Abstract
The role of cytochrome P450 drug metabolizing enzymes in the efficacy of tamoxifen treatment of breast cancer is subject to substantial interest and controversy. CYP2D6 have been intensively studied, but the role of CYP2C19 is less elucidated, and we studied the association of CYPC19 genotype and recurrence of breast cancer. We used outcome and genotyping data from the large publicly available International Tamoxifen Pharmacogenomics Consortium (ITPC) dataset. Cox regression was used to compute the hazard ratios (HRs) for recurrence. CYP2C19 genotype data was available for 2 423 patients and the final sample cohort comprised 2 102 patients. CYP2C19*2 or *19 alleles did not influence DFS. For the CYP2C19*2 allele, the HR was 1.05 (CI 0.78–1.42) and 0.79 (CI 0.32–1.94) for hetero- and homozygote carriers, respectively. The corresponding HR for hetero- and homozygote carriers of the CYP2C19*17 allele were 1.02 (CI 0.71–1.46) and 0.57 (CI 0.26–1.24), respectively. Accounting for CYP2D6 genotype status did not change these estimates. We found no evidence to support a clinically meaningful role of CYP2C19 polymorphisms and response to tamoxifen in breast cancer patients and, consequently, CYP2C19 genotype status should not be included in clinical decisions on tamoxifen treatment.
Similar content being viewed by others
Introduction
Breast cancer is, excluding skin cancers, the most common malignancy among women in the United States and caused about 571.000 deaths world-wide in 20151, 2. Tamoxifen is the standard treatment for premenopausal women with estrogen receptor (ER) positive breast cancer, and five years of adjuvant tamoxifen therapy reduces recurrences by nearly 50%3,4,5. In tumor cells, tamoxifen and its metabolites impede the binding of estrogen to the ER to inhibit expression of estrogen-responsive genes, thereby preventing tumor cell growth and angiogenesis6, 7. Patient responses to tamoxifen vary, and around 20–30% of patients receiving tamoxifen therapy in accordance with guidelines still suffer a breast cancer recurrence8.
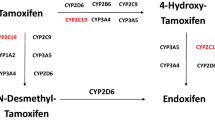
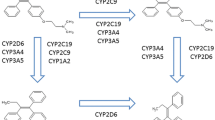
The complex tamoxifen metabolism (Fig. 1) is primarily catalyzed by cytochrome P450 (CYP) enzymes, which are subject to substantial differences in inter-individual expression and activity9,10,11,12. Endoxifen, the 4-hydroxy-N-desmethyl metabolite of tamoxifen, is central to mechanism of action and efficacy of tamoxifen, and concentrations thereof varies substantially between patients6, 13, 14. CYP2D6 is the principal enzyme catalyzing the conversion of tamoxifen to endoxifen, and the association of genomic variants in the CYP2D6 gene to outcome of tamoxifen treatment has been extensively studied15, 16. Two of the largest datasets reported a null-association, but contradictory findings have led to ongoing controversy over the value of using CYP2D6 genotyping to guide the prescription of tamoxifen17,18,19,20,21,22.
CYP2C19 catalyzes the formation of a proportion of tamoxifen metabolites, including the conversion of 4-OH-TAM to endoxifen (Fig. 1)23. The CYP2C19 gene is highly polymorphic. Loss of enzyme activity results from the CYP2C19*2 (681 G > A, rs4244285)24, 25 and CYP2C19*3 (636 G > A, rs4986893) alleles26, 27. The *2 allele is found in approximately 23–39% of Asians, 10–20% of Caucasians, and 15% of Africans23, 28. The *3 allele occurs in 5–10% of Asians (17). CYP2C19*17 (−806C > T, rs12248560 or −3402C > T, rs11188072) has been implicated in enhanced gene transcription26, 29, 30. The *17 allele is found in about 4% of Asians and 18–24% of Caucasians and Africans23, 28, 31, 32.
Studies differ with respect to the associations observed between CYP2C19 genotypes and clinical outcomes as well as to corresponding levels of tamoxifen and its metabolites33,34,35,36,37,38. Counterintuitively, the presence of a *2 allele has been associated with longer relapse-free time or better survival in tamoxifen-treated women in some39,40,41, but not all studies32, 42,43,44,45,46. In some studies the CYP2C19*17 allele is associated with more favorable outcomes in breast cancer patients treated with tamoxifen32, though null results have also been found40, 42, 43, 47. Contradictory results were obtained in the context of tamoxifen monotherapy in advanced breast cancer, where an association between the *17 allele and shorter time to treatment failure has been reported39, 41.
Given these contradictory findings, we used a large, publicly available dataset to investigate the association of CYP2C19*2 and CYP2C19*17 variants with breast cancer recurrence in both pre- and postmenopausal women treated with adjuvant tamoxifen therapy for ER-positive breast cancer.
Subjects and Methods
Data source and study population
The ITPC comprises research from 12 sites representing 9 countries, all designed to prospectively assess the contribution of genetic variation in tamoxifen metabolism and transport pathways to breast cancer recurrence risk. We required that patients had been prescribed 20 mg/day tamoxifen for an intended duration of 2 or 5 years, had not previously received systemic therapy for breast cancer prevention, had no known history of invasive or in situ breast cancer, had not used any other adjuvant therapy before tamoxifen, and initiated tamoxifen therapy within 182 days of breast cancer surgery. We included patients with non-metastatic, ER-positive tumors who had data on at least one CYP2C19 variant, whether a recurrence occurred, and follow-up time (Fig. 2).
Analytic variables
Disease-free survival time (DFS) was the number of months from diagnosis until breast cancer recurrence, defined as an ipsilateral local or regional recurrence (invasive or non-invasive), a distant recurrence, or a contralateral breast cancer (invasive or non-invasive). Patients who did not experience a recurrence were censored on the date of death from another cause or on the day of last disease-free evaluation.
Genotype exposures were CYP2C19*2 and CYP2C19*17, with reference to the wild-type CYP2C19*1. Various methods of genotyping were used in the seven studies comprising the data, with the majority of genotypes (60.5%) ascertained by the AmpliChip test platform (Roche Molecular Diagnostics, California, USA). In three instances where multiple methods were used for a single individual and the AmpliChip blood genotype did not match the CYP2C19*2 genotype obtained with another method, preference was given to the AmpliChip data due to the high sensitivity and specificity of this test49. The CYP2C19*3 allele was not assessed in this study because no variants were detected in the included data.
Covariables
Potential covariates of interest were: age, ethnicity, menopausal status, tumor grade and stage, progesterone receptor (PR) status, use of other adjuvant therapies (radiation and chemotherapy), and CYP2D6 metabolizer phenotype. Age (as a continuous variable), menopausal status (pre-, post-, or peri-), PR status, use of other adjuvant therapies, and Nottingham tumor grades were recorded directly in the ITPC dataset. Perimenopausal women (n = 57) were combined with post-menopausal women for all analyses. Categories as defined by the Office of Management and Budget50 were used to divide patients into three ethnic groups: Caucasian, Asian or Pacific Islander, and any other ethnicity (which included African-Americans, mixed ethnicity individuals, and individuals of any other ethnicity).
Tumor stage was derived from information on both tumor diameter and the number of positive lymph nodes. Missing information on in situ tumors and distant metastases prohibited use of the TNM staging system; however, the primary tumor and pathologic guidelines of the TNM system were used to classify tumors into five stages51.
A variable encoding individuals’ CYP2D6 metabolizer phenotype (ultra- UM, extensive- EM, intermediate- IM, or poor- PM) was available in the ITPC data, and accounted for both genetic factors and the use of CYP2D6-inhibiting drugs. We generated a variable designating overall tamoxifen metabolic activity (high, intermediate or low) by combining CYP2D6 phenotypes and CYP2C19 genotypes according to Schroth (Table 1)32.
Statistical analyses
Descriptive analyses including all covariates of interest were computed for all women analyzed. Cox regression was used to compute the hazard ratios (HRs) for recurrence and associated 95% confidence intervals (CIs). The tumor grade variable violated the proportional hazards assumption when assessed using log-log survival curves and were therefore excluded from all models. Models containing all possible variable subsets were analyzed using the change-in-estimate approach, with confounding indicated in models where the variable subset removed led to a hazard ratio changed by greater than 10% compared with the hazard ratio for the full model52. Final Cox proportional hazards models included age at diagnosis of primary breast cancer (as a continuous variable), tumor stage, and ethnicity (Caucasian or Asian, for CYP2C19*2 only) as covariates. Supplemental analyses stratified by CYP2D6 phenotype and menopausal status were also performed.
For multivariable analyses, individuals with missing values for any modeled variable were excluded. To assess the potential for bias due to the use of complete case analyses, imputation of missing values for CYP2C19*2 genotype, CYP2C19*17 genotype, ethnicity, age at breast cancer diagnosis, and tumor stage was done in a supplemental analysis.
All analyses were carried out in SAS version 9.4 (Cary, NC).
Data availability statement
Data were obtained from the International Tamoxifen Pharmacogenomics Consortium (ITPC) which are publicly available48.
Results
Study population
The seven sites containing eligible patients provided 2 102 women for analysis (Fig. 2). Of these, 296 women experienced a breast cancer recurrence. One woman who did not have a recurrence and was missing data on the last disease-free evaluation was censored on the date she was last known to be alive. Patient characteristics for the sample and source population by study site are presented in Table 2 and Supplementary Table S1, respectively. Characteristics of the sample and source data stratified by recurrence are presented in Supplementary Table S2. The median DFS was 61 months for all women, 45 months for women experiencing a recurrence, and 63 months for women with no recurrence.
Genotypes
Data on CYP2C19*2 were available from all seven study sites for 2 055 women, and data on CYP2C19*17 were reported from three sites for 1 253 women. Distributions and Hardy-Weinberg chi-squared statistics within each study site for the CYP2C19*2 and CYP2C19*17 genotypes and DNA sources are provided for sample and source populations in Supplementary Tables S3 and S4. Both variants were in Hardy-Weinberg equilibrium for each study site, except for CYP2C19*17 at site 8 (p = 0.02) and CYP2D6*2 at site 12 (p = 0.005)
CYP2C19 genotypes and DFS
For the CYP2C19*2 allele, adjusted hazard ratios for associations between variant heterozygotes and homozygotes with DFS were 1.05 (95% CI: 0.78, 1.42) and 0.79 (95% CI: 0.32, 1.94), respectively (Table 3). For the CYP2C19*17 allele, adjusted hazard ratios for associations between variant heterozygotes and homozygotes with DFS were 1.02 (95% CI: 0.71, 1.46) and 0.57 (0.26, 1.24), respectively (Table 3). Stratification by menopausal status and CYP2D6 phenotype did not yield any notable associations between CYP2C19 genotype and DFS (Supplementary Table S5). Results based on imputed data sets were not substantially different from the complete case analysis, but in general tended to be closer to the null (Supplementary Table S6).
CYP2D6 phenotype/CYP2C19 genotype combinations
Results are provided in Table 4. For the CYP2D6 phenotype/CYP2C19*2 genotype combinations, multivariate DFS hazard ratios for the phenotypically designated “high” and “intermediate” tamoxifen metabolic activity groups were 0.86 (95% CI: 0.45–1.66) and 0.89 (95% CI: 0.46–1.74), respectively, compared with the “low” metabolic activity group. The corresponding adjusted DFS hazard ratios for the CYP2D6 phenotype/CYP2C19*17 genotype combinations were 1.19 (95% CI: 0.73–1.94) and 1.21 (95% CI: 0.80–1.85).
Discussion
We found no evidence of a clinically meaningful association between CYP2C19*2 or CYP2C19*17 genotypes and DFS in tamoxifen-treated breast cancer patients in a large dataset. A secondary analysis of CYP2C19 genotype accounting for CYP2D6 phenotypes resulted in little change to the observation.
This study has the largest overall sample size of work on this topic to date and includes a larger number of CYP2C19 variants compared with prior studies. Even so, our estimates come with confidence intervals that suggest some limitation with respect to sample-size This study also benefits from the inclusion of a substantial number of premenopausal patients, permitting stratification of the association by menopausal status. Only two other studies have examined the association between CYP2C19 genotype and breast cancer recurrence within strata of menopausal status42, 44, and those studies included a combined total of only 85 premenopausal patients. The inclusion of a large premenopausal cohort is especially relevant as tamoxifen is the guideline endocrine therapy for these women5.
Combining CYP2D6 and CYP2C19*2 or CYP2C19*17 into singular phenotypes (as suggested by Schroth et al.32) did not suggest that this is of clinical relevance (Table 4). While this finding should be interpreted with caution, as confidence intervals are somewhat wide, this analysis indirectly lends further weight against the heavily discussed clinically meaningful role of CYP2D6 itself. Our results differ somewhat from those reported by Schroth, who reported a statistically significant inference of the CYP2C19*17 allele on event-free survival32. Our sample size is several orders of magnitude larger though, which we believe explains this apparent discrepancy.
At ITPC sites not testing for the CYP2C19*17 allele, misclassification of tamoxifen metabolic phenotype could have occurred, but a stratified sensitivity analysis restricted to sites testing for CYP2C19*17 did not provide substantially different results. CYP2C19*3 allele misclassification is unlikely to influence the overall result as this allele would only be expected to be common at site 1223. Allele distribution was reasonably consistent across study sites and compared well to reported literature frequencies. This suggest that errors from genotyping are less likely to present a main issue within our dataset. The CYP2C19*2 allele was assessed across 10 study sites. The allele frequencies were reasonably comparable, 23–31% for heterozygosity, bar two sites (project site #6 and #12) which yielded frequencies of 18 and 35% for *2 heterozygosity, respectively. These frequencies are within reason of the expected given the variability related to sample size and ethnicity composition of the respective populations. Site 12 had a relative low sample size and site 6 a high degree of missing values (32%). The latter diluted the frequency as, among those tested, 26% were heterozygous for the *2 allele. The CYP2C19*17 was only assessed at three sites that yielded homogenous and comparable allele frequencies with one gain of function allele frequencies between 32–37%. Allele distributions per ethnicity (Supplementary Table S5) compared well to reported literature frequencies, though we could not meaningfully compare the *17 allele frequency in Asian subjects to literature data as very few of Asian origin were tested in our sample. The lack of data on CYP2C19 inhibitor use could have biased our estimates towards the null. CYP2C19 genotyping using tumor-derived DNA (at three sites) may introduce misclassification due to potential loss-of-heterozygosity in tumor cells53, 54.Results of chi-squared tests for Hardy-Weinberg equilibrium indicate that loss-of-heterozygosity had a minor impact on observed CYP2C19 genotypes in this study. A minor violation of HWE was observed at study site 8, which accounted for the majority of samples assessed for CYP2C19*17. This minor violation represents a weakness even if misclassification due to loss-of-heterozygosity appears less likely to result in significant bias of overall study estimates20. A violation of HWE for CYP2C19*2 was observed at study site 12, but the sample of 240 subjects contributed little to the overall analysis.
While previous reports have found the presence of CYP2C19*2 to be associated with superior efficacy of tamoxifen treatment39,40,41, our results support other studies reporting no such association32, 42,43,44,45,46. The hazard ratio for the association of CYP2C19*17 homozygotes with a favorable DFS (HR = 0.57, 95% CI: 0.26, 1.24) is similar to the ratio found previously for the association of carrying CYP2C19*17 with relapse-free time (HR = 0.45, 95% CI: 0.21, 0.92) (31). About 40% of the patient population and the majority of *17 allele data in the ITPC dataset were from the latter study, so our study should not be viewed as independent evidence. Our findings for the *17 allele are consistent with results from a smaller, similar study, which reported a hazard ratio of 0.93 (95% CI = 0.64, 1.37) and found a near-null association among those with impaired CYP2D647. Despite the biologic plausibility of CYP2C19 playing an important role in patients with reduced CYP2D6 function, our stratified analyses do not support this hypothesis. The complex metabolism of tamoxifen, which include catalytic activity of CYP2C19, CYP1A2, CYP3A4/5, CYP2D6, CYP2B6 and CYP2C9, may explain the null-association found in this study. The formation of active tamoxifen metabolites in patients carrying reduced or increased CYP2C19 function alleles may be sufficiently compensated through parallel and serial metabolic pathways catalyzed by other P450 enzymes. This would mitigate the net overall clinical consequence of genomic CYP2C19 variants and result in a statistical inference toward the null.
A key limitation is that the ITPC dataset does not allow for differentiation between predictive and prognostic markers, since studies did not include women diagnosed with ER-negative tumors who were not treated with tamoxifen. Several studies indicate that CYP2C19 variants are associated with differences in baseline breast cancer risk, likely due to the inherent role of CYP2C19 in the metabolism of estrogen. However, this association has not been consistently observed, and the fact that the minor allele frequencies observed here match population-wide benchmarks argues against CYP2C19 genotype as a selection force. On the other hand, breast cancer etiology or survival is usually only relevant after childbirth in most women, which would render selection pressure less relevant.
Province et al. analyzed the ITPC dataset and reported poorer disease-free survival among CYP2D6 poor metabolizers and a weak association between poor metabolizer status and a shorter breast cancer-free interval15. These associations were not robust to variations in inclusion criteria, and this study has been heavily criticized for its reliance on statistical interpretations of ad hoc subset analyses and this issue remains highly controversial55,56,57,58. In light of these criticisms, the criteria for inclusion in our study were defined a priori. Province et al. also described the heterogeneity of results between the study sites, which is an additional challenge in interpreting the results of our study15.
In conclusion, we found no evidence to support a clinically meaningful role of CYP2C19 polymorphisms and response to tamoxifen in breast cancer patients. Given the complexity of tamoxifen pharmacodynamics and metabolism and the divergent results on the importance of genomic variants, it appears unlikely that a clinically useful simple predictive set of genomic variables will be identified.
References
American Cancer Society. Breast cancer at a glance, https://cancerstatisticscenter.cancer.org/#/cancer-site/Breast (Accessed 22/06/2017) (2017).
WHO Media Centre: Cancer fact sheet. http://www.who.int/mediacentre/factsheets/fs297/en/ (Accessed 22/06/2017) (2017).
Early Breast Cancer Trialists’ Collaborative Group. Tamoxifen for early breast cancer: an overview of the randomised trials. Lancet 351, 1451–67 (1998).
Early Breast Cancer Trialists’ Collaborative, G.. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365, 1687–717 (2005).
Burstein, H. J. et al. Adjuvant endocrine therapy for women with hormone receptor-positive breast cancer: american society of clinical oncology clinical practice guideline focused update. J Clin Oncol 32, 2255–69 (2014).
Johnson, M. D. et al. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat 85, 151–9 (2004).
Osborne, C. K. Tamoxifen in the treatment of breast cancer. N Engl J Med 339, 1609–18 (1998).
Hackshaw, A. et al. Long-term benefits of 5 years of tamoxifen: 10-year follow-up of a large randomized trial in women at least 50 years of age with early breast cancer. J Clin Oncol 29, 1657–63 (2011).
Boocock, D. J. et al. Identification of human CYP forms involved in the activation of tamoxifen and irreversible binding to DNA. Carcinogenesis 23, 1897–901 (2002).
Coller, J. K. et al. The influence of CYP2B6, CYP2C9 and CYP2D6 genotypes on the formation of the potent antioestrogen Z-4-hydroxy-tamoxifen in human liver. Br J Clin Pharmacol 54, 157–67 (2002).
Crewe, H. K., Ellis, S. W., Lennard, M. S. & Tucker, G. T. Variable contribution of cytochromes P450 2D6, 2C9 and 3A4 to the 4-hydroxylation of tamoxifen by human liver microsomes. Biochem Pharmacol 53, 171–8 (1997).
Crewe, H. K., Notley, L. M., Wunsch, R. M., Lennard, M. S. & Gillam, E. M. Metabolism of tamoxifen by recombinant human cytochrome P450 enzymes: formation of the 4-hydroxy, 4’-hydroxy and N-desmethyl metabolites and isomerization of trans-4-hydroxytamoxifen. Drug Metab Dispos 30, 869–74 (2002).
Furr, B. J. & Jordan, V. C. The pharmacology and clinical uses of tamoxifen. Pharmacol Ther 25, 127–205 (1984).
Stearns, V. et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst 95, 1758–64 (2003).
Province, M. A. et al. CYP2D6 genotype and adjuvant tamoxifen: meta-analysis of heterogeneous study populations. Clin Pharmacol Ther 95, 216–27 (2014).
Cronin-Fenton, D. P., Damkier, P. & Lash, T. L. Metabolism and transport of tamoxifen in relation to its effectiveness: new perspectives on an ongoing controversy. Future Oncol 10, 107–122 (2014).
Brauch, H. et al. Tamoxifen use in postmenopausal breast cancer: CYP2D6 matters. J Clin Oncol 31, 176–80 (2013).
Hertz, D. L., McLeod, H. L. & Irvin, J. W. J. Tamoxifen and CYP2D6: a contradiction of data. Oncologist 17, 620–30 (2012).
Lash, T. L., Lien, E. A., Sorensen, H. T. & Hamilton-Dutoit, S. Genotype-guided tamoxifen therapy: time to pause for reflection? Lancet Oncol 10, 825–33 (2009).
Ahern, T. P. et al. Cytochrome P-450 2D6 (CYP2D6) Genotype and Breast Cancer Recurrence in Tamoxifen-Treated Patients: Evaluating the Importance of Loss of Heterozygosity. Am. J. Epidemiol., doi:10.1093/aje/kww178 (2016).
Regan, M. M. et al. CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the breast international group 1-98 trial. J Natl Cancer Inst 104, 441–51 (2012).
Rae, J. M. et al. CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J Natl Cancer Inst 104, 452–60 (2012).
Kiyotani, K., Mushiroda, T., Nakamura, Y. & Zembutsu, H. Pharmacogenomics of tamoxifen: roles of drug metabolizing enzymes and transporters. Drug Metab Pharmacokinet 27, 122–31 (2012).
Scott, S. A. et al. PharmGKB summary: very important pharmacogene information for cytochrome P450, family 2, subfamily C, polypeptide 19. Pharmacogenet Genomics 22, 159–65 (2012).
de Morais, S. M. et al. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem 269, 15419–22 (1994).
CYP2C19 allele nomenclature. http://www.cypalleles.ki.se/cyp2c19.htm (2017)
De Morais, S. M. et al. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol 46, 594–8 (1994).
Scott, S. A. et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin Pharmacol Ther 90, 328–32 (2011).
Sim, S. C. et al. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther 79, 103–13 (2006).
Powers, J. L. et al. Multi-gene and Drug Interaction Approach for Tamoxifen Metabolite Patterns Reveals Possible Involvement of CYP2C9, CYP2C19 and ABCB1. J Clin Pharmacol. doi:10.1002/jcph.771 (2016).
Li-Wan-Po, A., Girard, T., Farndon, P., Cooley, C. & Lithgow, J. Pharmacogenetics of CYP2C19: functional and clinical implications of a new variant CYP2C19*17. Br J Clin Pharmacol 69, 222–230 (2010).
Schroth, W. et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol 25, 5187–93 (2007).
Desta, Z., Ward, B. A., Soukhova, N. V. & Flockhart, D. A. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther 310, 1062–75 (2004).
Gjerde, J. et al. Associations between tamoxifen, estrogens, and FSH serum levels during steady state tamoxifen treatment of postmenopausal women with breast cancer. BMC Cancer 10, 313 (2010).
Lim, J. S. L. et al. Impact of CYP2D6, CYP3A5, CYP2C9 and CYP2C19 polymorphisms on tamoxifen pharmacokinetics in Asian breast cancer patients. Br J Clin Pharmacol 71, 737–750 (2011).
Murdter, T. E. et al. Activity levels of tamoxifen metabolites at the estrogen receptor and the impact of genetic polymorphisms of phase I and II enzymes on their concentration levels in plasma. Clin Pharmacol Ther 89, 708–17 (2011).
Lim, J. S. L. et al. Association of CYP2C19*2 and associated haplotypes with lower norendoxifen concentrations in tamoxifen-treated Asian breast cancer patients. Br J Clin Pharmacol 81, 1142–1152 (2016).
Binkhorst, L., Mathijssen, R. H., Jager, A. & van Gelder, T. Individualization of tamoxifen therapy: Much more than just CYP2D6 genotyping. Cancer Treat Rev. doi:10.1016/j.ctrv.2015.01.002 (2015).
Ruiter, R. et al. CYP2C19*2 polymorphism is associated with increased survival in breast cancer patients using tamoxifen. Pharmacogenomics 11, 1367–1375 (2010).
Beelen, K. et al. CYP2C19 2 predicts substantial tamoxifen benefit in postmenopausal breast cancer patients randomized between adjuvant tamoxifen and no systemic treatment. Breast Cancer Res. Treat. 139, 649–655 (2013).
van Schaik, R. H. N. et al. The CYP2C19*2 genotype predicts tamoxifen treatment outcome in advanced breast cancer patients. Pharmacogenomics 12, 1137–1146 (2011).
Mwinyi, J. et al. Impact of variable CYP genotypes on breast cancer relapse in patients undergoing adjuvant tamoxifen therapy. Cancer Chemother. Pharmacol. 73, 1181–1188 (2014).
Serrano, D. et al. Efficacy of tamoxifen based on cytochrome P450 2D6, CYP2C19 and SULT1A1 genotype in the Italian Tamoxifen Prevention Trial. Pharmacogenomics J. 11, 100–107 (2011).
Chamnanphon, M. et al. Association of CYP2D6 and CYP2C19 polymorphisms and disease-free survival of Thai post-menopausal breast cancer patients who received adjuvant tamoxifen. Pharmgenomics Pers Med 6, 37–48 (2013).
Dezentje, V. O. et al. CYP2D6 genotype in relation to tamoxifen efficacy in a Dutch cohort of the tamoxifen exemestane adjuvant multinational (TEAM) trial. Breast Cancer Res Treat 140, 363–73 (2013).
Okishiro, M. et al. Genetic polymorphisms of CYP2D6 10 and CYP2C19 2, 3 are not associated with prognosis, endometrial thickness, or bone mineral density in Japanese breast cancer patients treated with adjuvant tamoxifen. Cancer 115, 952–961 (2009).
Moyer, A. M. et al. SULT1A1, CYP2C19 and disease-free survival in early breast cancer patients receiving tamoxifen. Pharmacogenomics 12, 1535–1543 (2011).
Whirl-Carrillo, M. et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 92, 414–7 (2012).
de Leon, J. AmpliChip CYP450 test: personalized medicine has arrived in psychiatry. Expert Rev Mol Diagn 6, 277–86 (2006).
Management of Office & Budget. Revisions to the Standards for the Classification of Federal Data on Race and Ethnicity. https://obamawhitehouse.archives.gov/omb/fedreg_1997standards (Accessed 22/06/2017) (2003).
American Cancer Society. Stages of breast cancer, https://www.cancer.org/cancer/breast-cancer/understanding-a-breast-cancer-diagnosis/stages-of-breast-cancer.html (Accessed 22/06/2017) (2017).
Kleinbaum, D. G., Kupper, L. L. & Morgenstern, H. Epidemiologic research: principles and quantitative methods. (Lifetime Learning Publications, 1982).
Johnson, J. A., Hamadeh, I. S. & Langaee, T. Y. Loss of heterozygosity at the CYP2D6 locus in breast cancer: implications for tamoxifen pharmacogenetic studies. J Natl Cancer Inst 107 (2015).
Ratain, M. J., Nakamura, Y. & Cox, N. J. CYP2D6 genotype and tamoxifen activity: understanding interstudy variability in methodological quality. Clin Pharmacol Ther 94, 185–7 (2013).
Berry, D. A. Response. J Natl Cancer Inst 106, djt380 (2014).
Berry, D. A. CYP2D6 genotype and adjuvant tamoxifen. Clin. Pharmacol. Ther. 96, 138–140 (2014).
Goetz, M. P. & Ingle, J. N. CYP2D6 genotype and tamoxifen: considerations for proper nonprospective studies. Clin. Pharmacol. Ther. 96, 141–144 (2014).
Province, M. A., Altman, R. B. & Klein, T. E. Interpreting the CYP2D6 results from the International Tamoxifen Pharmacogenetics Consortium. Clin. Pharmacol. Ther. 96, 144–146 (2014).
Acknowledgements
Rebecca A. Silliman, Department of Medicine, Section of Geriatrics, Boston University School of Medicine, Boston, Massachusetts, USA, is acknowledged for constructive commentaries to the manuscript. US National Institutes of Health, National Cancer Institute (R01CA166825) (TLL). The Lundbeck Foundation (R167-2013-15861) (DCF). Susan G. Komen for the Cure (CCR13264024) (TPA).
Author information
Authors and Affiliations
Contributions
T.L.L., D.C.F. and K.A.B. conceived and designed the study. P.D., K.A.B. and T.L.L. drafted the main manuscript. P.D., A.K., K.A.B., D.C.F., A.C., Y.H., E.A.M.J., C.L., T.P.A. and T.L.L. critically reviewed the manuscript and approved the final version. A.K., K.A.B., T.L.L., T.P.A. and D.C.E. performed the data analysis. P.D., A.K., K.A.B., D.C.F., A.C., Y.H., E.A.M.J., C.L., T.P.A. and T.L.L. contributed to interpretation of the analyses. Tables were prepared by P.D., A.K. and K.A.B. Figure 1 was prepared by K.A.B., T.L.L. and D.C.F.; Fig. 2 was prepared by P.D., K.A.B. and T.L.L.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Damkier, P., Kjærsgaard, A., Barker, K.A. et al. CYP2C19*2 and CYP2C19*17 variants and effect of tamoxifen on breast cancer recurrence: Analysis of the International Tamoxifen Pharmacogenomics Consortium dataset. Sci Rep 7, 7727 (2017). https://doi.org/10.1038/s41598-017-08091-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-08091-x
- Springer Nature Limited
This article is cited by
-
Effect of CYP2C19 genotypes on tamoxifen metabolism and early-breast cancer relapse
Scientific Reports (2021)