Abstract
The present study aimed to assess the toxic effects of pendimethalin herbicide and protective role of curcumin using the Allium test on cytological, biochemical and physiological parameters. The effective concentration (EC50) of pendimethalin was determined at 12 mg/L by the root growth inhibition test as the concentration reducing the root length by 50%. The roots of Allium cepa L. was treated with tap water (group I), 5 mg/L curcumin (group II), 10 mg/L curcumin (group III), 12 mg/L pendimethalin (group IV), 12 mg/L pendimethalin + 5 mg/L curcumin (group V) and 12 mg/L pendimethalin + 10 mg/L curcumin (group VI). The cytological (mitotic index, chromosomal abnormalities and DNA damage), physiological (rooting percentage, root length, growth rate and weight gain) and oxidative stress (malondialdehyde level, superoxide dismutase level, catalase level and glutathione reductase level) indicators were determined after 96 h of treatment. The results revealed that pendimethalin treatment reduced rooting percentage, root length, growth rate and weight gain whereas induced chromosomal abnormalities and DNA damage in roots of A. cepa L. Further, pendimethalin exposure elevated malondialdehyde level followed by antioxidant enzymes. The activities of superoxide dismutase and catalase were up-regulated and glutathione reductase was down-regulated. The molecular docking supported the antioxidant enzymes activities result. However, a dose-dependent reduction of pendimethalin toxicity was observed when curcumin was supplied with pendimethalin. The maximum recovery of cytological, physiological and oxidative stress parameters was recorded at 10 mg/L concentration of curcumin. The correlation studies also revealed positive relation of curcumin with rooting percentage, root length, weight gain, mitotic activity and glutathione reductase enzyme level while an inverse correlation was observed with chromosomal abnormalities, DNA damage, superoxide dismutase and catalase enzyme activities, and lipid peroxidation indicating its protective effect.
Similar content being viewed by others
Introduction
The enhancement of agricultural yield is accomplished by using agrochemicals including fertilizers and pesticides to fulfill the food requirement of the population. Because the availability of agricultural land is limited and also the major threat to global food security is weeds that compete with agricultural crops for water, sunlight and nutrition, thus decreasing agricultural production1. Herbicides play a key role in controlling weeds and consequently contribute to global food production. In 1941, Robert Pokorny introduced the first synthetic herbicide 2,4-D. After this, several herbicides have been discovered. Although, the production of vegetables has been enhanced by the implication of herbicides, their continuous use for many years may cause various environmental issues. They may be adsorbed by the soil and influence the quality and yield of the next crop. Herbicides may pollute surface water and groundwater through irrigation, spray drift, run-off and leaching. In addition, plants may absorb stable herbicides and convert them into unwanted residues2. The environmental factors influence the transformation of herbicides applied in the field. The persistence of herbicides may give rise to several health-related problems. Herbicides can also affect non-targeted organisms. However, the fate of herbicides in the soil is determined by several processes including application rate, agricultural practices, crop variety, transportation, transformation, adsorption, and climatic conditions3. Thus, the application of persistent herbicides requires knowledge of their dissipation and movement in the soil.
Pendimethalin (N-(1-ethylpropyl)-2,6-dinitro-3,4-xylidine), a dinitroaniline group-selective herbicide, is widely used to control a wide variety of weeds and broadleaf plants4. Generally, it is applied to soil before the emergence of seedlings, planting and sometimes early post-emergence. In light sandy loam soil, pendimethalin can penetrate to root zone and exert phytotoxic effects in presence of adequate moisture5. Pendimethalin can inhibit cell division causes chromosomal aberrations and interfere in the formation of cell wall6. It also retards root and shoot growth of plants7. Pendimethalin is a low mobile and low volatile dinitroaniline compound having poor water solubility8. Previously, numerous experiments have been conducted to study the phytotoxicity of pendimethalin in different crop species such as rice9, cotton10, pigweed species, common lambsquarters, barnyardgrass11, paspalum cultivars12 and various summer season flora13.
It has been reported that pendimethalin and other dinitroaniline herbicides might be adsorbed and degraded in the soil14,15,16, its lipophilicity, stability and soil adsorption characteristics pose a potential risk to the environment17,18. Dinitroaniline compounds are also toxic to non-targeted invertebrates and aquatic organisms. In addition, dinitroaniline can form a carcinogenic compound nitrosamine that is harmful to humans19.
Pendimethalin can cause oxidative stress and inhibit the antioxidant system of cells20. The excess production of reactive oxygen species (ROS) resulting protein oxidation, cell membrane damage, inactivation of enzymes, RNA and DNA damage21. ROS interacts with biomolecules and damages cellular processes that result in the reduction of yield and plant growth. ROS can damage the cell membrane causing lipid peroxidation that is a highly destructive process. Lipid peroxidation may alter the structure and function of cell membranes by oxidizing polyunsaturated fatty acids (PUFAs). Impairment of PUFAs may lead to loss of fluidity and secretary functions. Malondialdehyde (MDA), an important indicator of lipid peroxidation, may be produced as a by-product of PUFA oxidation22,23.
Plants have numerous enzymatic and non-enzymatic defense mechanisms to overcome the impact of ROS. They accumulate different metabolites such as osmoprotectants and amino acids to assure from stress. In addition, enzymatic antioxidants are one of the most important mechanisms of plants to counter stress conditions. However, toxic compounds can perturb the activity of antioxidant enzymes such as catalase (CAT), superoxide dismutase (SOD), glutathione reductase (GR), ascorbate peroxidase (APX) and glutathione-S-transferase (GST) reflecting the toxicity level and plant stress tolerance capability. The protein interactions with compounds using molecular docking tool provide information about the predominant mode of interaction and binding efficacy of protein and ligand presenting protein–ligand complexes as 3D-crystal structures24,25. Similarly, pendimethalin herbicide may disrupt the natural structure of enzymes/proteins by interacting with their residues.
Turmeric (Curcuma longa L.) is a spice that has increased an interest toward both the medical/scientific and culinary communities. Turmeric is a rhizomatous herbaceous perennial plant of the ginger (Zingiberaceae) family26. Turmeric has an important component curcumin a curcuminoid as well as essential oils. Curcuminoids are phenolic chemicals derived from the roots of the C. longa L. and other species of Curcuma. Curcumin (diferuloylmethane) is a low molecular weight polyphenol that was first chemically identified in 1910. It is often regarded as the most active component, comprising about 2–8% of most turmeric formulations27. Curcumin (1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione), a polyphenolic compound, is a brightly yellow color pigment and the main derivative of Curcuma spp. Curcumin has been demonstrated to target several signaling molecules while also displaying action at the cellular level, lending evidence to its numerous health advantages28. It has the ability to alter Stat3 phosphorylation and DNA binding activity in human cancer cells29. Several in vitro and in vivo studies have shown that the therapeutic potential of curcumin does not cause side effects in animal models or humans, even at very high doses30. Curcumin has been linked to several therapeutic effects, including the regulation of cancer cell proliferation via different biological pathways such as apoptosis, mutagenicity, cell cycle regulation, angiogenesis, invasion, and tumorigenesis31. It has also antioxidant, anti-microbial, anti-parasitic and anti-inflammatory properties. It has been reported that curcumin scavenges ROS by regulating antioxidant enzyme activities in hyperglycemia32. All the effects of curcumin have been reported in animal models whereas no any such evidences are explored in case of plant system so far.
Allium cepa L. is one of the most suitable plant models for investigating the toxic effects of environmental pollutants. It is a very sensitive assay to study chromosome aberrations induced by several toxic chemicals33,34. The accuracy of this test is indicated by the similarity in the toxicity results revealed in different experiments using A. cepa L., cell culture (in vitro) and animal tests (in vivo)35,36,37,38. Allium test has also been used to explore the harmful effects of environmental pollutants on normal cell division. This test is a suitable technique to elaborate information on toxic substances induced chromosomal alterations, inhibition of mitotic activity and DNA damage with a detoxification mechanism39,40.
The present study investigated the effects of pendimethalin on root development (cell division and elongation kinetics) and antioxidant enzymes (SOD, CAT and GR) level as well as the interactive potential of pendimethalin molecule in A. cepa L. In addition, the protective role of curcumin against these effects was also assessed.
Materials and methods
Test organisms
The equal-sized (25–35 mm diameter, untreated) A. cepa L. bulbs purchased from a local market were used as the test material. A. cepa L. (Amaryllidaceae) (2n = 16) was defined using taxonomic characters and approved at the Department of Botany, Faculty of Arts and Sciences, Giresun University. Bulbs were stored in a cool and dry environment. Before application, the external scales as well as brownish base plates were removed from the bulbs without damaging the root primordia.
Determination EC50 concentration
The root inhibition test was carried out by the modified method of Fiskesjö41. The half-maximal effective concentration (EC50) of pendimethalin was calculated by the Allium root growth inhibition test as the concentration that reduced the root length of A. cepa L. bulbs by 50% compared to the control group. Bulbs were treated with distilled water (negative control) and increasing concentrations of pendimethalin (by creating a group for each 1 mg/L between 1 and 20 mg/L) for 96 h at room temperature. The solutions applied to the bulb roots were renewed every 24 h. After 96 h of application, EC50 was calculated by the length of an average of 50 roots from 6 onions in each group. The mean root length of the control groups was considered to be 100% and the point of 50% growth point was identified as the EC50 dose depending on the test concentration. The EC50 value for pendimethalin was determined as 12 mg/L by root inhibition test and was used as the application dose in the study.
Experimental design
The experiments based on plant were performed in accordance with international guidelines and regulations. The experiments had been comprised into in six groups. The control group was treated with tap water, application groups were treated with curcumin (5 and 10 mg/L), EC50 concentration of pendimethalin (12 mg/L) and their combination at 24 °C for 96 h (Table 1). The roots were exposed to the treatment solution directly in 60 × 42 mm beakers. About 40 bulbs from each group were used for physiological study and 10 bulbs were selected randomly for cytological and biochemical studies.
Physiological parameters
About 50 bulbs from each group were used for the determination of root length based on radical formation using a millimeter ruler. The weight gain was measured using precision scales before and after treatment. The rooting percentage and relative injury rate was calculated using Eqs. (1) and (2)42.
Determination of chromosomal abnormalities (CAs), micronucleus (MN) and mitotic index (MI)
The root tips were fixed for 2 h in Clarke solution (ethanol, glacial acetic acid, 3:1) followed by 96% ethanol for 15 min. Then samples were processed in 70% ethanol at 4 °C. For cytological studies, the roots were hydrolyzed in 1 N HCl for 17 min at 60 °C, incubated with 45% acetic acid for 30 min and stained for 24 h in acetocarmine. Preparations of mitotic cells were analyzed under a binocular microscope at ×500 magnification43. The evaluation of the MN involvement has been carried out according to Fenech et al44.
For each group, 10 slides were prepared from randomly selected bulbs, 1000 cells for MN and CAs frequency and 10,000 cells for MI were counted in each slide (Eq. 3).
Comet assay
Comet assay (alkaline single-cell gel electrophoresis) was performed according to the modified method of Tice et al.45 The nuclei were isolated from fresh root tips in 600 µL ice-cold nuclear isolation buffer (400 mM 6H2O–MgCl2, 0.5% w/v Triton X-100, 0.4 M Tris, pH 7.5) using a petri dish with a razor blade and root tips were centrifuged at 1200 rpm for 7 min after being passed through a nylon mesh filter. A mixture of 1:1 nuclear suspension with 1% low melting point (LMPA) in phosphate-buffered saline (PBS) was placed on a pre-coated slide with 1% normal melting point agarose (NMPA). Electrophoresis was performed in chilled electrophoresis buffer at 0.7 V/cm at 4 °C (20 V, 300 mA) for 15 min using a power supply. Slides were rinsed three times with filtered water and neutralized with Tris buffer (0.4 M Tris, pH 7.5). The nuclei were stained for 5 min with ethidium bromide after immersion in cold water for 5 min. The preparations were washed with cold water and eliminated residual stain and coverslip sealed. These steps were taken with low light to avoid DNA degradation and examined with a fluorescence microscope. Comets were analyzed using Comet Assay software version 1.2.3b46. About 100 cells per slide were analyzed for DNA damage. The extent of DNA damage was scored from 0 to 4 depending upon the level of DNA damage. The cells were classified into five categories based on tail DNA (%): 0—undamaged (0–2%), 1—low damage (< 2–25%), 2—moderate damage (< 25–45%), 3—high damage (< 45–70%) and 4—extreme damage (< 70%)47. The total DNA damage per sample, expressed as arbitrary units, was calculated using Eq. (4).
[i: degree of damage (0, 1, 2, 3, 4), Ni: the number of cells in i degree].
Evaluation of antimutagenic effects
The antimutagenic effect of curcumin was calculated for each slide by Eq. (5) and mean ± SD (standard deviation) values were calculated48. In order to assess the antimutagenic effect, chromosomal abnormalities (CAs) and the arbitrary unit of the comet assay was evaluated.
Lipid peroxidation
Lipid peroxidation was evaluated by measuring the quantity of MDA according to the method of Ünyayar et al.49 Approximately 0.5 g of root tissue were homogenized with 5% trichloroacetic acid (TCA) and centrifuged at 12,000 rpm at 24 °C for 15 min. The supernatant, TCA solution (20%) and thiobarbituric acid (0.5%) has been transferred to the new tube and incubated at 96 °C for 25 min. The tubes were taken into the ice bath and centrifuged at 10,000 rpm for 5 min. The absorbance was recorded at 532 nm, the extinction coefficient was 155 mM/cm has been used to determine the quantity of MDA content.
Antioxidant enzyme assays
Superoxide dismutase
SOD level was assessed according to the method of Beauchamp and Fridovich50. About 0.5 g of roots was homogenized in 5 mL of 50 mM (pH 7.8) chilled sodium phosphate buffer. The homogenates were centrifuged at 10,500 rpm for 20 min and the supernatant was used for the enzyme assay. The reaction mixture containing 1.5 mL 0.05 M sodium phosphate buffer (pH 7.8), 0.3 mL 130 mM methionine, 0.3 mL 0.1 mM EDTA-Na2,0.3 mL 750 μM nitroblue tetrazolium chloride (NBT), 0.3 mL of 20 μM riboflavin, 0.01 mL of 4% (w/v) insoluble polyvinylpyrrolidone, 0.01 mL of enzyme extract, and 0.28 mL of deionized water was prepared. The reaction began with putting the tubes under two 15 W fluorescent lamps for 10 min and ending by keeping the tubes in the dark for 10 min. Absorbance was measured at 560 nm and a unit SOD enzyme activity was determined as the amount of SOD enzyme required for 50% inhibition of NBT reduction under treatment conditions.
Catalase
CAT activity was determined by using the method of Beers and Sizer51. The reaction mixture containing 2.8 mL of 0.3 mL 0.1 M H2O2, 1 mL deionized water and 200 mM sodium phosphate (1.5 mL) was formulated just before use to evaluate the reaction combination. The reaction was triggered by adding 0.2 mL of supernatant and CAT activity was measured by monitoring the absorbance decrease (240 nm) as a result of H2O2 consumption. Units of CAT-activity were determined by units per minute per g fresh weight; one unit of CAT-activity was defined for change of 0.1 at an absorbance of 240 nm and values are taken from the measurements of three independent samples and expressed as OD240nm/min g FW.
Glutathione reductase
Glutathione reductase (GR) levels were determined by the modified method of Carlberg and Mannervik52. The root tips (0.5 g) were homogenized in 0.2 M EDTA (pH 4.7). The GR level was measured in a 2 mL reaction mixture containing 1 M oxidized glutathione (GSSG), 0.1 mM nicotinamide adenine dinucleotide phosphate (NADPH), 0.05 M potassium phosphate buffer (pH 7.0) and 3 mM EDTA. The supernatant absorbances were recorded at 340 nm and values are taken from the measurements of three independent samples. The levels of GR were expressed as μmol NADPH/min.g FW.
Molecular docking
Molecular docking was performed to analyze molecular interactions of pendimethalin with antioxidant enzymes (superoxide dismutase, catalase and glutathione reductase) and DNA molecules. The crystallographic 3D structure of the SOD (PDB ID: 1ba9)53, CAT (PDB ID: 5gkn)54, GR (PDB ID: 2hqm)55, B-DNA dodecamer (PDB ID: 1bna)56, B-DNA dodecamer d (PDB ID: 195d)57 and DNA (PDB ID: 1cp8)58 molecules were obtained from the protein data bank. The 3D structure of pendimethalin (PubChem CID: 38479) was retrieved from PubChem. Enzymes and DNA molecules were prepared using Biovia Discovery Studio 2020 Client for docking. During the preparation process, the active sites of enzymes were determined by removing the water molecules and co-crystal ligands, further polar hydrogen atoms were added to enzymes. Energy minimization of enzymes was done with Gromos 43B1 using Swiss-PdbViewer (v.4.1.0)59 software whereas energy minimization of the 3D structure of pendimethalin was accomplished with the uff-force field employing Open Babel v.2.4.0 software60. The molecular docking process was started with the grid box containing the active sites of enzymes and the entire structure for DNA. Then docking was performed using Autodock 4.2.6 software61 based on Lamarckian genetic algorithm (LGA) and the LGA was run for 10 runs with an initial population size of 150 individuals for both enzymes and DNA. The docking analysis and 3D visualizations were performed with Biovia Discovery Studio 2020 Client.
Dose–response effects of curcumin
The evaluation of the dose–response effects of curcumin against pendimethalin toxicity was performed by calculating the percentage curative effect of curcumin against the changes caused by pendimethalin toxicity in all parameters. The recovery percentage caused by curcumin in the calculation was calculated by proportioning with the pendimethalin application group and control group data. For this, Eq. (6) was used and evaluated with the logarithmic values of the doses62.
Data analysis
Root elongation kinetics was characterized using logistic, log-logistic, Gompertz and Weibull models. The selection of the model was based on the best fit in Bayesian Information Criterion (BIC) and Akaike information criterion (AIC). The free parameters namely maximum growth (A), growth rate (µ) and length of lag phase (λ) shared by models were employed as descriptors of the kinetics. The functions of “tidyverse” and “drc” packages for R programming language were applied in root elongation kinetics63.
SPSS Statistics v22.0 (IBM Corp., USA, 2015) package program was used to perform statistical analyzes. Data were expressed as mean ± SD (standard deviation) in the tables and mean ± SEM (standard error of means) in the graphs. The statistical significance between the means was determined by the method of One-way ANOVA and Duncan's test and the p < 0.05 was deemed statistically significant. Since there were two independent variables in the analysis of mutagenicity inhibitions, independent samples t-test was used and p < 0.05 was deemed statistically significant.
Results
Effect of pendimethalin on root growth and protective role of curcumin
The effects of pendimethalin were evaluated on root length, rooting percentage, injury rate and weight gain (Table 2). About 100% rooting was recorded in group I (control), group II (5 mg/L curcumin) and group III (10 mg/L curcumin). However, the application of 12 mg/L pendimethalin (group IV) had decreased the rooting percentage to 30%. Further, combination of pendimethalin with curcumin in group V (12 mg/L pendimethalin + 5 mg/L curcumin) and group VI (12 g/L pendimethalin + 10 mg/L curcumin) had recovered rooting percentage to 47.50% and 57.50% respectively.
The relative injury rate represented the severity of the damage. The highest relative injury rate was observed in group IV (0.69). The relative injury rate was low in group VI (0.41). A concentration-dependent improvement in relative injury rate was recorded due to the application of curcumin at different concentrations.
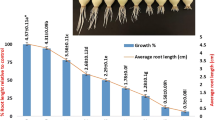
The toxicity of pendimethalin is indicated by retarded root growth. After 96 h of treatment, the mean root length was 16.92, 17.15 and 17.30 cm in group I, group II and group III respectively. About 50% reduction in root growth (8.45 cm) was recorded in pendimethalin-treated roots in group IV. However, the application of curcumin at different doses recovered root growth to 10.21 and 12.22 cm in group V and group VI respectively.
Table 3 shows the effects of pendimethalin and curcumin on weight gain. The highest weight gain was observed in group III (10.78 g) followed by group I (10.72 g) and group II (10.26 g). Pendimethalin treatment decreased the weight gain to 3.09 g in group IV. Further, the application of curcumin recovered weight gain to 4.92 and 6.37 g in group V and group VI respectively.
The kinetics of root elongation revealed significant decreases in root length (A), lag phase (λ) and growth rate (µ) of pendimethalin-treated roots in comparison to control (Fig. 1). No significant difference in λ and A of group I, group II and group III was observed. However, a significant difference in µ was noted in all groups.
Impact of pendimethalin on mitotic index and chromosomal abnormalities and protective role of curcumin
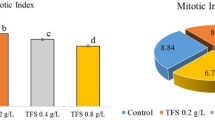
The cytogenetic effects of pendimethalin application and the protective role of curcumin were investigated by mitotic index (MI), chromosomal aberrations (CAs) and micronucleus (MN) formation. According to the data in Table 4, there was no significant difference among the MI rate of group I (9.09%), group II (9.14%) and group III (9.34%). The lowest MI (4.67%) was observed in group IV treated roots. However, a concentration-dependent increase in MI rate with the administration of curcumin (5 mg/L and 10 mg/L) and pendimethalin was recorded in group V (6.00%) and group VI (6.89%).
Different types of chromosomal abnormalities were shown in Table 5. A few abnormal cells were reported in control (group I) showing fragment, sticky chromosome and micronucleus formation. No chromosomal abnormalities were noted in roots in group II and group III treated with 5 mg/L and 10 mg/L curcumin, respectively. There was no significant difference in chromosomal abnormalities observed in group I, group II and group III. A range of chromosomal abnormalities such as fragment, sticky chromosome, bridge, binucleated cells, vagrant chromosome, unequal distribution of chromatin and micronucleus (Tables 4, 5) were recorded (Fig. 2). In addition, micronucleus formation usually indicated chromosomal instability and genotoxic effects. The frequency of chromosomal abnormalities was high in group IV treated root with pendimethalin. Administration of curcumin at 5 mg/L and 10 mg/L in group V and group VI decreased chromosomal abnormalities induced by pendimethalin. This dose-dependent decline in chromosomal abnormalities indicated the antimutagenic effect of curcumin. Mutagenicity inhibition was ascertained as 27.91% and 46.66% in group V and group VI respectively. The results indicated that chromosomal abnormalities were decreased with increasing mutagenicity inhibition.
DNA damage induced by pendimethalin in the roots of A. cepa L. and protecting role of curcumin were determined using single-cell gel electrophoresis test with three scales (head DNA, tail DNA and arbitrary unit) (Table 6). In the calculation of arbitrary units, cells were analyzed by classifying them into five categories (Fig. 3) according to tail DNA. There was no significant difference in head DNA (%), tail DNA (%) and arbitrary unit between control (group I) and curcumin alone applied groups (group II and group III). However, pendimethalin treatment (group IV) decreased head DNA percentage to 32.43, increased tail DNA percentage to 67.57% and arbitrary unit to 291.90. Further, administration of different doses of curcumin with pendimethalin in group V and group VI reduced the tail DNA percentage and arbitrary unit whereas increased head DNA percentage.
Effect of pendimethalin on lipid peroxidation and antioxidant enzymes and protective role of curcumin
MDA level is an important indicator of lipid peroxidation. MDA levels in group I, group II and group III was not statistically significant and were recorded as 5.75, 5.83 and 5.86 µmol/g FW respectively. However, the application of pendimethalin (group IV) increased MDA level (28.75 µmol/g FW). The application of curcumin along with pendimethalin in group V and VI recovered MDA levels as 19.36 and 17.25 µmol/g FW respectively. A significant difference in MDA level was observed among group IV, group V and group VI (Fig. 4a).
The antioxidant enzymes activities mitigate and repair ROS damage in plants. GR maintains the level of reduced glutathione in the cell. The reduced form of glutathione plays important role in the control of ROS. GR levels in group I, group II and group III was insignificant and noted as 55.16, 54.45 and 53.75 µmol NADPH/g FW respectively. GR levels in group IV, group V and group VI were significantly different from the above groups and recorded as 18.15, 22.48 and 28.56 µmol NADPH/g FW respectively (Fig. 4b).
SOD activity was affected by the production of superoxide radicals. SOD levels in control and curcumin alone applied groups were recorded as 35.46, 36.65 and 34.72 U/mg FW respectively. SOD levels in other groups were significantly higher than in the control. SOD levels in group IV, group V and group VI were recorded as 71.56, 59.25 and 46.5 U/mg FW respectively (Fig. 4c).
CAT neutralizes the generated hydrogen peroxide in plant cells by detoxifying it into water and oxygen. CAT levels in group I (0.59 OD240nm/min.gFW), group II (0.55 OD240nm/min.g FW) and group III (0.52 OD240nm/min.g FW) were insignificant. However, CAT levels in group IV (2.41 OD240nm/min.g FW), group V (1.96 OD240nm/min.gFW) and group VI (1.65 OD240nm/min.g FW) were significantly different from group I, group II and group III (Fig. 4d).
In addition, the results of molecular docking based on binding energy revealed that pendimethalin has capable to interact with antioxidant enzymes (Fig. 5) as well as DNA molecules (Fig. 6).
Table 7 explains the binding potential of pendimethalin with different antioxidant enzymes. The pendimethalin made hydrophobic interaction to residues (LYS70 and LYS136) of SOD with binding energy (− 1.83 kcal/mol) and inhibition constant (Ki: 45.34 mM). The residues (VAL73, ARG71, ARG111 (×2), HIS74) of CAT have interacted with pendimethalin through H-bond and other residues ARG71, ALA132, VAL145, TRY357 (×2), HIS361: made hydrophobic interaction which involved binding energy (− 8.23 kcal/mol) and inhibition constant (930.77 nM). The hydrophobic interaction by residues: CYS239 (×2), LYS407, ARG423 and H-bond interaction by residues LYS407 and ARG423 (×2) were participated in making complexation with GR by involving binding energy (− 3.15 kcal/mol) and inhibition constant (4.89 mM).
The B-DNA Dodecamer(1BNA) had made complex with pendimethalin through its bases (G4 and G10) of nucleic acid interaction using binding energy (− 3.97 kcal) and inhibition constant (1.24 mM). The pendimethalin has integrated with bases (G4, G10, and C11) of B-DNA Dodecamer D (195D) dissipating binding energy (− 4.71 kcal/mol) with inhibition constant (Ki 354.55 μM). The bases (C6 and A7) of DNA (1CP8) have complexed with pendimethalin by involving binding energy (− 4.34 kcal/mol) with inhibition constant (Ki 7.81 mM). The above explanation about the interactive efficiencies of pendimethalin toward DNA has been tabulated in Table 8.
Dose-depended protective effects of curcumin and its correlation with physiological, genetical and antioxidant enzymes
The dose-dependent protecting effects of curcumin against pendimethalin has been shown in Fig. 7. A concentration-dependent recovery in all parameters was observed after synergistic use of curcumin with pendimethalin. The highest protecting effect of curcumin was observed at 10 mg/L concentration. This concentration of curcumin recovered physiological parameters from 40.74 to 42.99%, genetic parameters 46.66–51.19%, antioxidant enzymes levels along lipid peroxidation from 28.13 to 69.42%.The highest recovery (69.42%) was noted in SOD enzyme level. The results indicated a direct or indirect relation among all the parameters.
The correlation study of all the parameters has been given in Fig. 8. Positive correlations are indicated by blue color and negative correlations by red color. The intensity of color and size of the circle is related to correlation coefficients. A positive correlation of pendimethalin was observed with CAs, micronucleus, DNA damage, SOD and CAT enzyme activities, and lipid peroxidation whereas a negative correlation was noted with rooting percentage, root length, weight gain, MI and GR enzyme activity. Similarly, the positive correlation of curcumin with rooting percentage, root length, weight gain, MI and GR enzyme activity was recorded while a negative correlation was observed with CAs, micronucleus, DNA damage, SOD and CAT enzyme activities, and lipid peroxidation indicating its protective effect.
Correlation of pendimethalin and curcumin with cytological, physiological and biochemical analysis (PDM: pendimethalin, MN: micronucleus, SOD: superoxide dismutase, DNA: DNA arbitrary unit bridge, CAT: catalase, CAs: chromosomal abnormalities, MDA: malondialdehyde, CCN: curcumin, RL: root length, MI: mitotic index, GR: glutathione reductase, WG: weight gain, RP: rooting percentage. Pearson correlation analysis (two-sided) was performed and visualized with Rstudio software. Positive correlations are shown in blue and negative correlations in red. The color intensity and the size of the circle are proportional to the correlation coefficients).
Discussion
The extensive use of pesticides like pendimethalin has usually contaminated the environment. Therefore, the assessment of action mechanisms of pendimethalin and its effect on mitotic activity, growth chromosomes, genetic material and antioxidant enzymes becomes important. The protective role of curcumin, as performed in this study, is imperative in providing useful information about the recovery of injured roots.
Pendimethalin caused significant decrease in root length, rooting percentage and weight gain but curcumin at 5 mg/L and 10 mg/L showed protecting effect on these parameters when treated alone or in combination with pendimethalin. The root elongation kinetics also supported decreases in lag phase (λ), root length (A) and growth rate (µ) in pendimethalin-treated roots as well as curcumin treatment-induced recovery in µ and A. The elongation kinetics-related parameters are sensitive endpoints to assess phytotoxic responses due to physical interaction between root and pesticide. Earlier studies explained pendimethalin-induced inhibition of germination64, reduced seedlings65 and root growth66. The MI may be a reliable endpoint generally used for cytotoxicity measurement. The inhibition of mitotic activity described the cytotoxic potential of pendimethalin17,67. In this study, pendimethalin-treated roots of A. cepa showed MI inhibition. However, at a higher concentration of curcumin (10 mg/L), mitotic activity of the cells was improved when applied alone or in combination with pendimethalin. Several studies have been reported mitodepressive effects of herbicides66,68,69,70. Like, pendimethalin exposure inhibits cell division and disturbs the formation of microtubules in root meristems of A. cepa. Microtubules are important for the development of cell wall and spindle fiber leading to cell division and differentiation71,72.
Although various types of CAs such as fragrant, sticky chromosome, bridge, binucleated cells, unequal distribution of chromatin and vagrant chromosome were monitored in pendimethalin treated roots, at a higher concentration of curcumin (10 mg/L) reduction in CAs were observed. Stickiness is a chromatid type abnormality and is associated with the effect of pollutants on depolymerization/degradation of DNA73. The appearance of fragments, bridges and micronuclei indicates the clastogenic potentiality of chemicals74,75. The arrest of cytokinesis in the cell cycle results in binucleated cells76. Interference of chemicals in spindle formation may lead to chromosomal anomalies like unequal distribution of chromatin and vagrant chromosome. Similar to the results, pendimethalin induced CAs was reported by Verma and Srivastava17 and Ahmad et al.67 The findings of this study are in agreement with the reports of other pesticides including spirodiclofen77, diniconazole78, prometryne69 and clopyralid79.
Environmental contaminants can induce DNA damage that is either repaired or unrepaired. These unrepaired damages may cause alterations in DNA. Presently, the comet assay is extensively used for toxicological studies of environmental contaminants80. The results of the comet assay revealed that pendimethalin could induce high levels of DNA damage associated with high toxic effects in roots of A. cepa. However, curcumin having a protective effect when supplied with pendimethalin could reduce DNA damage. Previous studies suggested that the antioxidant system is unable to control the activities of ROS and free radicals produced during the treatment of chemicals, in turn that may induce DNA damage81,82. Similar response was observed in other organisms Chinese hamster over cells83, Biomphalaris alexandrina snails84 and freshwater fish Clarias batrachus L85.
The present study ascertained a positive correlation of antioxidant enzyme activities and DNA damage with pendimethalin. However, curcumin treatment increased antioxidant enzymes in the roots when supplied with pendimethalin indicating its protective role. Enhancement in SOD and CAT levels was observed in pendimethalin and curcumin-treated roots. Further, curcumin supply with pendimethalin reduced SOD and CAT levels. In comparison to control, these SOD and CAT levels were still higher. The increased SOD level shows the activation of the antioxidant system. SOD, CAT and GR plays important role in thwarting oxidative stress. SOD, the first line of defense against ROS, catalyzes free radicals to generate hydrogen peroxide and oxygen. Meanwhile, CAT is required to catalyze hydrogen peroxide. In addition, GR utilizes reduced glutathione to play a key role in the defense mechanism86,87. This study suggested a reduction in GR level pointed towards the sensitivity of the plants to pendimethalin stress. Similarly, SOD and CAT are less sensitive to pendimethalin stress in comparison to GR. Similar responses were observed under the treatment of some herbicides in hairy fleabane88, rice89, wheat and maize86.
The molecular docking studies showed that the 3-D structure of pendimethalin has the interactive capability to antioxidant enzymes and DNA molecules. The higher binding energy (− 8.23 kcal/mol) for CAT-complex indicated that the pendimethalin could highly perturb the 3-D structure of CAT irrespective to other SOD and GR. In contrary to molecular docking, the biochemical analysis showed that the enhanced activity of antioxidant enzymes may lead to increase more enzyme synthesis for reducing oxidative stress.
The nucleic acid interactions showed that the pendimethalin could also alter the DNA structure which was attached between adjacent DNA bases (G10-G11 and C6-A7). Such a binding mode of pendimethalin between bases of nucleic acids may lead to DNA intercalation (Snyder et al. 2004) causing genotoxicity90,91. It has been determined that pendimethalin selectively binds to G and C nucleotides in the B-DNA structure. The GC-rich sections of DNA can intercalate by minor groove binders and act as a DNA topoisomerase-I toxin92.
MDA level reflects the extent of membrane peroxidation resulting from ROS activity. The results revealed high MDA levels in roots treated with pendimethalin indicating an imbalance between ROS production and antioxidant defense leading to lipid peroxidation and oxidative stress93. The severity of lipid peroxidation was reduced when curcumin was supplied with pendimethalin. Similarly, exposure of watermelon to bensulfuron-methyl and penoxsulam herbicides enhanced lipid peroxidation94.
Conclusion
The present study elucidates that pendimethalin induced cytotoxic and genotoxic effects, thus inhibited the growth of A. cepa roots. Further, pendimethalin disturbed the normal function of the cell and increased lipid peroxidation and activity of antioxidant enzymes. Molecular docking results suggested that pendimethalin could control the activity of antioxidant enzymes by interacting with their residues. However, the application of curcumin with pendimethalin alleviated the toxicity of herbicide by regulating antioxidant enzyme defense. The correlation study also suggests that curcumin can reduce chromosomal abnormalities, DNA damage, antioxidant enzyme activities (SOD and CAT) and lipid peroxidation indicating its ability to minimize oxidative stress. Hence, the synergistic use of curcumin with herbicide: pendimethalin could minimize the deteriorating impact in plants. However, a detailed study is required to unravel the exact mechanism of curcumin action in the amelioration of herbicide toxicity.
References
Qaim, M. Role of new plant breeding technologies for food security and sustainable agricultural development. Appl. Econ. Perspect. Policy 42, 129–150 (2020).
Rana, S. S., & Rana, M. C. Principles and practices of weed management. 29–138 (Palampur, 2018).
Sondhia, S. Dissipation of pendimethalin in soil and its residues in chickpea (Cicer arietinum L.) under field conditions. Bull. Environ. Contam. Toxicol. 89, 1032–1036 (2012).
Grey, T. & Webster, T. Cotton (Gossypium hirsutum L.) Response to pendimethalin formulation, timing, and method of application in Herbicides-current research and case studies in use (ed. Price, A. & Kelton, J.) 27–46 (InTech, 2013).
Bandyopadhyay, S. & Choudhury, P. P. Leaching behavior of pendimethalin causes toxicity towards different cultivars of Brassica juncea and Brassica campestris in sandy loam soil. Interdiscipl. Toxicol. 2, 250–253 (2009).
Ansari, S. M. et al. Pendimethalin induces oxidative stress, DNA damage, and mitochondrial dysfunction to trigger apoptosis in human lymphocytes and rat bone-marrow cells. Histochem. Cell Biol. 149, 127–141 (2018).
Zain, S., Dafaallah, A. & Zaroug, M. Efficacy and selectivity of pendimethalin for weed control in soybean (Glycine max (L.) Merr.), Gezirastate, Sudan. Agric. Sci. Pract. 7, 59–68 (2020).
Sondhia, S. Harvest time residues of pendimethalin in tomato, cauliflower, and radish under field conditions. Toxicol Environ Chem 95, 254–259 (2013).
Ahmed, S. & Chauhan, B. S. Efficacy and phytotoxicity of different rates of oxadiargyl and pendimethalin in dry-seeded rice (Oryza sativa L.) in Bangladesh. Crop Prot. 72, 169–174 (2015).
Stephenson, D. O., Bond, J. A., Landry, R. L. & Edwards, H. M. Effect of coapplied glyphosate, pyrithiobac, pendimethalin, or S-metolachlor on cotton injury, growth, and yield. Weed Technol. 27, 305–309 (2013).
Soltani, N., Shropshire, C. & Sikkema, P. H. Weed management in white beans with soil-applied grass herbicides plus halosulfuron. Am. J. Plant Sci. 11, 1998–2011 (2020).
Vinothini, G. & Arthanari, P. M. Pre emergence herbicide application and hand weeding for effective weed management in irrigated kodo millet (Paspalum scrobiculatum L.). Int. J. Chem. Stud. 5, 366–369 (2017).
Brunharo, C. A., Watkins, S. & Hanson, B. D. Season-long weed control with sequential herbicide programs in California tree nut crops. Weed Technol. 34, 834–842 (2020).
Jursík, M., Kočárek, M., Suchanová, M., Kolářová, M. & Šuk, J. Effect of irrigation and adjuvant on residual activity of pendimethalin and metazachlor in kohlrabi and soil. Plant Soil Environ. 65, 387–394 (2019).
Mendes, K. F., Olivatto, G. P., de Sousa, R. N., Junqueira, L. V. & Tornisielo, V. L. Natural biochar effect on sorption–desorption and mobility of diclosulam and pendimethalin in soil. Geoderma 347, 118–125 (2019).
Trivedi, N. & Dubey, A. Degradation studies of pendimethalin by indigenous soil bacterium Pseudomonas strain PD1 using spectrophotometric scanning and FTIR. Arch. Microbiol. 203, 4499–4507 (2021).
Verma, S. & Srivastave, A. Morphotoxicity and cytogenotoxicity of pendimethalin in the test plant Allium cepa L.—A biomarker based study. Chemosphere 206, 248–254 (2018)
Huang, B. et al. Soil fumigation alters adsorption and degradation behavior of pesticides in soil. Environ. Pollut. 246, 264–273 (2019).
Singh, B. & Singh, K. Microbial degradation of herbicides. Crit. Rev. Microbiol. 42, 245–261 (2016).
Ahmad, M. I., Zafeer, M. F., Javed, M. & Ahmad, M. Pendimethalin-induced oxidative stress, DNA damage and activation of anti-inflammatory and apoptotic markers in male rats. Sci. Rep. 8, 17139 (2018).
Singh, D. & Roy, B. K. Evaluation of malathion-induced cytogenetical effects and oxidative stress in plants using Allium test. Acta Physiol. Plant. 39, 92 (2017).
Tuteja, N., Ahmad, P., Panda, B. B. & Tuteja, R. Genotoxic stress in plants: shedding light on DNA damage, repair and DNA repair helicases. Mutat. Res. 681, 134–149 (2009).
Sharma, P., Jha, A. B., Dubey, R. S. & Perrarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defence mechanism in plants under stressful conditions. J. Bot. 2012, 217037 (2012).
Srivastava, A. K. & Singh, D. Assessment of malathion toxicity on cytophysiological activity, DNA damage and antioxidant enzymes in root of Allium cepa model. Sci. Rep. 10, 886 (2020).
Kurt, D., Acar, A., Çavuşoğlu, D., Yalçin, E. & Çavuşoğlu, K. Genotoxic effects and molecular docking of 1,4-dioxane: combined protective effects of trans-resveratrol. Environ. Sci. Pollut. Res. 28, 54922–54935 (2021).
Gupta, S. C., Patchva, S. & Aggarwal, B. B. Therapeutic roles of curcumin: Lessons learned from clinical trials. AAPS J 15, 195–218 (2013).
Zhou, H., Beevers, C. S. & Huang, S. The targets of curcumin. Curr. Drug Targets 12, 332–347 (2011).
Gopi, S., Amalraj, A., Kunnumakkara, A. & Thomas, S. Inflammation and natural products. (Academic Press, 2021).
Alexandrow, M. G. et al. Curcumin: A novel Stat3 pathway inhibitor for chemoprevention of lung cancer. Eur. J. Canc. Prev. 21, 407–412 (2012).
Anand, P. et al. Biological activities of curcumin and its analogues (congeners) made by man and mother nature. Biochem Pharmacol 76, 1590–1611 (2008).
Shetty, N. P., Prabhakaran, M. & Srivastava, A. K. Pleiotropic nature of curcumin intargeting multiple apoptotic-mediated factors and relatedstrategies to treat gastric cancer: A review. Phytother. Res. 35, 5397–5416 (2021).
Sharma, A., Dikshit, M., Tiwari, R. C. & Sharma, V. B. Haridra-curcumin the active principle. World J. Pharm. Res. 10, 357–362 (2021).
Khanna, N. & Sharma, S. Allium cepa root chromosomal aberration assay: A review. Indian J. Pharm. Biol. Res. 1, 105–119 (2013).
Sabeen, M. et al. Allium cepa assay based comparative study of selected vegetables and the chromosomal aberrations due to heavy metal accumulation. Saudi J. Biol. Sci. 27, 1368–1374 (2020).
Turkez, H. & Aydın, E. Anti-genotoxic role of eicosapentaenoic acid against imazalil-induced DNA damage in vitro. Toxicol. Ind. Health 29, 584–590 (2013).
Acar, A., Çavuşoğlu, K., Türkmen, Z., Çavuşoğlu, K. & Yalçın, E. The investigation of genotoxic, physiological and anatomical effects of paraquat herbicide on Allium cepa L. Cytol. 80, 343–351 (2015).
Çıldır, D. S. & Liman, R. Cytogenetic and genotoxic assessment in Allium cepa exposed to imazalil fungicide. Environ. Sci. Pollut. Res. 27, 20335–20343 (2020).
Oladokun, E. I., Sogbanmu, T. O., & Anikwe, J. C. Sublethal concentrations of dichlorvos and paraquat induce genotoxic and histological effects in the Clarias gariepinus. Environ. Anal. Health Toxicol. 35, e2020013 (2020).
Acar, A. In vivo toxicological assessment of diquat dibromide: Cytotoxic, genotoxic, and biochemical approach. Environ. Sci. Pollut. Res. 28, 47550–47561 (2021).
Bonciu, E. et al. An evaluation for the standardization of the Allium cepa test as cytotoxicity and genotoxicity assay. Caryologia 71, 191–209 (2018).
Fiskesjö, G. The Allium test as a standard in environmental monitoring. Hereditas 102, 99–112 (1985).
Praveen, A. & Gupta, M. Nitric oxide confronts arsenic stimulated oxidative stress and root architecture through distinct gene expression of auxin transporters, nutrient related genes and modulates biochemical responses in Oryza sativa L. Environ. Pollut. 240, 950–962 (2018).
Staykova, T. A., Ivanova, E. N. & Velchevai, I. G. Cytogenetic effect of heavy metal and cyanide in contamined waters from the region of southwest Bulgaria. J. Cell. Mol. Biol. 4, 41–46 (2005).
Fenech, M. et al. Human Micronucleus project. HUMN project: detailed description of the scoring criteria for the cytokinesis-block micronucleus assay using isolated human lymphocyte cultures. Mutat. Res. 534, 65–75 (2003)
Tice, R. R. et al. Singlecell gel/cometassay: guidelinesfor in vitroand in vivogenetic toxicology testing. Environ. Mol. Mutagen. 35, 206–221 (2000).
Końca, K. et al. A cross-platform public domain PC image-analysis program for the comet assay. Mutat. Res. 534, 15–20 (2003).
Garcia, O., Romero, I., González, J. E. & Mandina, T. Measurements of DNA damage on silver stained comets using free Internet software. Mutat. Res. 627, 186–190 (2007).
Acar, A. Ameliorative effects of cape gooseberry (Physalis peruviana L.) against monosodium glutamate (MSG)–induced toxicity: Genetic and biochemical approach. Environ Sci Pollut Res 28, 18035–18049 (2021).
Unyayar, S., Celik, A., Cekic, F. O. & Gozel, A. Cadmium-induced genotoxicity, cytotoxicity and lipid peroxidation in Allium sativum and Vicia faba. Mutagenesis 21, 77–81 (2006).
Beauchamp, C. & Fridovich, I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44, 276–287 (1971).
Beers, R. F. & Sizer, I. W. Colorimetric method for estimation of catalase. J. Biol. Chem. 195, 133–139 (1952).
Carlberg, I. & Mannervik, B. Purification and characterization of the flavoenzyme glutathione reductase from rat liver. J. Biol. Chem. 250, 5475–5480 (1975).
Banci, L. et al. Solution structure of reduced monomeric Q133M2 copper, zinc superoxide dismutase (SOD). Why is SOD a dimeric enzyme? Biochemistry 37, 11780–11791 (1998).
Yonekura, K. & Maki-Yonekura, S. Refinement of cryo-EM structures using scattering factors of charged atoms. J. Appl. Crystallogr. 49, 1517–1523 (2016).
Yu, J. & Zhou, C. Z. Crystal structure of glutathione reductase Glr1 from the yeast Saccharomyces cerevisiae. Proteins 68, 972–979 (2007).
Drew, H. R. et al. Structure of a B-DNA dodecamer: Conformation and dynamics. Proc. Natl. Acad. Sci. 78, 2179–2183 (1981).
Balendiran, K., Rao, S. T., Sekharudu, C. Y., Zon, G. & Sundaralingam, M. X-ray structures of the B-DNA dodecamer d (CGCGTTAACGCG) with an inverted central tetranucleotide and its netropsin complex. Acta Crystallogr. D Biol. Crystallogr. 51, 190–198 (1995).
Katahira, R. et al. Solution structure of the novel antitumor drug UCH9 complexed with d (TTGGCCAA) 2 as determined by NMR. Nucleic Acids Res. 26, 744–755 (1998).
Guex, N. & Peitsch, M. CSWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (2005).
O’Boyle, N. M. et al. Open Babel: An open chemical toolbox. J. Cheminform. 3, 33 (2011).
Morris, G. M. et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem 30, 2785–2791 (2009).
Acar, A. Therapeutic effects of royal jelly against sodium benzoate-induced toxicity: cytotoxic, genotoxic, and biochemical assessment. Environ. Sci. Pollut. Res. 28, 34410–34425 (2021).
R Core Team. R: A language and environment for statistical computing. https://www.R-project.org/. (R Foundation for Statistical Computing, Vienna, 2018).
Siddiqui, S., Meghvansi, M. K., Khan, S. S. & Aali, N. S. Mutagenic effect of herbicide (Maleic hydrazide) on seed germination and radicle length on Trigonella foenum-graecum L.. Indian J Appl. Pure Biol. 23, 103–106 (2008).
Singh, H. P., Batish, D. R. & Kohli, R. K. Handbook of sustainable weed management. 892 (The Haworth press, USA, 2006).
Singh, N. B., Yadav, K. & Amit, N. Mitigating effects of salicylic acid against herbicidal stress. J. Stress Physiol. Biochem. 8, 27–35 (2012).
Ahmad, M. I., Zafeer, M. F., Javed, M., & Ahmad, M. Pendimethalin-induced oxidative stress, DNA damage and activation of anti-inflammatory and apoptotic markers in male rats. Sci. Rep. 8, 17139. https://doi.org/10.1038/s41598-018-35484-3 (2018).
Mesi, A. & Kopliku, D. Cytotoxic and genotoxic potency screening of two pesticides on Allium cepa L.. Proc. Technol. 8, 19–26 (2013).
Karaismailoglu, M. C. Investigation of the potential toxic effects of prometryne herbicide on Allium cepa root tip cells with mitotic activity, chromosome aberration, micronucleus frequency, nuclear DNA amount and comet assay. Caryologia 68, 323–329 (2015).
Prasath, A., Panneerselvan, L., Provatas, A., Naidu, R. & Megharaj, M. Genotoxicity assessment of acute exposure of 2,4-dinitroanisole, its metabolites and 2,4,6-trinitrotoluene to Daphnia carinata. Ecotoxicology 25, 1873–1879 (2016).
De Storme, N. & Geelen, D. Cytokinesis in plant male meiosis. Plant Signal. Behav. 8, e23394 (2013).
Karsenti, E. & Vernos, I. The mitotic spindle: A self-made machine. Science 294, 543–547 (2001).
Badr, A., El-Shazly, H. H. & Mohamed, H. I. Plant responses to induced genotoxicity and oxidative stress by chemicals, in Induced genotoxicity and oxidative stress in plants (eds Khan, Z., Ansari, M. Y. K. & Shahwar, D.) 103–131. (Springer, 2021).
Luo, L. Z., Werner, K. M., Gollin, S. M. & Saunders, W. S. Cigarette smoke induces anaphase bridges and genomic imbalances in normal cells. Mutat. Res. 554, 375–385 (2004).
Saxena, P. N., Chauhan, L. K. S. & Gupta, S. K. Cytogenetic effects of commercial formulation of cypermethrin in root meristem cells of Allium sativum: Spectroscopic basis of chromosome damage. Toxicology 216, 244–252 (2005).
Ateeq, B., Farah, M. A., Ali, M. N. & Ahmad, W. Clastogenicity of pentachlorophenol, 2,4-D and butachlor evaluated by Allium root tip test. Mutat. Res. 514, 105–113 (2002).
Çavuşoğlu, D., Yalçin, E., Çavuşoğlu, K., Acar, A. & Yapar, K. Molecular docking and toxicity assessment of spirodiclofen: Protective role of lycopene. Environ. Sci. Pollut. Res. 28, 57372–57385 (2021).
Kalefetoğlu Macar, T., Macar, O., Yalçιn, E. & Çavuşoğlu, K. Preventive efficiency of cornelian cherry (Cornus mas L.) fruit extract in diniconazole fungicide-treated Allium cepa L. roots. Sci. Rep. 11, 2534 (2021).
Amaç, E. & Liman, R. Cytotoxic and genotoxic effects of clopyralid herbicide on Allium cepa roots. Environ. Sci. Pollut. Res. 28, 48450–48458 (2021).
Nan, P., Yan, S. G., Wang, Y. X., Du, Q. Y. & Chang, Z. J. Oxidative stress, genotoxicity and cytotoxicity of 1-methyl-3-octylimidazolium chloride on Paramisgurnus dabryanus. Environ. Toxicol. Pharmacol. 47, 1–5 (2016).
Rucińska, R., Sobkowiak, R. & Gwóźdź, E. A. Genotoxicity of lead in lupin root cells as evaluated by the comet assay. Cell. Mol. Biol. Lett. 9, 519–528 (2004).
Aitken, R. J. & Koppers, A. J. Apoptosis and DNA damage in human spermatozoa. Asian J. Androl. 13, 36–42 (2011).
Demir, N., Aydin, S. & Bucurgat, Ü. Ü. Assessment of genotoxic effects of pendimethalin in chinese hamster over cells by the single cell gel electrophoresis (comet) assay. Turk. J. Pharm. Sci. 14, 185–190 (2017).
Ibrahim, A. M., Ahmed, A. K., Bakry, F. A., Rabei, I. & Abdel-Ghaffar, F. Toxicological impact of butralin, glyphosate-isopropylammonium and pendimethalin herbicides on physiological parameters of Biomphalaria alexandrina snails. Mollusc. Res. 39, 224–233 (2019).
Gupta, P. & Verma, S. K. Evaluation of genotoxicity induced by herbicide pendimethalin in fresh water fish Clarias batrschus (linn.) and possible role of oxidative stress in induced DNA damage. Drug Chem. Toxicol. https://doi.org/10.1080/01480545.2020.1774603 (2020).
Nemat Alla, M. M., Badawi. A. M., Hassan. N. M., El-Bastawisy, Z. M. & Badran, E. G. Induction of glutathione and glutathione-associated enzymes in butachlor-tolerant plant species. Am. J. Plant Physiol. 2, 195–205 (2007).
Wang, H. M. et al. Effects of salt stress on antioxidant defence system in the root of Kandelia candel. Bot. Stud. 55, 57 (2014).
Piasecki, C. et al. Oxidative stress and differential antioxidant enzyme activity in glyphosate-resistant and sensitive hairy fleabane in response to glyphosate treatment. Bragantia 78, 379–396 (2019).
Jung, H. I., Kuk, Y. I., Back, K. & Burgos, N. R. Resistance pattern and antioxidant enzyme profiles of protoporphyrinogen oxidase (PROTOX) inhibitor-resistant transgenic rice. Pestic. Biochem. Physiol. 91, 53–65 (2008).
Snyder, R. D. A review and investigation into the mechanistic basis of the genotoxicity of antihistamines. Mutat. Res. 411, 235–248 (1998).
Snyder, R. D., Ewing, D. E. & Hendry, L. B. Evaluation of DNA intercalation potential of pharmaceuticals and other chemicals by cell-based and three-dimensional computational approaches. Environ. Mol. Mutagen. 44, 163–173 (2004).
Miskovic, K., Bujak, M., Lončar, M. B. & Glavaš-Obrovac, L. Antineoplastic DNA-binding compounds: Intercalating and minor groove binding drugs. Arh. Hig. Rada. Toksikol. 64, 593–602 (2013).
Özdemir, F., Bor, M., Demiral, T. & Türkan, I. Effects of 24-epibrassinolide on seed germination, seedling growth, lipidperoxidation, proline content and antioxidative system of rice (Oryza sativa L.) under salinity stress. Plant Growth Regul. 42, 203–211 (2004) .
Ali, A. et al. Strigolactone alleviates herbicide toxicity via maintaining antioxidant homeostasis in watermelon (Citrullus lanatus). Agriculture 11, 419–437 (2021).
Author information
Authors and Affiliations
Contributions
A. A. carried out the experimental design and all laboratory research, and contributed to article writing. D. S. and A. K. S. performed statistical analyzes and molecular docking, and contributed to article writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Acar, A., Singh, D. & Srivastava, A.K. Assessment of the ameliorative effect of curcumin on pendimethalin-induced genetic and biochemical toxicity. Sci Rep 12, 2195 (2022). https://doi.org/10.1038/s41598-022-06278-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-06278-5
- Springer Nature Limited