186/s12985-022-01759-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-022-01759-5