Abstract
A growing body of evidence suggests a relationship between olfactory dysfunction and the pathogenesis of mental disorders. Our previous studies indicated that chronic nasal inflammation caused loss of olfactory sensory neurons and gross atrophy of the olfactory bulb, which may lead to olfactory dysfunction. Simultaneously, increasing numbers of reports have elucidated the importance of gut microbiota to maintain brain function and that dysbiosis may be associated with neuropsychiatric disorders. Here we examined whether chronic nasal inflammation perturbed gut microbiota and whether there were sex differences in this pattern. Eight-week-old C57BL/6 mice repeatedly received bilateral nasal administration of lipopolysaccharide (LPS) 3 times/week to cause chronic nasal inflammation or saline as a control. At 9 weeks, cecal feces were used for 16S metagenomic analysis and whole blood and fresh tissue of spleen were used for ELISA analyses. Microbiome analysis demonstrated a remarkable change of the gut microbiota in male mice with chronic nasal inflammation which was different from that in female mice. In both mice, systemic inflammation did not occur. This has shown for the first time that chronic nasal inflammation correlates with sex-dependent changes in the gut microbiota. The detailed mechanism and potential alteration to brain functions await further studies.
Similar content being viewed by others
Introduction
Mental disorders include depression, bipolar disorder, schizophrenia and other psychoses, dementia, and developmental disorders including autism. The number of patients with a mental disorder is increasing and about 300,000,000 people experience depression in the world (WHO, 2018). This may correlate to the increase in the number of suicides associated with mental disorders, which is a worldwide problem. Given that many patients with mental disorders such as depression, autism, schizophrenia, and Alzheimer’s disease are reported to have impairment of sense of smell1,2,3 and that chronic rhinitis and rhinosinusitis are a risk factor for depression and anxiety4,5, the olfactory system is purported to be associated with the pathogenesis of mental disorders. However, the mechanisms connecting the nose to the brain are still an enigma.
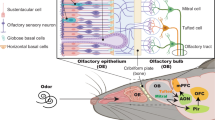
Nasal inflammation may be a trigger for brain tissue damage and impairment of function. In our previous studies, we demonstrated that chronic nasal inflammation causes loss of olfactory sensory neurons (OSNs) and gross atrophy of the olfactory bulb (OB), the first relay station of the olfactory neurocircuit in the central nervous system. In the OB, glial cells were activated, pro-inflammatory cytokines were elevated, and subsequently tufted cell-related neurocircuits underwent degeneration in a mouse model with chronic nasal inflammation6,7,8. These results suggest that an olfactory neural pathway is a route through which nasal inflammation damages the brain. In fact, patients with mental disorders, as well as chronic rhinosinusitis, have smaller OB volume9,10.
Microbiota resides in various sites throughout the body, such as the nasal and oral cavities, lung, gut and even the brain, with the biggest colony within the gut11. The gut microbiota interacts with the host through the immune, metabolic and nervous systems and influences the brain function12,13. Therefore, an unbalanced composition of microbiota, termed dysbiosis, breaks down an individual’s homeostasis and is associated with a variety of disorders including inflammatory bowel disease, metabolic syndrome, obesity, and diabetes14,15. Recent reports indicate that neurodegenerative and neuropsychiatric diseases such as Alzheimer’s disease, Parkinson’s disease, multiple sclerosis, autism spectrum disorder and depression are also related to dysbiosis16,17, indicating the strong connection between the gut and the brain. Antibiotics are a trigger to perturb gut microbiota, which induces a loss of taxonomic and functional diversity combined with a reduced colonization resistance against invading pathogens18. The mode of delivery (cesarean or vaginal birth) and of feeding (breast milk or synthetic milk) can also influence the gut microbiota in the process of development in a newborn baby19. However, other factors perturbing adult gut microbiota are not well documented.
In the present study, we addressed the question of whether chronic nasal inflammation would cause perturbation of gut microbiota. We also compared the results between sexes.
Results
Nasal inflammation
After repeated administration of lipopolysaccharide (LPS) to bilateral nostrils, inflammatory cells such as macrophages (F4/80-positive), neutrophils (Ly-6G-positive), T cells (CD3e-positive) and B cells (CD45R-positive) locally infiltrated some regions of the olfactory mucosa (OM) in male and female mice, while these cells were not observed in the OM of saline-treated mice (Supplementary Figure S1). Some macrophages (CD11b-positive) produced interleukin (IL)-1β, a proinflammatory cytokine, in the LPS-treated OM in male and female mice (Supplementary Figure S1). During repeated administration of LPS, numbers of mature and immature OSNs (OMP- and GAP43-positive, respectively) gradually decreased similarly in male and female mice (Supplementary Figure S2).
Gut microbiota
Diversity
First, we examined Chao1 and Shannon indices to examine the α-diversity. As shown in Fig. 1A, the Chao1 index did not change in LPS-treated mice compared to saline-treated control in male or female mice. The value was also similar in male and female mice. In contrast, Shannon index significantly increased in the male LPS-treated compared to male saline-treated control mice. The value was significantly higher in female than in male saline-treated control mice (Fig. 1B).
Diversity of gut microbiota. (A) Chao1 index. (B) Shannon index. The middle line in the box plot represents the median value, and the box is drawn indicating the 25 to 75% quartiles. Whiskers show minimum and maximum values and the ends of the whiskers represent the non-outlier range. Mean values are shown as ×. Data were analyzed with a 2-way ANOVA (sex × treatment) with Tukey’s HSD post hoc test. **p < 0.01. (C) Principle component analysis (PCA) based on Bray–Curtis dissimilarities among all sample sets. Dots with different symbols indicate samples collected from different groups. PC1: first principal coordinate, percent variation 24.01%; PC2: second principal coordinate, percent variation 16.44%. n = 5 for each group.
Next, we performed Bray–Curtis PC (principle component) plots to compare the β-diversity amongst the 4 groups. The values of the male saline group distributed apart from those of the male LPS group, while values were more accumulated in female mice (Fig. 1C).
Phylum analysis
At the phylum level of the taxonomy, Bacteroidetes and Firmicutes were the two predominant phyla in the gut and accounted for more than 90% of all bacteria in mice (Fig. 2A). The ratio of Bacteroidetes increased with a relative decrease in Firmicutes in male LPS-treated compared to male saline-treated mice, whereas the ratio did not change in female mice. The ratio of Firmicutes to Bacteroidetes significantly decreased in male LPS-treated compared to male saline-treated mice, while the value did not change in female mice (Fig. 2B). The ratio was significantly higher in male saline-treated compared to female saline-treated mice (Fig. 2B).
Phylum level analysis of gut microbiota. (A) The relative abundance of gut microbiota at phylum level. (B) The relative ratio of Firmicutes/Bacteroidetes in gut microbiota. The ratio significantly decreased in male LPS-treated compared to male saline-treated mice. *p < 0.05, **p < 0.01. n = 5 for each group.
Family and genus analysis
At the family level of gut microbiota, five bacterial families significantly increased and two families significantly decreased after LPS treatment in male mice, whereas no bacterial family significantly changed in female mice amongst the 20 bacterial families analyzed (see “Materials and methods” section). The average relative abundances of Bacteroidaceae, Paraprevotellaceae (k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_Paraprevotellaceae), Porphyromonadaceae, Rikenellaceae and Ruminococcaceae increased and Erysipelotrichaceae and Lactobacillaceae decreased in male LPS-treated compared to male saline-treated mice (Fig. 3, Table 1). At the genus level, four bacterial genera increased and two decreased in male LPS-treated compared to male saline-treated mice, while no bacterial genera significantly changed in female LPS-treated mice amongst the 20 bacterial genera analyzed (see “Materials and methods” section). The abundance of Bacteroides, Oscillospira, Parabacteroides and Prevotella (k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_Paraprevotellaceae; g_Prevotella) significantly increased and Allobaculum and Lactobacillus significantly decreased in male LPS-treated mice (Fig. 4, Table 2).
The relative abundance of gut microbiota at family level. Significant increases in the abundances of Bacteroidaceae (A), Paraprevotellaceae (k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_Paraprevotellaceae) (B), Porphyromonadaceae (C), Rikenellaceae (D), and Ruminococcaceae (E) and decreases in Erysipelotrichaceae (F) and Lactobacillaceae (G) in male LPS-treated compared to male saline-treated mice were detected. *p < 0.05, **p < 0.01. n = 5 for each group.
The relative abundance of gut microbiota at genus level. Significant increases in the abundances of Bacteroides (A), Oscillospira (B), Parabacteroides (C) and Prevotella (D) and decreases in Allobaculum (E) and Lactobacillus (F) in male LPS-treated compared to male saline-treated mice were detected. No significant changes in the abundance in female LPS-treated compared to female saline-treated mice were detected. *p < 0.05, **p < 0.01. n = 5 for each group.
Systemic cytokines
To address the possibility that the chronic nasal inflammation caused systemic inflammation, we examined the expression levels of representative pro-inflammatory cytokines, TNFα and IL-1β, using serum and spleen extraction by ELISA analysis. The amounts of TNFα in serum were 24.5 ± 3.4, 24.0 ± 2.5, 42.4 ± 12.5 and 26.9 ± 2.7 pg/mL in male saline, male LPS, female saline and female LPS group mice, respectively, and did not differ between saline-treated control and LPS-treated male or female mice (Fig. 5A). The amounts of IL-1β in serum were 152.7 ± 18.0, 73.7 ± 26.7, 238.6 ± 42.8 and 82.2 ± 31.3 pg/mL in male saline, male LPS, female saline and female LPS group mice, respectively, and did not increase in LPS-treated male or female mice compared to saline-treated controls (Fig. 5B). The amounts of TNFα or IL-1β in the spleen extract did not differ between control and LPS-treated male or female mice (Fig. 5C, D).
Discussion
This study is the first to draw a link between chronic nasal inflammation and perturbation of gut microbiota and that this is different in male and female mice. This may show that nasal inflammation affects the brain not only through the damage of the olfactory neural pathway but also by perturbing the gut-brain axis.
The Shannon index showed a lower heterogeneity of bacteria in male control mice compared to the other groups and the beta diversity in the Bray–Curtis dissimilarity showed a separation of plots in the male saline group compared to the other plots of different groups. Similarly, the Firmicutes/Bacteroidetes ratio significantly decreased only in male LPS-treated compared to male saline-treated mice but the ratio did not change in female mice and was similar to male LPS-treated mice. The abundances of higher numbers of bacteria changed after LPS treatment in male mice compared to female mice. In most cases, the abundance values for male LPS-treated mice approached those in female mice. These results suggest that male mice were more vulnerable to chronic nasal inflammation and tended to alter gut microbiota after LPS treatment. The fact that the heterogeneity (Shannon value) was lower in male mice than in female mice in the normal condition may explain the vulnerability in male mice. In addition, the results can be interpreted that the initial sex differences in the gut microbiota in saline-treated control mice were lost after chronic nasal inflammation, suggesting that the effects of nasal inflammation on the gut microbiota exceeded the intrinsic sex differences.
After long-term intranasal administration of LPS, the level of Bacteroidetes increased and Firmicutes decreased at the phylum level in male mice. At the genus level, Bacteroides, Oscillospira, Parabacteroides and Prevotella significantly increased, whereas Allobaculum and Lactobacillus decreased in LPS-treated male mice.
Higher Bacteroidetes and lower Firmicutes at the phylum level in the gut microbiota are known to be demonstrated in adult male mice after chronic restraint stress for 3 weeks20. Similarly, in human studies, Bacteroidetes, Proteobacteria and Actinobacteria significantly increase and Firmicutes significantly decrease in major depressive disorder patients compared to healthy controls21. Another study showed that severe stress is associated with lower relative levels of Firmicutes at the phylum level and higher Bacteroides, Parabacteroides, Rhodococcus, Methanobrevibacter and Roseburia but lower Phascolarctobacterium at the genus level in Belgian children22. In an older report, Bacteroides was shown to increase in astronauts during confinement in the Skylab test chamber with Skylab food, but not in those in the normal condition with Skylab food23. These reports suggest a relationship between chronic stress and increased Bacteroidetes at the phylum level and Bacteroides at the genus level.
In contrast, chronic stress induces the reduction of Lactobacillus at the genus level. The mice that have been exposed to chronic unexpected mild stress and those that received the microbiota from the stressed mice both show high levels of anxiety- and depressive-like behavior and change in the composition of gut microbiota. In these mice, Lactobacillus was lower in abundance, whereas Akkermansia was higher24. In another study, the genus Lactobacillus was reduced in the male mice exposed to the stressor25. Also, Lactobacillus as well as Bifidobacterium has been shown to be reduced in cosmonauts after space missions26. Since Lactobacillus has been shown to decrease in response to a psychological challenge, it has been used as a probiotic in humans and experimental animals to improve behavioral abnormalities27. For example, Lactobacillus johnsonii has been shown to attenuate stress-induced and intraperitoneal injection of LPS-induced anxiety-like behavior in male mice28; Lactobacillus rhamnosus reduced stress-induced anxiety- and depression-related behavior in male mice29; Supplementation of Lactobacillus reuteri prevented longstanding LPS-induced changes in anxiety-like behavior and stress-induced brain activation in male mice30. Together with these reports, our results demonstrated that male, but not female mice with chronic nasal inflammation are likely to suffer from chronic stress. The inflammatory state in the OB in LPS-treated mice may be transmitted via the olfactory neurocircuit to the higher brain regions, such as piriform cortex, hypothalamus, amygdala, hippocampus and prefrontal cortex, which are brain regions associated with the stress response. In fact, mental stress induces neuroinflammation in these brain regions, suggesting that brain responses to the peripheral inflammation are similar to those to the mental stress31,32. On the other hand, mice with chronic nasal inflammation may suffer from impaired sense of smell due to the loss of OSNs and the atrophy of the OB. This anosmia-like situation may cause the mice much stress, leading to changes in the gut and possibly the brain as a stress response. Examining the changes at the cellular and molecular levels in the higher brain regions could result in better understanding of the psychiatric condition of the mice and the mechanisms underlying chronic nasal inflammation-induced dysbiosis.
In the present study, the abundance of Allobaculum was also found to decrease after LPS treatment in male mice. The abundance of Allobaculum is reported to be negatively correlated to leptin concentrations in the serum33. Since leptin is a hormone that is secreted mainly by white adipose tissue and strongly suppresses food intake by acting at the hypothalamus, LPS-treated male mice in the present study may reduce their food intake due to the decreased abundance of Allobaculum. In fact, we observed an inhibition in the normal increase in body weight during the observation period in LPS-treated mice (Supplementary Figure S3), although the increase in body weight was also suppressed in female mice with no decrease in Allobaculum. The relationship between nasal inflammation, perturbation of gut microbiota, serum leptin level and amount of food intake is an avenue for future research.
Several studies have reported sex differences in the composition of gut microbiota under physiological conditions in humans and animals34. In this study, we also found sex differences in the gut microbiota in the normal condition (saline-treated mice). The ratios of Clostridiaceae, Christensenellaceae, Coriobacteriaceae and Erysipelotrichaceae were higher in males, while those of Bacteroidaceae, Dehalobacteriaceae, Paraprevotellaceae, Porphyromonadaceae and Rikenellaceae were higher in females at the family level (Supplementary Table S1). Similarly, at the genus level, the ratios of Adlercreutzia, Allobaculum and Clostridium were higher in male mice, whereas those of Bacteroides, Dehalobacterium, Parabacteroides and Prevotella were higher in female mice (Supplementary Table S2).
In addition, we found sex-dependent differences in the pattern of perturbation of gut microbiota after chronic nasal inflammation. A greater number of bacteria were perturbed by the nasal inflammation in male mice compared to female mice. However, it remains unclear whether chronic nasal inflammation causes the sex-dependent differences in the perturbation of gut microbiota or whether the initial differences in the gut microbiota may contribute to the different responses to nasal inflammation. Further experiments using male and female mice that display the same initial microbiota would give us more information on the sex-dependent differences in the response. In addition, it may be another limitation of this study that feces from male and female mice cannot be normalized since they should be housed separately. Taking this into consideration, accumulating data on the sex differences in gut microbiota may bring new findings related to the gut-brain interaction and provide an explanation of sex differences in the incidence of psychiatric disorders.
Materials and methods
Animals
Male and female C57BL/6J JmsSlc mice (Sankyo lab, Tokyo, Japan) were purchased at 6 weeks, acclimated for 2 weeks at our animal facility and entered the study at 8 weeks. Mice were consistently housed in groups separated by gender. The intranasal administration was performed according to our previous study6,7,8. Briefly, the mice were anesthetized with isoflurane and 10-µL physiologic saline (saline-treated mice) or LPS from Escherichia coli O55:B5 (1 mg/mL; Sigma) (LPS-treated mice) was administered to bilateral nostrils three times per week for 9 weeks. All protocols were approved by, and all methods were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Kyorin University Faculty of Health Sciences (Protocol ‘I17-09-02’). In addition, this study was carried out in compliance with the ARRIVE guidelines.
Experimental procedure
All experiments were performed following the schedule in Fig. 6. Mice with bilateral nasal administration of saline or LPS (3 times/week) were sacrificed at 9 weeks after the first administration and 2 days after the last administration. At that time, mice were deeply anesthetized with ketamine and xylazine, whole blood was taken from the right ventricle and a piece of spleen was obtained and fresh frozen for enzyme-linked immunosorbent assay (ELISA) analysis. The feces from the cecum was obtained and fresh frozen by liquid nitrogen for 16S rRNA analysis. Other groups of mice were transcardially perfused with phosphate buffered saline followed by 4% paraformaldehyde after whole blood was taken from the right ventricle for histological analyses.
Experimental protocol. Mice received repeated bilateral intranasal administration of LPS or saline 3 times per week and were sacrificed at 9 weeks after the first administration. Feces, blood, fresh tissue of spleen were obtained for 16S rRNA and ELISA analyses. Olfactory mucosa (OM) was used for histological analysis.
Histological analysis
Frozen sections of OM were used for histological analysis. Briefly, post-fixation with the same fixative at 4 °C overnight, the rostral half of the calvaria and the nasal bone were decalcified by 2 × K-CX (Falma, Tokyo, Japan) for 2.5 h at room temperature. The OM was cryoprotected with 20% sucrose in PBS (wt/vol) at room temperature overnight, embedded in OCT compound (Sakura Finetek USA Inc., Torrance, CA), and maintained at − 80 °C until use.
Olfactory mucosa was coronally cut on a cryostat into 20 μm slices and was rehydrated with TBST (10 mmol/L Tris-HCl [pH 7.4] and 100 mmol/L NaCl with 0.1% Tween 20), blocked with blocking buffer (5% normal donkey serum [vol/vol] in TBST) at room temperature for 1 h. It was then incubated with primary antibodies diluted in blocking buffer overnight. The antibodies used in the present study were rat anti-F4/80- (1:200, Abcam, Cambridge, UK), rat anti-Ly-6G- (1:200, AdipoGen, Liestal, Switzerland), rabbit anti-CD3e- (1:150, Thermo Fisher Scientific, Waltham, MA), rat anti-CD45R (1:200, Abcam), goat anti-olfactory marker protein- (1:1000, Wako, Osaka, Japan), rabbit anti-GAP43- (1:1000, Novus Biologicals, Littleton, CO), rabbit anti-CD11b- (1:500, Abcam) and goat anti-IL-1β (1:200, R&D Systems, Minneapolis, MN) antibodies. Sections were incubated with host-matched secondary antibodies: Alexa Fluor 568- or 488-conjugated donkey anti-species IgGs (1:300, Thermo Fisher Scientific). Nuclei were counterstained with DAPI. The sections were coverslipped with fluorescence mounting medium (Dako Agilent, Santa Clara, CA) and imaged using a fluorescence microscope with structured illumination (BZ-X710; Keyence, Osaka, Japan).
16S metagenomic analysis
Genomic DNA was extracted from fecal samples and purified using Genomic DNA from stool samples (Macherey-Nagel GmbH & Co. KG, Germany). Each DNA specimen from feces was amplified using the Ion 16S Metagenomics Kit (Thermo Fisher Scientific, Bremen, Germany) as described in the previous study35. The amplicons were purified and prepared for the sequencing library by using the Ion Plus Fragment Library Kit (Thermo Fisher Scientific) and the Ion Personal Genome Machine (PGM) Hi-Q sequencing kit following the protocol of the kit. The sequencing runs were performed on the Ion PGM platform (Thermo). Unmapped bam files were converted to a fastq format using Torrent server (Thermo) by File Exporter v5.0.3.1.
Bioinformatic analysis of bacterial 16S rRNA amplicon data was conducted using the QIIME v1.9.1 software pipeline. Sequences were aligned and clustered into operational taxonomic units (OTU) based on the de novo OTU picking algorithm using the QIIME implementation of UCLUST with Greengenes database. OTUs with 97% similarity level were selected for taxonomical assignment and employed for the alpha diversity, richness (Chao 1) and evenness (Shannon) analysis. Bray–Curtis index was calculated as beta diversity. The counts in per sample were 32,476, 43,435, 53,816, 57,830, 62,106, 63,217, 64,877, 66,416, 71,516, 76,788, 79,871, 128,837, 136,371, 151,462, 159,118, 166,430, 169,062, 309,313, 347,304, and 660,519 and the depth of coverage was determined to be 32,476, the minimum value. Among sixty-eight families and one hundred and eighteen genera of bacteria found by the machine, 45 families and 46 genera were identified. We further analyzed 20 families and 20 genera that had more than 0.1% of existence in any one group (male saline, male LPS, female saline or female LPS).
ELISA assay
Blood samples were stored at 4 °C overnight and centrifuged at 3500 rpm at 4 °C for 20 min. The supernatant serum was collected and stored at − 80 °C until use. The pieces of spleen were homogenized in 20 times volume of Tissue Protein Extraction Reagent (T-PER; Thermo Fisher Scientific, Waltham, MA) containing Halt Protease Inhibitor Cocktail, EDTA-free (Thermo Fisher Scientific) and centrifuged at 15,000 rpm at 4 °C for 5 min. The supernatant extract was collected and stored at − 80 °C until use.
The levels of TNFα and IL-1β in the serum and spleen extraction were determined by ELISA using DuoSet ELISA (R&D Systems, Minneapolis, MN) according to the manufacturer’s instructions. All samples were measured in duplicate. Values were compared between male saline, male LPS, female saline and female LPS group mice (n = 5).
Statistical analysis
Microbiota and cytokine levels were compared statistically by 2-way analysis of variance, followed by Tukey’s HSD post hoc tests for multiple comparisons using Statistica software (Dell Software, Round Rock, TX). A p < 0.05 represented a significant difference. Values are reported as means ± SEMs.
References
Kohli, P., Soler, Z. M., Nguyen, S. A., Muus, J. S. & Schlosser, R. J. The association between olfaction and depression: a systematic review. Chem. Senses 41, 479–486. https://doi.org/10.1093/chemse/bjw061 (2016).
Marin, C. et al. Olfactory dysfunction in neurodegenerative diseases. Curr. Allergy Asthma Rep. 18, 42. https://doi.org/10.1007/s11882-018-0796-4 (2018).
Rochet, M., El-Hage, W., Richa, S., Kazour, F. & Atanasova, B. Depression, olfaction, and quality of life: a mutual relationship. Brain Sci. https://doi.org/10.3390/brainsci8050080 (2018).
Bedolla-Barajas, M., Morales-Romero, J., Pulido-Guillen, N. A., Robles-Figueroa, M. & Plascencia-Dominguez, B. R. Rhinitis as an associated factor for anxiety and depression amongst adults. Braz. J. Otorhinolaryngol. 83, 432–438. https://doi.org/10.1016/j.bjorl.2016.05.008 (2017).
Kim, D. H., Han, K. & Kim, S. W. Relationship between allergic rhinitis and mental health in the general Korean adult population. Allergy Asthma Immunol. Res. 8, 49–54. https://doi.org/10.4168/aair.2016.8.1.49 (2016).
Hasegawa-Ishii, S., Imamura, F., Nagayama, S., Murata, M. & Shimada, A. Differential effects of nasal inflammation and odor deprivation on layer-specific degeneration of the mouse olfactory bulb. eNeuro https://doi.org/10.1523/ENEURO.0403-19.2020 (2020).
Hasegawa-Ishii, S., Shimada, A. & Imamura, F. Lipopolysaccharide-initiated persistent rhinitis causes gliosis and synaptic loss in the olfactory bulb. Sci. Rep. 7, 11605. https://doi.org/10.1038/s41598-017-10229-w (2017).
Hasegawa-Ishii, S., Shimada, A. & Imamura, F. Neuroplastic changes in the olfactory bulb associated with nasal inflammation in mice. J. Allergy Clin. Immunol. 143, 978e973-989e973. https://doi.org/10.1016/j.jaci.2018.09.028 (2019).
Croy, I. & Hummel, T. Olfaction as a marker for depression. J. Neurol. 264, 631–638. https://doi.org/10.1007/s00415-016-8227-8 (2017).
Thomann, P. A. et al. Reduced olfactory bulb and tract volume in early Alzheimer’s disease—a MRI study. Neurobiol. Aging 30, 838–841. https://doi.org/10.1016/j.neurobiolaging.2007.08.001 (2009).
Bell, J. S. et al. Invited review: from nose to gut—the role of the microbiome in neurological disease. Neuropathol. Appl. Neurobiol. 45, 195–215. https://doi.org/10.1111/nan.12520 (2019).
Clemente, J. C., Ursell, L. K., Parfrey, L. W. & Knight, R. The impact of the gut microbiota on human health: an integrative view. Cell 148, 1258–1270. https://doi.org/10.1016/j.cell.2012.01.035 (2012).
Fung, T. C., Olson, C. A. & Hsiao, E. Y. Interactions between the microbiota, immune and nervous systems in health and disease. Nat. Neurosci. 20, 145–155. https://doi.org/10.1038/nn.4476 (2017).
Belizario, J. E. & Faintuch, J. Microbiome and gut dysbiosis. Exp. Suppl. 109, 459–476. https://doi.org/10.1007/978-3-319-74932-7_13 (2018).
Butel, M. J. Probiotics, gut microbiota and health. Med. Mal. Infect. 44, 1–8. https://doi.org/10.1016/j.medmal.2013.10.002 (2014).
Collins, S. M., Surette, M. & Bercik, P. The interplay between the intestinal microbiota and the brain. Nat. Rev. Microbiol. 10, 735–742. https://doi.org/10.1038/nrmicro2876 (2012).
Cryan, J. F. & Dinan, T. G. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 13, 701–712. https://doi.org/10.1038/nrn3346 (2012).
Lange, K., Buerger, M., Stallmach, A. & Bruns, T. Effects of antibiotics on gut microbiota. Dig. Dis. 34, 260–268. https://doi.org/10.1159/000443360 (2016).
Backhed, F. et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 852. https://doi.org/10.1016/j.chom.2015.05.012 (2015).
Guo, Y. et al. Prophylactic effects of bifidobacterium adolescentis on anxiety and depression-like phenotypes after chronic stress: a role of the gut microbiota-inflammation axis. Front. Behav. Neurosci. 13, 126. https://doi.org/10.3389/fnbeh.2019.00126 (2019).
Jiang, H. et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 48, 186–194. https://doi.org/10.1016/j.bbi.2015.03.016 (2015).
Michels, N. et al. Gut microbiome patterns depending on children’s psychosocial stress: reports versus biomarkers. Brain Behav. Immun. 80, 751–762. https://doi.org/10.1016/j.bbi.2019.05.024 (2019).
Holdeman, L. V., Good, I. J. & Moore, W. E. Human fecal flora: variation in bacterial composition within individuals and a possible effect of emotional stress. Appl. Environ. Microbiol. 31, 359–375 (1976).
Li, N. et al. Fecal microbiota transplantation from chronic unpredictable mild stress mice donors affects anxiety-like and depression-like behavior in recipient mice via the gut microbiota-inflammation-brain axis. Stress 22, 592–602. https://doi.org/10.1080/10253890.2019.1617267 (2019).
Galley, J. D. et al. Exposure to a social stressor disrupts the community structure of the colonic mucosa-associated microbiota. BMC Microbiol. 14, 189. https://doi.org/10.1186/1471-2180-14-189 (2014).
Liz’ko, N. N., Shilov, V. M., Syrykh, G. D. & Legen’kov, V. I. Intestinal microflora makeup of astronauts before and after space flights. Kosm Biol. Aviakosm Med. 13, 9–13 (1979).
Sarao, L. K. & Arora, M. Probiotics, prebiotics, and microencapsulation: a review. Crit. Rev. Food Sci. Nutr. 57, 344–371. https://doi.org/10.1080/10408398.2014.887055 (2017).
Jang, H. M., Lee, K. E., Lee, H. J. & Kim, D. H. Immobilization stress-induced Escherichia coli causes anxiety by inducing NF-kappaB activation through gut microbiota disturbance. Sci. Rep. 8, 13897. https://doi.org/10.1038/s41598-018-31764-0 (2018).
Bravo, J. A. et al. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. U. S. A. 108, 16050–16055. https://doi.org/10.1073/pnas.1102999108 (2011).
Murray, E. et al. Pubertal probiotic blocks LPS-induced anxiety and the associated neurochemical and microbial outcomes, in a sex dependent manner. Psychoneuroendocrinology 112, 104481. https://doi.org/10.1016/j.psyneuen.2019.104481 (2020).
Kim, Y. K. & Won, E. The influence of stress on neuroinflammation and alterations in brain structure and function in major depressive disorder. Behav. Brain Res. 329, 6–11. https://doi.org/10.1016/j.bbr.2017.04.020 (2017).
Zhu, Y., Klomparens, E. A., Guo, S. & Geng, X. Neuroinflammation caused by mental stress: the effect of chronic restraint stress and acute repeated social defeat stress in mice. Neurol. Res. 41, 762–769. https://doi.org/10.1080/01616412.2019.1615670 (2019).
Ravussin, Y. et al. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity (Silver Spring) 20, 738–747. https://doi.org/10.1038/oby.2011.111 (2012).
Markle, J. G. et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339, 1084–1088. https://doi.org/10.1126/science.1233521 (2013).
Osaki, T. et al. Influence of intestinal indigenous microbiota on intrafamilial infection by Helicobacter pylori in Japan. Front. Immunol. 9, 287. https://doi.org/10.3389/fimmu.2018.00287 (2018).
Acknowledgements
This study was supported by the Grant-in-Aid for Scientific Research KAKENHI 19K17870 (to Y.M.), and 18K07395 (to S.H.-I.) and a Grant from Kyorin University. We thank Mr. Seiya Higashi and Dr. Eriko Nozaki for their contribution to metagenome analysis, Mr. Mizuki Naruse, Ms. Mai Hirano and Ms. Ami Wakabayashi for their contributions to animal experiments and Ms. Yumi Hanazato, Ms. Mikiko Sakaguchi and Ms. Hinami Asano for their contribution to the lab work.
Author information
Authors and Affiliations
Contributions
Y.M. and S.H.I. designed the research. Y.M., T.O. and S.H.I. performed all experiments and analyzed data and wrote the main manuscript text. A.S. and S.K. interpreted the results and modified the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mishima, Y., Osaki, T., Shimada, A. et al. Sex-dependent differences in the gut microbiota following chronic nasal inflammation in adult mice. Sci Rep 11, 4640 (2021). https://doi.org/10.1038/s41598-021-83896-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-83896-5
- Springer Nature Limited