Abstract
Clusterin (CLU) is considered one of the most important roles for pathogenesis of Alzheimer’s Disease (AD). The early genome-wide association studies (GWAS) identified the CLU rs11136000 polymorphism is significantly associated with AD in Caucasian. However, the subsequent studies are unable to replicate these findings in different populations. Although two independent meta-analyses show evidence to support significant association in Asian and Caucasian populations by integrating the data from 18 and 25 related GWAS studies, respectively, many of the following 18 studies also reported the inconsistent results. Moreover, there are six missed and a misclassified GWAS studies in the two meta-analyses. Therefore, we suspected that the small-scale and incompletion or heterogeneity of the samples maybe lead to different results of these studies. In this study, large-scale samples from 50 related GWAS studies (28,464 AD cases and 45,784 controls) were selected afresh from seven authoritative sources to reevaluate the effect of rs11136000 polymorphism to AD risk. Similarly, we identified that the minor allele variant of rs11136000 significantly decrease AD risk in Caucasian ethnicity using the allele, dominant and recessive model. Different from the results of the previous studies, however, the results showed a negligible or no association in Asian and Chinese populations. Collectively, our analysis suggests that, for Asian and Chinese populations, the variant of rs11136000 may be irrelevant to AD risk. We believe that these findings can help to improve the understanding of the AD’s pathogenesis.
Similar content being viewed by others
Introduction
Alzheimer’s Disease (AD) is a commonest kind of neurodegenerative disorders with a complex pathogenesis, and has become one of the leading causes of death in elderly people1,2. It is characterized by accumulation and toxic effect of the amyloid β-peptide (Aβ) deposits and neurofibrillary tangles in brain3. Previous studies predict that the newly diagnosed AD patients are expected to reach as many as 135 million by 2050 from about 35 million in 2009 around the world if lack of the effective preventive measures4,5.
Clusterin (CLU) is considered one of the most important roles for pathogenesis of AD by influencing the structure and neurotoxic effects of Aβ deposits6,7,8, and some of the variants at CLU can affect its expression level in brain9,10. Two early genome-wide association studies (GWAS) identified a single nucleotide polymorphism (SNP) rs11136000 (T < C) significantly associated with AD in the CLU gene by analyzing the large-scale Caucasian populations11,12. In particular, Harold et al.11 and Lambert et al.12 analyzed 11,756 and 14,490 individuals from USA, UK, Ireland, Germany, France, Italy, Spain, Belgium and Finland, respectively, and both of them found that the minor allele variant of rs11136000 can reduce the risk of AD (95% confidence interval (CI) of odds ratio (OR) less than the value 1).
However, the subsequent studies report consistent13,14,15,16,17,18 and inconsistent19,20,21,22,23,24,25,26,27,28 results involved in Caucasian, Asian and African populations. For example, by analyzing 268 AD cases and 389 controls from China, Lin et al. find that the participants carrying 2 copies of minor allele in rs11136000 are associated with a decreased risk of AD17. The consistent result in North American Caucasian population is also identified by Carrasquillo et al.18. While in Canadian and Korean populations, the rs11136000 is found not associated with AD according to the studies of Bettens et al.24 and Chung et al.27, respectively. Then, two independent meta-analysis studies re-assess the results of these GWAS studies published before June 20, 2013 (18 studies) and August 31, 2014 (25 studies), respectively, and both of them found this SNP is significantly associated with AD in populations of Asian and Caucasian29,30. But among the subsequent 18 GWAS studies published after August 31, 2014, many of them report inconsistent results in the corresponding populations31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47. Moreover, by comparing the selected GWAS articles published before June 20, 2013 in the two meta-analysis studies, we find the selection is incomplete for both of them. In particular, Liu et al.29 miss two GWAS articles about Caucasian populations16,24, and Du et al.30 miss a GWAS article about Asian population27. In fact, through our further investigation, a total five related GWAS articles published before August 31, 2014 are not collected in the two meta-analysis studies48,49,50,51,52. In addition, a GWAS study about American and German populations is misclassified to the Asian ethnicity subgroup in Du et al.’s study22.
We suspected that the small-scale and incompletion or heterogeneity of the samples maybe lead to different results of these studies. In this study, we selected 50 related GWAS studies with large-scale samples from 40 articles (28,464 cases and 45,784 controls, about 40.3% increase over the total number of the previous two meta-analysis studies29,30) by searching the PubMed, ClinicalKey, AlzGene, Google Scholar, CNKI, Wanfang and VIP databases, and reevaluated the association between AD and rs11136000 polymorphism in Caucasian, Asian and Chinese population using the method of meta-analysis as previously described53,54,55,56,57,58,59,60,61,62,63. The use of more complete and larger scale samples would make the results more reliable.
Methods and Materials
Selection of literatures and GWAS studies
All of the possible studies were selected by searching the databases of PubMed (http://www.ncbi.nlm.nih.gov/pubmed, ClinicalKey (https://www.clinicalkey.com/), Wanfang (http://www.wanfangdata.com.cn/), CNKI (http://www.cnki.net/) and VIP (http://www.cqvip.com/) using the keywords: “Alzheimer’s disease”, “rs11136000”, “Clusterin” or “CLU”. The CNKI, Wanfang and VIP are very authoritative and reliable Chinese database. And then, we consulted the related studies collected in AlzGene database (http://www.alzgene.org/) which was a publicly available resource providing the information of AD genetic variants from 1,395 GWAS studies (updated April 18, 2011)64. In addition, we further queried references of these identified GWAS studies in previous steps and the articles citing them using the Google Scholar (http://scholar.google.com/).
After that, the appropriate studies were identified by the following criteria: (1) The study is a GWAS to analysis the association of rs11136000 polymorphism and AD. (2) It is a case-control design study. (3) The study provides both of the numbers of cases and controls. (4) The study provides the information about the ethnicity of each individual. (5) The detailed data for rs11136000 genotypes are available in the study.
Extraction of the related data
We extracted the related data for subsequent analysis from these identified studies: (1) each study’s publication date. (2) The first the author’s name in each of these studies. (3) The numbers of AD patients and controls of each study. (4) The sample’s ethnicity of each study. (5) The detailed genotype data of rs11136000 polymorphism both in AD patients and controls. (6) The types of genotyping platforms. (7) The key results of each study (i.e. the OR value and its 95% CI, as well as the corresponding P value). Moreover, if these results are not provided in the study directly, we would calculate them by the genotype data using the R program (http://www.r-project.org/).
Genetic model choice
The rs11136000 polymorphism contains two types of variants (T and C). T is the minor allele and C is the major allele. We assumed that they are the lower and high risk factor for AD, respectively. Then, the dominant model (TT + TC allele versus CC allele), allele model (T versus C) and recessive model (TT versus TC + CC) were used in this study. According to Table 1, all these studies were meta-analyzed using allele model, while only the studies offering CC, CT and TT genotypes data were analyzed using dominant or recessive model.
Hardy–Weinberg equilibrium (HWE) test
The HWE test of the rs11136000 polymorphism in AD patient and control groups was performed using a non-continuity correction chi-squared method with the significance level P < 0.01 as previously described65. Briefly, for the SNP in each case and control group, the simulated P values were calculated to measure the deviation from HWE based on 10,000 iterations. The R package ‘Genetics’ was used to perform the HWE test (https://cran.r-project.org/web/packages/genetics/index.html).
Heterogeneity test
In this study, the heterogeneity among the kinds of populations was measured by the two parameters, I2 value and Cochran’s Q. I2 value range from 0 to 100%, and it is calculated by Cochran’s Q according to the formula \({I}^{2}=\frac{Q-(k-1)}{Q}\times 100 \% \). The Cochran’s Q is based on a chi-squared distribution with k − 1 degrees of freedom, and k means the number of studies. Usually, the extreme, high, moderate and low heterogeneity was considered corresponding to the I 2 value of >75%, 50–75%, 25–50%, and <25%, respectively. In this study, the threshold of significant heterogeneity was set as I2 > 50% and P < 0.01 according to previous studies53,54,55,56.
Meta-analysis in entirety and subgroup
According to the results of heterogeneity test, the random and the fixed effect model were performed when the heterogeneity was significant or not, respectively66. We used the R package ‘meta’ to perform the meta-analysis, and determine the significance level of association between rs11136000 and AD through the pooled OR value and its 95% CI, as well as the corresponding P value (http://cran.r-project.org/web/packages/meta/index.html). And then, the original samples were further split into Caucasian, Asian, East Asian and Chinese populations, and the meta-analysis was performed in these subgroups.
Publication bias analysis and sensitivity analysis
We first evaluated the publication bias of the studies used in dominant, allele and recessive model, respectively, by the two common checking methods, the Begg’s test67 and Egger’s test68. The threshold of significant publication bias was set as P < 0.05. Then, we used the asymmetry of the funnel plots to describ the results of the publication bias analysis. Finally, for sensitivity analyses, we excluded each study in turn from the whole sample to measure the influence of each study.
Data availability
All the datasets used in this are available from the corresponding author.
Results
Study acquisition and data extraction
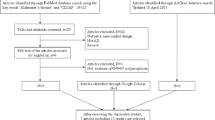
By a keyword search in the publicly available databases and a screening according to the criteria, a total 46 studies from 36 articles were identified which mainly involved in Caucasian and Asian populations. Moreover, a study about Sweden population was selected from AlzGene database, and three studies involved in Asian populations were identified by the citation check using Google Scholar.
Figure 1 showed the workflow of selection. Then, the related data of these 50 studies were extracted, and the main information was described in Table 1 (the detailed genotype data, the OR value and its 95% CI, as well as the corresponding P value were shown in Supplementary Table S1).
Hardy–Weinberg equilibrium test
We calculated the P value of HWE to assess the genotype distribution of rs11136000 polymorphism in AD patients and controls separately. Using a significance level of P < 0.01, we observed that a few of the samples deviated from HWE, including the case samples from the study of Yu et al. (P = 9.0 × 10−3) and Gu et al. (P = 2.0 × 10−4), and the control samples from the study of Rezazadeh et al. (P = 1.0 × 10−4), Gu et al. (P = 1.0 × 10−4) and Lin et al. (P = 9.0 × 10−3). More detailed information about the results of the HWE test was described in Supplementary Table S2.
Heterogeneity Test and Meta-analysis
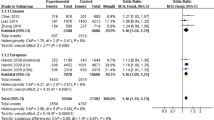
After the test, we found that there is no the significant genetic heterogeneity of rs11136000 polymorphism among all of the 50 selected studies using the dominant (I2 = 0% and P = 0.60), allele (I2 = 10% and P = 0.28) and recessive model (I2 = 33% and P = 0.04). Therefore, the meta-analysis with fixed effect model was performed to assess the association between rs11136000 and the risk of AD, and we found significant results in all the three models. In particular, the significant association between the minor allele (T) of rs11136000 and a decreased risk of AD was identified in the allele (OR = 0.875, 95% CI = 0.854–0.896, P < 0.0001) (Fig. 2), dominant (OR = 0.848, 95% CI = 0.817–0.879, P < 0.0001) and recessive model (OR = 0.822, 95% CI = 0.779–0.868, P < 0.0001) (Supplementary Figs S1 and S2).
Forest plot for the meta-analysis of rs11136000 polymorphism using allele model. All the 50 selected studies are used to meta-analysis of the allele contrast (T versus C) by the fixed effect model (Mantel-Haenszel) because the genetic heterogeneity is not significant. The minor allele (T) of rs11136000 was significantly associated with a decreased risk of AD.
Subgroup Analysis
We further performed the meta-analysis in the subgroups to assess the association between rs11136000 and the risk of AD in different ethnicities. Among all the 50 selected studies, the great majority of them involved in Caucasian or Asian ethnicity, except two studies about African and American population, respectively, and four mixed population studies (Table 1). Therefore, we first divided these studies into Caucasian or Asian ethnicity subgroups. We found a significant association between the minor allele (T) of rs11136000 and a decreased risk of AD in Caucasian ethnicity using the allele (OR = 0.864, 95% CI = 0.842–0.888, P < 0.0001), dominant (OR = 0.829, 95% CI = 0.796–0.864, P < 0.0001) and recessive model (OR = 0.819, 95% CI = 0.774–0.867, P < 0.0001) (Supplementary Figs S3–S5). For the Asian ethnicity, however, only a weak association was observed in allele model (OR = 0.921, 95% CI = 0.871–0.973, P = 0.0034) (Fig. 3a), but not the dominant (OR = 0.922, 95% CI = 0.846–1.005, P = 0.0649) (Fig. 3b) and recessive model (OR = 0.747, 95% CI = 0.511–1.092, P = 0.1326) (Fig. 3c).
The Asian population in this study was composed of the Indian, Iranian, Korean and Japanese individuals separately from a GWAS study, and the Chinese individuals from 12 GWAS studies. Therefore, we then assessed the association between this SNP and risk of AD in East Asian and Chinese populations. Interestingly, the results of meta-analysis in East Asian population were similar to these in Asian population (Supplementary Figs S6–S8). However, the association was not significant in Chinese population using the allele (OR = 0.939, 95% CI = 0.878–1.004, P = 0.0654) (Fig. 4a), dominant (OR = 0.988, 95% CI = 0.887–1.101, P = 0.8270) (Fig. 4b) and recessive model (OR = 0.615, 95% CI = 0.355–1.068, P = 0.0841) (Fig. 4c), which was different from the findings in the previous studies29,30.
Moreover, given that a few samples from four GWAS studies (three Asian populations and a mixed population) deviated from HWE, we further tested whether they affected the accuracy of the results by removing these studies from whole sample, Asian, East Asian and Chinese subgroups, respectively. The results were consistent with what we had been observed previously in whole sample and the subgroups using allele, dominant and recessive model. Table 2 showed the detailed information of the results.
Publication bias analysis and sensitivity analysis
As the funnel plots show (Fig. 5), we did not identify the significant publication bias in the three genetic models. In particular, the P value of Begg’s and Egger’s test is 0.80 and 0.24, respectively, for dominant model. Similarly, the P value is 0.43 (Begg’s test) and 0.21 (Egger’s test) for the allele model, and 0.22 (Begg’s test) and 0.61 (Egger’s test) for the recessive model. Moreover, through the sensitivity analysis, for all the three genetic models, we did not found a significant change of the association level between rs11136000 and AD when excluding any of the studies. Supplementary Tables S3–S5 described the related information in detailed.
Discussion
AD was characterized by accumulation and toxic effect of the Aβ deposits in brain3, and previous studies reported that the CLU could markedly influence the fibrillary Aβ formation and accumulation to mediate its toxicity in vivo, and likely as one of the most important roles for pathogenesis of AD6,7. Then, the subsequent GWAS studies found some variants in CLU were differently distributed between AD patients and controls11,12,13,14,15,16,17,18. Among these variants, a significant association was found between the minor allele (T) of rs11136000 and a decreased risk of AD by Harold et al.11, Lambert et al.12, Carrasquillo et al.13 and Seshadri et al.14. However, these results could not be repeated in other populations by the following studies19,20,21,22,23,24,25,26,27,28.
Although the two independent meta-analyses found a significant association between the minor allele (T) of rs11136000 and a decreased risk of AD in Caucasian and Asian ethnicities by integrating the data from related GWAS studies published before June 20, 2013 (18 studies) and August 31, 2014 (25 studies), respectively29,30, many of the following studies also reported the inconsistent results31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47. Moreover, according to our further investigation, the two meta-analyses missed out a total six related GWAS studies published before August 31, 201448,49,50,51,52, and a GWAS study about American and German populations is misclassified to the Asian ethnicity subgroup in Du et al.’s meta-analysis22. Therefore, we suspected that the small-scale and incompletion or heterogeneity of the samples maybe lead to different results of these studies.
In this study, 50 related GWAS studies (including the 6 missing and 18 novel studies) were selected afresh from seven authoritative sources, and the association level between rs11136000 and risk of AD in Caucasian, Asian and Chinese ethnicity was re-evaluated. We also found a significant association between rs11136000 polymorphism and the decreased risk of AD in Caucasian ethnicity using the dominant (OR = 0.829, 95% CI = 0.796–0.864, P < 0.0001), allele (OR = 0.864, 95% CI = 0.842−0.888, P < 0.0001) and recessive model (OR = 0.819, 95% CI = 0.774−0.867, P < 0.0001). Different from the results of the previous studies, however, rs11136000 polymorphism was found not associated with the risk of AD in Asian ethnicity using the dominant (OR = 0.922, 95% CI = 0.846–1.005, P = 0.0649) and recessive model (OR = 0.747, 95% CI = 0.511−1.092, P = 0.1326), as well as in Chinese population using the dominant (OR = 0.988, 95% CI = 0.887−1.101, P = 0.8270), allele (OR = 0.939, 95% CI = 0.878–1.004, P = 0.0654) and recessive model (OR = 0.615, 95% CI = 0.355−1.068, P = 0.0841).
As far as we know, our meta-analysis about the association of the CLU rs11136000 polymorphism with the risk of AD is by far the largest scale study. The results reveal a significant association between them in Caucasian ethnicity but not Chinese ethnicity, which is consistent with the findings of most of the corresponding GWAS studies. In summary, we believe that these findings can help to improve the understanding of the AD’s pathogenesis.
References
Burns, A. & Iliffe, S. Alzheimer’s disease. Brit Med J 338 (2009).
Querfurth, H. W. & LaFerla, F. M. Mechanisms Of Disease Alzheimer’s Disease. New Engl J Med 362, 329–344 (2010).
Hardy, J. & Selkoe, D. J. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297, 353–356 (2002).
Ittner, L. M. & Gotz, J. Amyloid-beta and tau–a toxic pas de deux in Alzheimer’s disease. Nature reviews. Neuroscience 12, 65–72 (2011).
Vaudano, E. et al. Boosting translational research on Alzheimer’s disease in Europe: The Innovative Medicine Initiative AD research platform. Alzheimer’s & dementia: the journal of the Alzheimer’s Association 11, 1121–1122 (2015).
Lambert, M. P. et al. Diffusible, nonfibrillar ligands derived from Abeta1-42 are potent central nervous system neurotoxins. Proceedings of the National Academy of Sciences of the United States of America 95, 6448–6453 (1998).
DeMattos, R. B. et al. Clusterin promotes amyloid plaque formation and is critical for neuritic toxicity in a mouse model of Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America 99, 10843–10848 (2002).
Desikan, R. S. et al. The role of clusterin in amyloid-beta-associated neurodegeneration. JAMA neurology 71, 180–187 (2014).
Allen, M. et al. Genetic association of variants with late-onset Alzheimer’s disease risk and brain gene expression. Alzheimer’s & Dementia: The Journal of the Alzheimer’s Association 8, P451 (2012).
Yu, J. T. & Tan, L. The role of clusterin in Alzheimer’s disease: pathways, pathogenesis, and therapy. Molecular neurobiology 45, 314–326 (2012).
Harold, D. et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nature genetics 41, 1088–1093 (2009).
Lambert, J. C. et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nature genetics 41, 1094–1099 (2009).
Carrasquillo, M. M. et al. Replication of CLU, CR1, and PICALM associations with alzheimer disease. Archives of neurology 67, 961–964 (2010).
Seshadri, S. et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. Jama 303, 1832–1840 (2010).
Ferrari, R. et al. Implication of common and disease specific variants in CLU, CR1, and PICALM. Neurobiology of aging 33(1846), e1847–1818 (2012).
Kamboh, M. I. et al. Association of CLU and PICALM variants with Alzheimer’s disease. Neurobiology of aging 33, 518–521 (2012).
Lin, Y. L. et al. Genetic polymorphisms of clusterin gene are associated with a decreased risk of Alzheimer’s disease. European journal of epidemiology (2012).
Carrasquillo, M. M. et al. Late-onset Alzheimer disease genetic variants in posterior cortical atrophy and posterior AD. Neurology 82, 1455–1462 (2014).
Giedraitis, V. et al. Genetic analysis of Alzheimer’s disease in the Uppsala Longitudinal Study of Adult Men. Dementia and geriatric cognitive disorders 27, 59–68 (2009).
Golenkina, S. A. et al. Analysis of clusterin gene (CLU/APOJ) polymorphism in Alzheimer’s disease patients and in normal cohorts from Russian populations. Molekuliarnaia biologiia 44, 620–626 (2010).
Yu, J. T. et al. Implication of CLU gene polymorphisms in Chinese patients with Alzheimer’s disease. Clinica chimica acta; international journal of clinical chemistry 411, 1516–1519 (2010).
Gu, H. et al. Association of clusterin gene polymorphisms with late-onset Alzheimer’s disease. Dementia and geriatric cognitive disorders 32, 198–201 (2011).
Ma, J. F. et al. Association study of clusterin polymorphism rs11136000 with late onset Alzheimer’s disease in Chinese Han population. American journal of Alzheimer’s disease and other dementias 26, 627–630 (2011).
Bettens, K. et al. Both common variations and rare non-synonymous substitutions and small insertion/deletions in CLU are associated with increased Alzheimer risk. Molecular neurodegeneration 7, 3 (2012).
Chen, L. H. et al. Polymorphisms of CR1, CLU and PICALM confer susceptibility of Alzheimer’s disease in a southern Chinese population. Neurobiology of aging 33(210), e211–217 (2012).
Ohara, T. et al. Association study of susceptibility genes for late-onset Alzheimer’s disease in the Japanese population. Psychiatric genetics 22, 290–293 (2012).
Chung, S. J. et al. Association of GWAS top hits with late-onset Alzheimer disease in Korean population. Alzheimer disease and associated disorders 27, 250–257 (2013).
Lu, S. J. et al. Clusterin variants are not associated with southern Chinese patients with Alzheimer’s disease. Neurobiology of aging 35, 2656 e2659–2656 e2611 (2014).
Liu, G. et al. The CLU gene rs11136000 variant is significantly associated with Alzheimer’s disease in Caucasian and Asian populations. Neuromolecular medicine 16, 52–60 (2014).
Du, W., Tan, J., Xu, W., Chen, J. & Wang, L. Association between clusterin gene polymorphism rs11136000 and late-onset Alzheimer’s disease susceptibility: A review and meta-analysis of case-control studies. Experimental and therapeutic medicine 12, 2915–2927 (2016).
Moreno, D. J. et al. Association of GWAS Top Genes With Late-Onset Alzheimer’s Disease in Colombian Population. American journal of Alzheimer’s disease and other dementias 32, 27–35 (2017).
Santos-Reboucas, C. B. et al. rs3851179 Polymorphism at 5′ to the PICALM Gene is Associated with Alzheimer and Parkinson Diseases in Brazilian Population. Neuromolecular medicine 19, 293–299 (2017).
Seripa, D. et al. Role of CLU, PICALM, and TNK1 Genotypes in Aging With and Without Alzheimer’s Disease. Molecular neurobiology (2017).
Shankarappa, B. M. et al. Effect of CLU and PICALM polymorphisms on AD risk: A study from south India. Asian journal of psychiatry 27, 7–11 (2017).
Jia, J. et al. Four Novel Susceptibility Variants Identified In Chinese are Associated With Sporadic Alzheimer’s Disease. Alzheimers & Dementia 13, P189–P191 (2017).
Alaylioglu, M. et al. The Association Between Clusterin and APOE Polymorphisms and Late-Onset Alzheimer Disease in a Turkish Cohort. Journal of geriatric psychiatry and neurology 29, 221–226 (2016).
Huang, F. et al. Lower Prevalence of Alzheimer’s Disease among Tibetans: Association with Religious and Genetic Factors. Journal of Alzheimer’s disease: JAD 50, 659–667 (2016).
Montanola, A. et al. ApoA1, ApoJ and ApoE Plasma Levels and Genotype Frequencies in Cerebral Amyloid Angiopathy. Neuromolecular medicine 18, 99–108 (2016).
Rezazadeh, M. et al. Genetic Factors Affecting Late-Onset Alzheimer’s Disease Susceptibility. Neuromolecular medicine 18, 37–49 (2016).
Wang, H. Z. et al. Validating GWAS-Identified Risk Loci for Alzheimer’s Disease in Han Chinese Populations. Molecular neurobiology 53, 379–390 (2016).
Luo, H. Y. et al. Association study of clusterin polymorphism rs11136000 with late-onset Alzheimer’s disease in Bai population. Chinese Journal of Behavioral Medicine and Brain Science 25, 240–243 (2016).
Belcavello, L. et al. Association of MTHFR and PICALM polymorphisms with Alzheimer’s disease. Molecular biology reports 42, 611–616 (2015).
Ferrari, R. et al. Screening of Early and Late Onset Alzheimer’s Disease Genetic Risk Factors in a Cohort of Dementia Patients from Liguria, Italy. Current Alzheimer research 12, 802–812 (2015).
Jiao, B. et al. Polygenic Analysis of Late-Onset Alzheimer’s Disease from Mainland China. PloS one 10, e0144898 (2015).
Sen, A. et al. Lack Of Associations Between Clu And Picalm Gene Polymorphisms And Alzheimer’s Disease In a Turkish Population. Ideggyogyaszati szemle 68, 113–120 (2015).
Sleegers, K. et al. A 22-single nucleotide polymorphism Alzheimer’s disease risk score correlates with family history, onset age, and cerebrospinal fluid Abeta42. Alzheimer’s & dementia: the journal of the Alzheimer’s Association 11, 1452–1460 (2015).
Xiao, Q. et al. Risk prediction for sporadic Alzheimer’s disease using genetic risk score in the Han Chinese population. Oncotarget 6, 36955–36964 (2015).
Corneveaux, J. J. et al. Association of CR1, CLU and PICALM with Alzheimer’s disease in a cohort of clinically characterized and neuropathologically verified individuals. Human molecular genetics 19, 3295–3301 (2010).
Mullan, G. M. et al. Plasma clusterin levels and the rs11136000 genotype in individuals with mild cognitive impairment and Alzheimer’s disease. Current Alzheimer research 10, 973–978 (2013).
Nizamutdinov, I. I. et al. Biochip for determination of genetic markers of sporadic Alzheimer’s disease in the Russian Slavic population. Molekuliarnaia biologiia 47, 949–958 (2013).
Pedraza, O. et al. Evaluation of memory endophenotypes for association with CLU, CR1, and PICALM variants in black and white subjects. Alzheimer’s & dementia: the journal of the Alzheimer’s Association 10, 205–213 (2014).
Roussotte, F. F. et al. Combined effects of Alzheimer risk variants in the CLU and ApoE genes on ventricular expansion patterns in the elderly. The Journal of neuroscience: the official journal of the Society for Neuroscience 34, 6537–6545 (2014).
Han, Z. et al. Analyzing large-scale samples confirms the association between the rs1051730 polymorphism and lung cancer susceptibility. Scientific reports 5, 15642 (2015).
Wang, Y. et al. Association between LRP1 C766T polymorphism and Alzheimer’s disease susceptibility: a meta-analysis. Scientific reports 7, 8435 (2017).
Zheng, J. Y. et al. Associations of rs3740677 within GAB2 Gene with LOAD in Chinese Han Population. Molecular neurobiology 54, 4015–4020 (2017).
Liu, G. et al. PICALM rs3851179 Variant Confers Susceptibility to Alzheimer’s Disease in Chinese Population. Mol Neurobiol 54, 3131–3136 (2017).
Liu, G. et al. Alzheimer’s Disease rs11767557 Variant Regulates EPHA1 Gene Expression Specifically in Human Whole Blood. J Alzheimers Dis 61, 1077–1088 (2018).
Cheng, L., Hu, Y., Sun, J., Zhou, M. & Jiang, Q. DincRNA: a comprehensive web-based bioinformatics toolkit for exploring disease associations and ncRNA function. Bioinformatics (2018).
Liu, G. Y. et al. Genetic Variants and Multiple Sclerosis Risk Gene SLC9A9 Expression in Distinct Human Brain Regions. Molecular Neurobiology 54, 6820–6826 (2017).
Liu, G. et al. Alzheimer’s disease rs11767557 variant regulates EPHA1 gene expression specifically in human whole blood. Journal of Alzheimer’s Disease 61 (2017).
Liu, G. et al. Integrating genome-wide association studies and gene expression data highlights dysregulated multiple sclerosis risk pathways. Mult Scler 23, 205–212 (2017).
Liu, G. et al. Multiple sclerosis risk pathways differ in Caucasian and Chinese populations. J Neuroimmunol 307, 63–68 (2017).
Jiang, Q. H. et al. Alzheimer’s Disease Variants with the Genome-Wide Significance are Significantly Enriched in Immune Pathways and Active in Immune Cells. Molecular Neurobiology 54, 594–600 (2017).
Bertram, L., McQueen, M. B., Mullin, K., Blacker, D. & Tanzi, R. E. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nature genetics 39, 17–23 (2007).
Anderson, L. N., Cotterchio, M., Cole, D. E. & Knight, J. A. Vitamin D-related genetic variants, interactions with vitamin D exposure, and breast cancer risk among Caucasian women in Ontario. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 20, 1708–1717 (2011).
Demets, D. L. Methods for combining randomized clinical trials: strengths and limitations. Statistics in medicine 6, 341–350 (1987).
Begg, C. B. & Mazumdar, M. Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101 (1994).
Egger, M., Davey Smith, G., Schneider, M. & Minder, C. Bias in meta-analysis detected by a simple, graphical test. Bmj 315, 629–634 (1997).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31360031); the Major Project of Guizhou Province (Qian Ke He Major Project [2016] 3022-07); the Youth Science and Technology Talent Project of Guizhou Province [2017]5617.
Author information
Authors and Affiliations
Contributions
Z.H. and X.Z. designed research, Z.H. J.Q., J.Z. and X.Z. selected data, Z.H. performed research, analyzed data, and wrote the paper. All authors discussed the results, and contributed to the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Han, Z., Qu, J., Zhao, J. et al. Analyzing 74,248 Samples Confirms the Association Between CLU rs11136000 Polymorphism and Alzheimer’s Disease in Caucasian But Not Chinese population. Sci Rep 8, 11062 (2018). https://doi.org/10.1038/s41598-018-29450-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-29450-2
- Springer Nature Limited