Abstract
We used palaeoproteomics and peptide mass fingerprinting to obtain secure species identifications of key specimens of early domesticated fauna from South Africa, dating to ca. 2000 BP. It can be difficult to distinguish fragmentary remains of early domesticates (sheep) from similar-sized local wild bovids (grey duiker, grey rhebok, springbok—southern Africa lacks wild sheep) based on morphology alone. Our analysis revealed a Zooarchaeology by Mass Spectrometry (ZooMS) marker (m/z 1532) present in wild bovids and we demonstrate through LC–MS/MS that it is capable of discriminating between wild bovids and caprine domesticates. We confirm that the Spoegrivier specimen dated to 2105 ± 65 BP is indeed a sheep. This is the earliest directly dated evidence of domesticated animals in southern Africa. As well as the traditional method of analysing bone fragments, we show the utility of minimally destructive sampling methods such as PVC eraser and polishing films for successful ZooMS identification. We also show that collagen extracted more than 25 years ago for the purpose of radiocarbon dating can yield successful ZooMS identification. Our study demonstrates the importance of developing appropriate regional frameworks of comparison for future research using ZooMS as a method of biomolecular species identification.
Similar content being viewed by others
Introduction
The transition from hunting and gathering to food production is one of the most important processes in human history. In parts of sub-Saharan Africa, domesticated animals appeared earlier than domesticated plants1. Domesticated sheep (Ovis aries) were introduced into North Africa from western Asia, spreading to eastern Africa by about 5000 BP and then southwards. There are no wild sheep in southern Africa, therefore the earliest animals were undoubtedly imported and were probably the earliest domesticates in southern Africa2,3. The timing is hotly debated. Domesticated sheep have long been thought to have reached southernmost Africa ca. 2000 BP4,5, but the initial studies relied on dating by association—at that time, only ‘conventional’ radiocarbon dating was possible, necessitating large samples. Most dates were derived from wood charcoal, and considered to apply also to faunal and other assemblages from the same stratigraphic levels. Subsequently, direct AMS radiocarbon dating of supposedly early sheep bones showed that many were actually younger than the layers in which they were found6,7,8, although a few did in fact date to ca. 2000 BP. A sheep astragalus from Toteng 1 in northern Botswana is directly dated by AMS to 2020 ± 40 BP (Beta-186669)9,10. Part of a sheep mandible and a calcaneum from Blombos Cave, on the south coast, are similarly dated to 1960 ± 50 BP and 1880 ± 55 BP (OxA-4543 and OxA-4544 respectively)11. The oldest date reported for cattle bone in southern Africa is also from Toteng: 2070 ± 40 (Beta-190488)9,10. One sheep phalanx from the site of Spoegrivier in Namaqualand (Fig. 1) is directly AMS dated to 2105 ± 65 BP (OxA-3862)7, which calibrates at two sigma to 351-308 BC [4.8% probability] or 197 BC- 110 AD [90.7% probability], using OxCal 4.4 and the SHCal20 calibration curve12,13). At present, this is the oldest secure date for domesticated animals in southern Africa. Taken as a whole, the chronology based on individually dated sheep or cattle bones supports the introduction of domesticated animals into southern Africa shortly before 2000 BP14. The number of specimens is small, so each example is critically important.
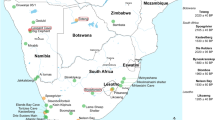
Location of archaeological sites and image of oldest sample investigated (P4859C) (a) Map showing locations of archaeological sites that yielded specimens analysed in this study. (b) First phalanx from Spoegrivier dated to 2105 ± 65 BP (centre) compared with modern sheep (left) and springbok (right).
There remains a question as to whether the species identifications of these specimens are secure. All those mentioned above have been identified only by their morphology, and confusion with several similar-sized species of wild bovids is possible, as highlighted by the mismatch between morphological and ancient DNA (aDNA) identifications of putative early domesticates at the site of Blydefontein in central South Africa15,16,17,18,19, at Sehonghong in Lesotho20,21,22 and elsewhere23. Several teeth from Leopard’s Cave in Namibia, dated to just over 2000 BP and originally thought to be from domesticated caprines (sheep and/or goat), have since been shown by Le Meillour et al. (2020) to derive from wild springbok24. This study, like Le Meillour et al. (2020), uses proteomic analysis to distinguish species based on species-specific protein sequences. The archaeological bones included in this study were originally identified as sheep by faunal expert Richard Klein, based on morphological criteria. These specimens were selected jointly by Richard Klein, Royden Yates and Judith Sealy for AMS radiocarbon dating to begin to establish a more secure chronology for the introduction of domesticated stock into southern Africa based on direct AMS dates rather than dating by association7,8. A key criterion for selection was the choice of skeletal elements most clearly diagnostic of sheep, as opposed to wild species. These specimens are unlikely to be goats, since domesticated goats have not been recorded from precolonial times in south-western South Africa, although they were present in the northern and eastern parts of the country5,25. There remains, however, a possibility that the morphological identifications as sheep may be wrong. In recent years, as biomolecular methods of species identification have developed, the expectation has arisen that these should be used to cross-check morphological identification of key early specimens of domesticates that form the cornerstones of our chronologies for the introduction of food production in different parts of the world17,26,27,28,29,30,31.
Biomolecular approaches to species identification augment morphological species assignment, especially when remains are fragmentary, poorly preserved and/or from non-diagnostic skeletal elements. A number of recent studies have utilised ancient protein (or palaeoproteomic) techniques to identify archaeological bone and ivory remains in sub-Saharan Africa. Le Meillour et al. developed a protocol for the identification of degraded bone fragments and applied it to the identification of remains from Toteng32 and from Leopard’s Cave in Namibia24. Zooarchaeology by Mass Spectrometry (ZooMS) has been used to identify animal species from which bone was selected to fashion into artefacts33, to distinguish East African sheep and goats2, to identify Asian fauna moving into eastern Africa30, and to investigate early farmers’ choice of elephant or hippo ivory in south-eastern Africa34. Proteomic techniques have also been used to identify the oldest authenticated proteins ever found, from eggshell at the site of Laetoli, Tanzania, 3.8 Ma35.
ZooMS is one of the most widely used and cost-effective ancient protein analytical strategies and is now well-established as a method of identifying hominid and zooarchaeological remains36. It uses matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry to obtain characteristic profiles of protein fragments (or peptides), typically from collagen, extracted from archaeological bone. Different taxa have slight variations in the amino acid sequences of their collagen so the resulting peptide mass ‘fingerprints’ provide markers that in many cases allow robust and objective species identification. New markers identified by ZooMS can subsequently be validated by Liquid Chromatography Tandem Mass Spectrometry (LC–MS/MS), the ‘gold standard’ of proteomic analysis. This powerful technique allows direct amino acid sequencing of proteomes and proteins, enabling verification of phylogenetically relevant sites. LC–MS/MS is not based simply on the total mass of the peptide, like ZooMS, but also on secondary fragmentation of the peptide to interrogate each amino acid. ZooMS and other palaeoproteomic methods also offer some advantages compared with aDNA. Collagen is a robust biomolecule, which has been shown to preserve longer than aDNA37. In addition, the amount of sample required is typically smaller than that needed for aDNA analysis, involving less destruction of precious specimens, and some methods of sampling, e.g. eraser rubbings, are effectively non-destructive38,39,40.
ZooMS has proven successful in distinguishing sheep from morphologically similar species, especially goats, in various parts of the world2,28,41,42. Successful application depends on the ZooMS fingerprint of sheep showing clear differences from those of local wild fauna and other domesticates. This is particularly important in Africa, home to many species of wild bovids that have not yet been fully investigated using proteomic approaches. In this paper, we focus on south-western Africa, where these techniques have been underutilised, comparing 8 archaeological specimens morphologically identified as sheep with the three species of similar-sized wild bovids with which sheep bones are most likely to be confused (Table 1). These are Antidorcas marsupialis (springbok), Pelea capreolus (grey rhebuck) and Sylvicapra grimmia (grey or common duiker). This last is rather smaller than sheep, so confusion is less likely, but we include it here for completeness. In other parts of Africa there are additional wild species (e.g. Aepyceros melampus, impala) that may be confused with sheep, but impala do not occur in our study area so are not included in our reference samples.
The aims of this research were, therefore (1) to identify ZooMS peptide markers capable of distinguishing sheep from wild south-west African bovids of similar size and morphology, (2) to search for these markers in faunal remains believed to be those of domesticated sheep, especially the earliest directly dated sheep specimen in southern Africa, from Spoegrivier, and (3) to assess the viability of minimally destructive collagen extraction for archaeological samples from these contexts. To achieve these aims we first utilised ZooMS and spectral comparison of modern reference material to identify candidate markers. We subsequently validated these candidates using LC–MS/MS.
Results
ZooMS and LC–MS/MS—modern reference samples
The modern domesticated sheep and goat samples from South Africa analysed here have collagen peptide mass fingerprints identical to those previously published for domestic sheep and goat in other parts of the African continent, and the world2,27,28. Specifically, these include the presence of distinguishing peptides at m/z 3033 in sheep and m/z 3093 in goat. The peptide mass fingerprint for grey rhebok differs from domesticated sheep and goat by the presence of peaks at m/z 1150, m/z 1166, m/z 1532 and m/z 1550. The peptide mass fingerprint for grey duiker differs from domesticated sheep and goat by the presence of peaks at m/z 1192, m/z 1208, m/z 1532. The peptide mass fingerprint for springbok differs from domesticated sheep and goat with the presence of peaks at m/z 1532, m/z 1550 and m/z 2553 (Supplementary Fig. S1 and Table S1).
The peaks at m/z 1550 and m/z 2553 did not always appear or show high intensity in the modern wild bovid spectra. It is a well-reported phenomenon that certain peptides are ‘poor flyers’ in MALDI-TOF43. We therefore decided to investigate only the peak at m/z 1532 further, since it was present in all three wild bovids (Fig. 2). We used LC–MS/MS of reference samples A (springbok), D (grey rhebok), G (Namaqua Afrikaner sheep), and M (grey duiker) to validate this candidate marker for discriminating domesticated sheep from similar size wild bovids in this region. LC–MS/MS confirmed the presence of this marker (GEPGPAGAVGPAGAVGPR (COL1A2 position 889–906 (from start of triple helix, see Brown et al. 202044), underline is hydroxyproline) in all three wild bovids, but not in the modern sheep where the corresponding peptide was GEPGPVGAVGPAGAVGPR (Fig. 3; Supplementary Fig. S2). We were unable to detect a peak corresponding to this sequence (m/z 1571 (single hydroxyproline residue) or m/z 1555 (proline only)) in our ZooMS data for the reference sheep sample, despite its presence in the LC–MS/MS data. According to the BLASTp search, this peptide is specific to caprines (sheep and goat). The recreation of the springbok sequence (see Supplementary material and supplementary Table S2) also showed various single amino acid polymorphisms (SAPs) present between springbok and sheep that could be used to distinguish them by proteomics.
Presence and absence of species-specific mass ions in four ZooMS spectra. (a–c) Reference spectra of modern springbok, grey rhebok and common duiker show the presence of peptide with a m/z of 1532.8 and related ions. (d) Modern sheep reference spectrum showing the absence of the peptide with a m/z of 1532.8.
ZooMS and LC–MS/MS—archaeological samples
Examination of the ZooMS spectra of the eight archaeological samples showed sheep-specific markers in all samples except Byneskranskop P4862, which had little protein and did not produce good enough spectra to enable identification (supplementary Table S1). All lack the m/z 1532 marker characteristic of wild bovids studied here. In addition, all have the marker at m/z 3033 seen in sheep and lack the marker at m/z 3093 seen in goats. Where both solution and minimally destructive (eraser or film) extraction methods were used, identifications were consistent (Table 2). The solution method was the most effective for species identification (23/24 samples were successfully assigned). All three minimally destructive methods had a success rate of > 50%, however aluminum film had the greatest success rate and PVC eraser had the lowest success rate (PVC eraser 6/11; aluminum film 9/11; diamond film 8/11). Of the six archaeological samples tested using the PVC eraser and the polishing films, only one (Byneskranskop P4862, mentioned above) failed to provide spectra good enough for identification; that sample also yielded insufficient protein for the solution method. The ZooMS identifications are thus consistent with the morphological identifications, indicating that the seven bone fragments from the archaeological sites of Spoegrivier, Kasteelberg A, Die Kelders and Byneskranskop 1 are sheep.
In order to further validate the ZooMS markers seen in the spectra of the archaeological samples, collagen from the oldest sample from the site of Spoegrivier, P4859C, was also analysed by LC–MS/MS to allow for direct protein sequencing. The presence of the caprine version of the marker identified by ZooMS further validated the morphological identification as sheep (Fig. 4). In addition, when searched against the whole sheep proteome, this sample contained sheep-specific peptides for four additional proteins (alpha-2-HS-glycoprotein, collagen V, vitronectin, and a protein with homology to prothrombin), as well as three other caprine-specific proteins (vitamin K-dependent protein S, collagen VI, and perlecan) (Supplementary Tables S3 and S4). While the sequences of these proteins are not known for the three wild bovids considered here, these proteins are likely more taxonomically varied than collagen, which is highly conserved for structural reasons45. Since the collagen sequences are distinct between these species, it is likely that the other proteins are as well. Recreating the sequences for these proteins was, however, beyond the scope of this study, since sheep could be distinguished from the wild species based on the differences seen in type 1 collagen.
LC–MS/MS spectra for caprine specific peptide GEPGPVGAVGPAGAVGPR in sample P4859C from Spoegrivier, South Africa, morphologically assigned to sheep. The corresponding peptide for springbok, grey rhebok, grey duiker was not detected. Hy on y16 indicates the presence of hydroxyproline, greyed out V is indicative of species-specific amino acid.
For additional validation, the deamidation levels of the peptides recovered by LC–MS/MS for P4859C and sample G (modern sheep) were calculated (Fig. 5). Asparagine and glutamine residues naturally deamidate over time into aspartic and glutamic acids, respectively. While the rates of these reactions depend on many factors (such as temperature, humidity, and pH46,47,48), higher levels of deamidation are generally associated with older proteins, and hence can help indicate whether the proteins recovered are authentically archaeological, or derive from modern contamination. P4859C shows higher rates of deamidation than the modern sheep sample G, especially for the slower (up to 10X49) glutamine reaction, indicating that we recovered and analysed authentically ancient proteins.
Deamidation levels of modern sheep sample G and Spoegrivier sheep P4859C: Overall percentage of deamidation for asparagine (N) and glutamine (Q) residues for tryptic peptides from archaeological sample P4859C compared with modern sheep bone G, excluding contaminants. Error bars represent standard deviations and the numbers above each bar represent the numbers of peptides on which the calculations are based.
Discussion
ZooMS is an important method to develop in contexts where there is poor preservation of organic remains, as proteins such as collagen can survive longer than DNA in archaeological sites35,37,50. Our results provide a framework for using ZooMS to distinguish sheep from the wild species springbok, grey rhebok, and grey duiker. We apply this to remains from archaeological sites in southern Africa with good bone collagen preservation. Using proteomics, our study confirmed that the earliest specimen identified as a domesticate on the basis of its morphology, a first phalanx directly dated to 2105 ± 65 BP, is indeed a sheep. This specimen has now been identified by three independent methods (morphology, ZooMS, LC–MS/MS). We can therefore have confidence that domesticated sheep were indeed introduced into South Africa by ca. 2000 BP.
Recent biomolecular studies at other archaeological sites in this part of the world have shown that some putative early domesticates are actually wild species24; see also16,17,18. The winter rainfall climate of south-western Africa precluded the spread of African grain crops (principally sorghum and millets) into the region, so that agriculture was practised only after European colonisation in the seventeenth century CE. The introduction of domesticated animals ca. 2000 years ago therefore marked the transition from a hunter-gatherer world to food production. The timing and mechanism of the arrival of herding is thus a key issue in archaeology, linguistics, population genetics and related fields14,51,52,53,54.
We successfully identified a ZooMS marker peak at m/z 1532 (GEPGPAGAVGPAGAVGPR, COL1A2 position 889–906) that was present in all three wild bovids (springbok, grey rhebok, and grey duiker) but not in sheep samples. This marker was validated by LC–MS/MS protein sequencing. ZooMS relies on whole peptide mass (MS1 data), whereas LC–MS/MS fragments this mass into the constituent amino acids (MS2) and determines their order. This allows for unambiguous distinction between peptide masses. We were therefore able to confirm the sequence differences between sheep and the wild bovids, allowing for distinction between the groups. Additionally, through validation of this candidate marker we also constructed COL1A1 and COL1A2 protein sequences for springbok. Thus, this ZooMS marker provides a complementary method to osteological identification.
The consensus springbok collagen sequences that we present differ from those shown in supplementary Fig. S1 of Le Meillour et al. (2020)24. That figure shows the GEPGPVGAVGPAGAVGPR (corresponding caprine ortholog of the m/z 1532 marker) as present in springbok alpha 2 chain of collagen type 1. If correct, this would invalidate our study. To address this issue we re-analysed the LC–MS/MS files provided by Le Meillour et al. (2020). The outcome of this analysis (Supplementary Fig. S3) shows that in fact the GEPGPVGAVGPAGAVGPR peptide is not present whilst the GEPGPAGAVGPAGAVGPR peptide (corresponding to wild bovids) is present in the raw data reported by Le Meillour et al. (2020) for their springbok reference material (SPR 89 and SPR 1670), matching our own findings. Note that we are not challenging the species identifications proposed by Le Meillour et al. (2020), which are based on different markers. We merely note an inconsistency between their LC–MS/MS files and their Fig. S1. This demonstrates the need to check every residue in a proposed sequence with at least two different peptide fragments, preferably using more than one software package, as we have done here.
We have also shown that it is possible to identify the ZooMS marker at m/z 1532 and others using minimally destructive sampling methods (Supplementary Table S1). Given that many archaeological bones are fragmentary, and specimens of particular interest may have been subjected to other types of destructive analyses, minimally destructive sampling is often desirable. All minimally destructive sampling techniques used in this study produced identifications. Because we tested only six archaeological specimens using these methods, we do not have enough data for a thorough comparison; however, our results show aluminum film gave the most identifications. Further testing is necessary to determine which minimally destructive sampling method is optimal. As PVC eraser is the cheapest and most available option it is likely that it will become the most popular in future studies. Importantly, the bone fragments in our study were not polished or worked, and therefore the collagen probably bound to the PVC eraser or polishing film better than that from highly worked, polished bone surfaces such as those of bone points, which when tested previously with these methods, have had low rates of success33. Based on our data, we suggest that continued investigation of minimally destructive sampling techniques is merited; they may in future supplant destructive sampling as the most common strategy. Our results also confirm that it is possible to use archived collagen extracts for ZooMS. Collagen extracted in 1992–3 for radiocarbon dating yielded ZooMS spectra comparable to freshly extracted collagen, and was suitable for LC–MS/MS protein sequencing. We were also able to detect other proteins (such as alpha-2-HS-glycoprotein and osteocalcin), further expanding the utility of archived collagen. Archived collagen may be available from important directly dated samples for which there is no more bone available, or on which curators are unwilling to allow further destructive analysis.
As applications of ZooMS expand in different parts of the world with different fauna, ongoing developmental work is required to build and validate reference peptide fingerprint libraries to investigate whether it is possible to distinguish species of interest using the peptide fingerprinting method. In this paper, we have taken a step towards this goal, focussing on domesticated sheep and co-occurring wild species in south-western Africa. Future studies will benefit from this diagnostic ZooMS marker to distinguish between sheep and wild African bovids of similar size. This type of work is essential to develop a reliable picture of the spread of herding in southern Africa and elsewhere.
Materials and methods
For a complete description of all methodology see supplemental methods.
Samples
For this investigation, we examined reference bone samples from South African sheep, goat, and three wild bovid species to obtain peptide fingerprints for comparison with archaeological samples from four sites (Fig. 1). We analysed both small fragments of bone (< 100 mg), the removal of which slightly alters the overall morphology of the specimen, and ‘minimally destructive samples’ by which we mean eraser/film rubbings, as described below. We define ‘minimally destructive’ as sampling that does not affect the overall morphology of the specimen(s). A list of samples is provided in Table 1.
Protein extraction
Protein extraction was performed on samples derived from destructive sampling according to the protocol described in Buckley et. al. (2009)36, with minor modification.
ZooMS
ZooMS analysis was performed on all destructive and minimally destructive samples following the protocols described in Buckley et al. (2009)36; Fiddyment et al. (2015)38; Kirby, Manick and Newman (2019)40. Where possible 20 µg of protein and a trypsin:protein ratio (µg) of 1:50 was used. Instrument and settings were typical for archaeological samples and the same as described in Jensen et al. (2020)55.
LC–MS/MS acquisition
The reference samples A (springbok), D (grey rhebok), G (Namaqua Afrikaner sheep), and M (grey duiker), as well as collagen from the earliest dated archaeological sample (P4859C) from Spoegrivier were further analysed by LC–MS/MS following methods previously published for palaeoproteomic samples55,56. Tryptic and elastase digestions for each sample were combined into a single mass spectrometry run.
LC–MS/MS data analysis
To validate the presence of the suspected ZooMS markers, the Thermo RAW files generated were then searched using the software MaxQuant (v.1.6.3.4)57. The database was prepared using previously published type 1 collagen sequences from several species including Ovis aries, Capra hircus, Bos taurus, Mus musculus, and Homo sapiens. For MaxQuant settings see supplemental methods. Proteins were considered confidently identified if at least two razor + unique peptides covering distinct areas of the sequence were recovered. MS/MS spectra were assessed manually for confident identification. Deamidation was assessed using publicly available code56, with the contaminant proteins filtered out. In addition, the archaeological sample P4859C was searched in MaxQuant against the sheep proteome database from Uniprot (downloaded 20/3/20) in order to identify other proteins besides COL1.
Construction of springbok collagen sequence
The raw file created from modern springbok bone (A) was also searched with PEAKS (v.7)58 against a database comprising COL1 sequences of the following species: Ovis aries, Capra hircus, Bos taurus, Sus scrofa, Mus musculus, and Homo sapiens. The searches (de novo, PEAKS DB, PEAKS PTM, and SPIDER) were performed with peptide mass tolerance + /− 10 ppm and fragment mass tolerance + /− 0.05 Da. SAPs were then authenticated by searching the sample of modern springbok bone (A) with MaxQuant (v 1.6.3.4) using the same database as described above, but with the added springbok predicted sequences. These trypsin and elastase searches were repeated and the springbok sequence(s) updated until a single predicted springbok sequence remained that incorporated all SAPs confidently detected.
The confidently matched spectra were then aligned using MAFFT’s online service59 (v.7) and a consensus sequence was then made from these peptides. An amino acid residue was only considered correctly assigned if it could be confidently identified in at least two overlapping peptides. All residues that did not reach this standard are marked as X in the final sequence. The final sequence is provided in the supplementary results.
Data availability
The mass spectrometry proteomics data (including the averaged MALDI-TOF TXT files) have been deposited to the ProteomeXchange Consortium via the PRIDE60 partner repository with the dataset identifier PXD021949.
References
Marshall, F. & Hildebrand, E. Cattle before crops: the beginnings of food production in Africa. J. World Prehist. 16, 99–143 (2002).
Prendergast, M. E., Janzen, A., Buckley, M. & Grillo, K. M. Sorting the sheep from the goats in the Pastoral Neolithic: morphological and biomolecular approaches at Luxmanda, Tanzania. Archaeol. Anthropol. Sci. 11, 3047–3062 (2018).
Sadr, K. The Neolithic of Southern Africa. J. Afr. Hist. 44, 195–209 (2003).
Schweitzer, F. R. & Scott, K. J. Early occurrence of domestic sheep in sub-Saharan Africa. Nature 241, 547–547 (1973).
Klein, R. G. The prehistory of stone age herders in the Cape province of South Africa. Goodwin Ser. 5, 5–12 (1986).
Inskeep, R. R. Nelson Bay Cave, Cape Province, South Africa: The Holocene Levels Vol. 1 and 2 (BAR Publishing, 1987).
Sealy, J. & Yates, R. The chronology of the introduction of pastoralism to the Cape, South Africa. Antiquity 68, 58–67 (1994).
Sealy, J. & Yates, R. Direct radiocarbon dating of early sheep bones: two further results. S. Afr. Archaeol. Bull. 51, 109–110 (1996).
Robbins, L. et al. The advent of herding in Southern Africa: early AMS dates on domestic livestock from the Kalahari Desert. Curr. Anthropol. 46, 671–677 (2005).
Robbins, L. H. et al. Recent archaeological research at Toteng, Botswana: early domesticated livestock in the Kalahari. J. Afr. Archaeol. 6, 131–149 (2008).
Henshilwood, C. A revised chronology for pastoralism in southernmost Africa: new evidence of sheep at c. 2000 b.p. from Blombos Cave, South Africa. Antiquity 70, 945–949 (1996).
Ramsey, C. B. Bayesian analysis of radiocarbon dates. Radiocarbon 51, 337–360 (2009).
Hogg, A. G. et al. SHCal20 southern hemisphere calibration, 0–55,000 Years cal BP. Radiocarbon 62, 759–778 (2020).
Sadr, K. Livestock first reached southern Africa in two separate events. PLoS ONE 10, e0134215 (2015).
Bousman, C. B. et al. The quest for evidence of domestic stock at Blydefontein Rock Shelter. South. Afr. Humanit. 28, 39–60 (2016).
Horsburgh, K. A. & Moreno-Mayar, J. V. Molecular identification of sheep at Blydefontein Rock Shelter, South Africa. South. Afr. Humanit. 27, 65–80 (2015).
Horsburgh, K. A., Orton, J. & Klein, R. G. Beware the springbok in sheep’s clothing: how secure are the faunal identifications upon which we build our models?. Afr. Archaeol. Rev. 33, 353–361 (2016).
Horsburgh, K. A., Moreno-Mayar, J. V. & Klein, R. G. Counting and miscounting sheep: genetic evidence for pervasive misclassification of wild fauna as domestic stock. South. Afr. Humanit. 30, 53–69 (2017).
Scott, K. & Plug, I. Osteomorphology and osteometry versus aDNA in taxonomic identification of fragmentary sheep and sheep/goat bones from archaeological deposits: Blydefontein Shelter, Karoo, South Africa. South. Afr. Humanit. 28, 61–79 (2016).
Horsburgh, K. A., Moreno-Mayar, J. V. & Gosling, A. L. Revisiting the Kalahari debate in the highlands: ancient DNA provides new faunal identifications at Sehonghong, Lesotho. Azania: Archaeol. Res. Afr. 51, 295–306 (2016).
Plug, I. Reply to Horsburgh et al. 2016: ‘Revisiting the Kalahari debate in the highlands’. Azania: Archaeol. Res. Afr. 53, 98–113 (2018).
Horsburgh, K. A. A reply to Plug 2017: science requires self-correction. Azania: Archaeol. Res. Afr. 53, 114–118 (2018).
Horsburgh, K. A. & Gosling, A. L. Systematic ancient DNA species identification fails to find late holocene domesticated cattle in southern Africa. Biology 9, 316 (2020).
Le Meillour, L. et al. Palaeoproteomics gives new insight into early southern African pastoralism. Nat. Sci. Rep. 10, 14427 (2020).
Badenhorst, S. Goats (Capra hircus), the Khoekhoen and pastoralism: current evidence from Southern Africa. Afr. Archaeol. Rev. 23, 45–53 (2006).
Orton, J., Mitchell, P., Klein, R., Steele, T. & Horsburgh, K. A. An early date for cattle from Namaqualand, South Africa: implications for the origins of herding in southern Africa. Antiquity 87, 108–120 (2013).
Birch, S. E. P., Scheu, A., Buckley, M. & Çakırlar, C. Combined osteomorphological, isotopic, aDNA, and ZooMS analyses of sheep and goat remains from Neolithic Ulucak, Turkey. Archaeol. Anthropol. Sci. 11, 1669–1681 (2018).
Buckley, M. et al. Distinguishing between archaeological sheep and goat bones using a single collagen peptide. J. Archaeol. Sci. 37, 13–20 (2010).
Buckley, M. & Collins, M. J. Collagen survival and its use for species identification in Holocene-lower Pleistocene bone fragments from British archaeological and paleontological sites. Antiqua 1, e1 (2011).
Prendergast, M. E. et al. Reconstructing Asian faunal introductions to eastern Africa from multi-proxy biomolecular and archaeological datasets. PLoS ONE 12, e0182565 (2017).
Sinet-Mathiot, V. et al. Combining ZooMS and zooarchaeology to study Late Pleistocene hominin behaviour at Fumane (Italy). Nat. Sci. Rep. 9, 12350 (2019).
Le Meillour, L. et al. Identification of degraded bone and tooth splinters from arid environments using palaeoproteomics. Palaeogeogr. Palaeoclimatol. Palaeoecol. 511, 472–482 (2018).
Bradfield, J., Forssman, T., Spindler, L. & Antonites, A. R. Identifying the animal species used to manufacture bone arrowheads in South Africa. Archaeol. Anthropol. Sci. 11, 2419–2434 (2019).
Coutu, A. N., Whitelaw, G., le Roux, P. & Sealy, J. Earliest evidence for the ivory trade in Southern Africa: isotopic and ZooMS analysis of seventh–tenth century ad ivory from KwaZulu-Natal. Afr. Archaeol. Rev. 33, 411–435 (2016).
Demarchi, B. et al. Protein sequences bound to mineral surfaces persist into deep time. Elife 5, e17092 (2016).
Buckley, M., Collins, M., Thomas-Oates, J. & Wilson, J. C. Species identification by analysis of bone collagen using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 23, 3843–3854 (2009).
Buckley, M. & Wadsworth, C. Proteome degradation in ancient bone: diagenesis and phylogenetic potential. Palaeogeogr. Palaeoclimatol. Palaeoecol. 416, 69–79 (2014).
Fiddyment, S. et al. Animal origin of 13th-century uterine vellum revealed using noninvasive peptide fingerprinting. Proc. Natl. Acad. Sci. 112, 15066–15071 (2015).
McGrath, K. et al. Identifying archaeological bone via non-destructive ZooMS and the materiality of symbolic expression: examples from Iroquoian bone points. Nat. Sci. Rep. 9, 11027 (2019).
Kirby, D. P., Manick, A. & Newman, R. Minimally invasive sampling of surface coatings for protein identification by peptide mass fingerprinting: a case study with photographs. J. Am. Inst. Conserv. https://doi.org/10.1080/01971360.2019.1656446 (2019).
Buckley, M. & Kansa, S. W. Collagen fingerprinting of archaeological bone and teeth remains from Domuztepe, South Eastern Turkey. Archaeol. Anthropol. Sci. 3, 271–280 (2011).
Taylor, W. et al. Early pastoral economies along the Ancient Silk Road: biomolecular evidence from the Alay Valley, Kyrgyzstan. PLoS ONE 13, e0205646 (2018).
Korzow Richter, K. et al. What’s the catch? Archaeological application of rapid collagen-based species identification for Pacific Salmon. J. Archaeol. Sci. 116, 105116 (2020).
Brown, S., Douka, K., Collins, M. & Richter, K. K. On the standardization of ZooMS nomenclature. J. Proteom. https://doi.org/10.1016/j.jprot.2020.104041 (2020).
Shoulders, M. D. & Raines, R. T. Collagen structure and stability. Annu. Rev. Biochem. 78, 929–958 (2009).
Schroeter, E. R. & Cleland, T. P. Glutamine deamidation: an indicator of antiquity, or preservational quality?. Rapid Commun. Mass Spectrom. 30, 251–255 (2016).
Welker, F. et al. Variations in glutamine deamidation for a Châtelperronian bone assemblage as measured by peptide mass fingerprinting of collagen. STAR: Sci. Technol. Archaeol. Res. 3, 15–27 (2017).
Ramsøe, A. et al. DeamiDATE 1.0: Site-specific deamidation as a tool to assess authenticity of members of ancient proteomes. J. Archaeol. Sci. 115, 105080 (2020).
Leo, G. et al. Deamidation at asparagine and glutamine as a major modification upon deterioration/aging of proteinaceous binders in mural paintings. Anal. Chem. 83, 2056–2064 (2011).
Welker, F. et al. Ancient proteins resolve the evolutionary history of Darwin’s South American ungulates. Nature 522, 81–84 (2015).
Breton, G. et al. Lactase persistence alleles reveal partial East African ancestry of southern African Khoe pastoralists. Curr. Biol. 24, 852–858 (2014).
Jerardino, A., Fort, J., Isern, N. & Rondelli, B. Cultural diffusion was the main driving mechanism of the Neolithic transition in southern Africa. PLoS ONE 9, e113672 (2014).
Lander, F. & Russell, T. The archaeological evidence for the appearance of pastoralism and farming in southern Africa. PLoS ONE 13, e0198941 (2018).
Macholdt, E. et al. Tracing pastoralist migrations to southern Africa with lactase persistence alleles. Curr. Biol. 24, 875–879 (2014).
Jensen, T. Z. T. et al. The biomolecular characterization of a finger ring contextually dated to the emergence of the Early Neolithic from Syltholm, Denmark. Roy. Soc. Open Sci. 7, 191172 (2020).
Mackie, M. et al. Palaeoproteomic profiling of conservation layers on a 14th century Italian wall painting. Angew. Chem. Int. Ed. 57, 7369–7374 (2018).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Han, X., He, L., Xin, L., Shan, B. & Ma, B. PeaksPTM: mass spectrometry-based identification of peptides with unspecified modifications. J. Proteom. Res. 10, 2930–2936 (2011).
Katoh, K., Rozewicki, J. & Yamada, K. D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166 (2019).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucl. Acids Res. 47, D442–D450 (2019).
Acknowledgements
Coutu was funded by the European Union’s Seventh Framework Programme (FP7/2007–2013) as a Marie Curie International Outgoing Fellow under Grant agreement PIOF-GA-2012-332165, “TEMBo”. Taurozzi, Mackie, and Collins are supported by the Danish National Research Foundation award PROTEIOS (DNRF128). Jensen is supported by the European Union’s EU Framework Programme for Research and Innovation Horizon 2020 under Grant Agreement no. 676154 (ArchSci2020). Prof. Jesper Velgaard Olsen at the Novo Nordisk Center for Protein Research is thanked for providing access and resources, where work is funded in part by a donation from the Novo Nordisk Foundation (Grant No. NNF14CC0001). Sealy is supported by the South African Research Chairs programme of the National Research Foundation and the Department of Science and Innovation (Grant no 84407). We thank Neil Rusch for taking the photograph in Fig 1.
Author information
Authors and Affiliations
Contributions
J.S., M.J.C., A.C. and A.J.T. conceived the study. J.S. and A.C. collected the samples. A.J.T. and A.C. prepared samples for MALDI-TOF and LC–MS/MS. M.M. performed the LC–MS/MS data acquisition. M.M., A.C., A.J.T., T.Z.T.J. and J.S. interpreted data. A.C., J.S., A.J.T. and M.M. wrote the manuscript with critical input from T.Z.T.J. and M.J.C.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Coutu, A.N., Taurozzi, A.J., Mackie, M. et al. Palaeoproteomics confirm earliest domesticated sheep in southern Africa ca. 2000 BP. Sci Rep 11, 6631 (2021). https://doi.org/10.1038/s41598-021-85756-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-85756-8
- Springer Nature Limited
This article is cited by
-
A label-free quantification method for assessing sex from modern and ancient bovine tooth enamel
Scientific Reports (2024)
-
Chemical evidence for milk, meat, and marine resource processing in Later Stone Age pots from Namaqualand, South Africa
Scientific Reports (2023)
-
Paleoproteomic profiling for identification of animal skin species in ancient Egyptian archaeological leather using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS)
Heritage Science (2022)
-
From quartz curvature to late Holocene mobility at Spring Cave, Western Cape, South Africa
Archaeological and Anthropological Sciences (2022)
-
Male-biased migration from East Africa introduced pastoralism into southern Africa
BMC Biology (2021)