Abstract
CYP2C19*2 and CYP2C19*17 might influence tamoxifen metabolism and clinical outcome. Our aim was to investigate the effect of CYP2C19 genotypes on tamoxifen concentrations and metabolic ratios (MRs) and breast cancer recurrence in a large cohort of Caucasian women. Genetic variants (CYP2D6 and CYP2C19 genotypes), tamoxifen and metabolites concentrations, baseline characteristics, and breast cancer recurrence from the CYPTAM study (NTR1509) were used. CYP2C19*2 and CYP2C19*17 genotypes were evaluated as alleles and as groups based on CYP2D6 genotypes (high, intermediate and low activity). Log-rank test and Kaplan–Meier analysis were used to evaluate differences in recurrence defined as relapse-free survival (RFS). Classification tree analyses (CTAs) were conducted to assess the levels of interactions per polymorphism (CYP2D6 and CYP2C19 genotypes) on endoxifen concentrations. No differences in mean concentrations and MRs were observed when comparing CYP2C19 genotypes (CYP2C19*1/*1; CYP2C19*1/*2; CYP2C19*2/*2; CYP2C19*1/*17; CYP2C19*17/*17; CYP2C19*2/*17). Only significant differences (p value < 0.05) in mean concentrations and MRs were observed when comparing tamoxifen activity groups (high, intermediate and low activity). A log-rank test did not find an association across CYP2C19 genotypes (p value 0.898). CTAs showed a significant relationship between CYP2D6 and endoxifen (p value < 0.0001), but no association with CYP2C19 genotypes was found. CYP2C19 polymorphisms do not have a significant impact on tamoxifen metabolism or breast cancer relapse.
Similar content being viewed by others
Introduction
Worldwide, breast cancer is still the most frequent malignity in women1,2, and accounted for 571,000 deaths in 20151. Since the majority of newly diagnosed breast cancer cases are estrogen-receptor positive1,2, endocrine therapy with tamoxifen or aromatase inhibitors is recommended3,4. For many years, tamoxifen has been prescribed as monotherapy or with subsequent switch to an aromatase inhibitor after 2 or 3 years of endocrine therapy3,4. In the adjuvant scenario, tamoxifen therapy decreases mortality and disease recurrences of breast cancer5, whilst in the metastatic setting prolonged survival outcomes has been observed6. Unfortunately, there is a high variability in tamoxifen response7, and about 30% of patients using adjuvant tamoxifen still will have a disease relapse5.
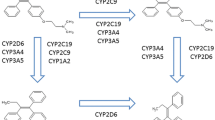
Tamoxifen is a competitive estrogen receptor antagonist8,9 and is metabolized into its primary metabolites, N-desmethyl-tamoxifen (NDM-tamoxifen; Supplementary Table 1) and 4-hydroxy-tamoxifen, followed by a second conversion into endoxifen (Fig. 1)8,9,10. Both 4-hydroxy-tamoxifen and endoxifen have similar anti-estrogenic potencies11, but endoxifen is reported as the active metabolite, as it is found in 5–10 times higher concentrations than 4-hydroxy-tamoxifen8.
In tamoxifen metabolism, the limiting step in the transformation to endoxifen is regulated by CYP2D6 enzyme8,9. Although many studies have associated genetic variants in CYP2D6 gene with clinical outcome12, many other researchers have reported null-association between survival outcome and decreased CYP2D6 enzyme activity13. Since CYP2D6 only partly contributes to explaining the 42.3% variability of endoxifen concentrations14 and 68.7% of endoxifen formation (metabolic ratio (MR) of NDM-tamoxifen/endoxifen)15, CYP2D6 genotyping has not been implemented in the daily clinical practice in order to predict tamoxifen efficacy3,4. However, other polymorphisms in other drug-metabolizing enzymes involved in tamoxifen metabolism might also have an impact in the endoxifen formation and potentially in clinical outcome8,16.
CYP2C19 gene is highly polymorphic17, and it plays multiple roles in the tamoxifen pathway (Fig. 1)8. Several polymorphisms in the gene encoding the CYP2C19 enzyme have been described. While CYP2C19*17 variant leads to an increased enzymatic activity8,16, other variants e.g. CYP2C19*2 and CYP2C19*317 genotypes have a decreased enzyme activity8,16. Regarding the role of these CYP2C19 genotypes and tamoxifen metabolism, several studies have been published. Lim and colleagues reported no association between tamoxifen and its metabolites concentration levels and CYP2C19 genotypes18. In line with these outcomes, Mürdter et al. failed to find an association regarding CYP2C19 genotypes and endoxifen, 4-hydroxy-tamoxifen and NDM-tamoxifen concentrations or MRs. In contrast, Gjerde et al. observed a higher 4-hydroxy-tamoxifen formation in CYP2C19*17 carriers19. Interestingly, Lim and colleagues reported in a recent study an association of CYP2C19*2 with norendoxifen, also named 4-hydroxy-N,N-didesmethyltamoxifen20. Norendoxifen is an active metabolite of tamoxifen, which is the result of the direct de-methylation of endoxifen. In contrast to endoxifen and tamoxifen, Lu et al. characterized this metabolite as dual aromatase inhibitor and selective estrogen-receptor modulator21 which may lead to an interesting novel drug22.
Also, the relationship between CYP2C19 genotypes and breast cancer recurrence has been examined, yet contradictory results have also been published. Schroth and colleagues described a more favorable survival outcome for CYP2C19*17 carriers (Hazard Ratio (HR):0.45; 95% Confidence Interval (CI) 0.21–0.92; p value 0.03)23. Similarly, a meta-analysis described improved survival outcomes in CYP2C19*17 carriers24. However, Moyer failed to find an association between clinical outcome and CYP2C19*17 genotype (HR: 0.93; 95% CI 0.64–1.37; p value 0.667)25. In line with Moyer, these results were recently ratified by Damkier and colleagues after analyzing the publicly available dataset of the International Tamoxifen Pharmacogenomics Consortium (ITPC)26. In this heterogeneous group, homo- and heterozygotes of the CYP2C19*17 variant were not associated with better survival outcome.
In the same manner, CYP2C19*2 genotype has been studied, and conflicting results were found again. Schaik and colleagues reported better clinical outcomes in the advanced setting (HR: 0.72; 95% CI 0.57–0.90; p value: 0.004) in a cohort of 499 patients27. In the same line, Beelen observed better survival results in adjuvant tamoxifen-treated group (HR: 0.26; 95% CI: 0.12–0.55; p value: 0.001)28, which is accordance with Ruiter and colleagues29. In contrast, Damkier showed again no association between CYP2C19*2 genotype and breast cancer outcomes in a larger group of patientsle 2.
Interestingly, another approach to evaluate the effect of CYP2C19 genotypes was also performed by Schroth and colleagues and later reproduced by Damkier and colleagues26. In their studies, patients were categorized according to their CYP2D6 and CYP2C19 genotypes in three tamoxifen activity groups (high, intermediate and low). While Schroth reported differences in clinical outcomes across these groups, Damkier failed to find any type of association. An important limitation in the majority of these studies might have been the analysis of each allele in isolation from the other one. Due to the particularities of the CYP2C19 gene, a better approach might be the use of the real CYP2C19 genotypes. For instance, a CYP2C19*2/*17 individual illustrates this relevant limitation. In case this patient were studied for CYP2C19*2 genotype only, the actual CYP2C19 effect would be masked by other genotypes, e.g. CYP2C19*1730,31.
Due to this large variety in results, we aimed to investigate the role of CYP2C19 genotypes on tamoxifen metabolism breast cancer survival outcomes in the large cohort of the prospective CYPTAM study32, which enrolled 667 Caucasian pre- and post-menopausal patients diagnosed with early-breast cancer receiving adjuvant tamoxifen.
Methods
Study objectives
The primary objective of this study was to investigate the impact of CYP2C19*2 and CYP2C19*17 on the concentrations and MRs of tamoxifen, NDM-tamoxifen, 4-hydroxy-tamoxifen and endoxifen. To this end, patients were classified according to their CYP2C19 genotypes in six possible groups: CYP2C19*1/*1; CYP2C19*1/*2; CYP2C19*2/*2; CYP2C19*1/*17; CYP2C19*17/*17; CYP2C19*2/*17. At the same time, we also performed another analysis in which we evaluated the effect of CYP2C19*2 (CYP2C19*1/*1 and CYP2C19*1/*2 versus CYP2C19*2/*2) and CYP2C19*17 (CYP2C19*1/*1 and CYP2C19*1/*17 versus CYP2C19*17/*17) separately. However, tamoxifen metabolism is complex and mainly determined by CYP2D6, and accounting only for CYP2C19*2 and CYP2C19*17 would not be of significant value, since these genotypes have a minor effect on tamoxifen variability8,16,33. Accordingly, tamoxifen activity groups based on the actual CYP2C19 and CYP2D6 genotypes were made, and individuals could be categorized in the low, intermediate or high activity group (Table 1).
The secondary objective was to assess the effect of these two CYP2C19 variants with breast cancer outcomes in a large cohort of Caucasian patients diagnosed with early-breast cancer receiving adjuvant tamoxifen. In the core CYPTAM study, the selected primary endpoint was relapse-free survival (RFS), which was defined as the time from enrolment to loco-regional or distant relapse or second breast cancer. In case of a switch to an aromatase inhibitor, patients were censored at the moment of tamoxifen discontinuation32.
Study design and population
To research the influence of CYP2C19 variants on tamoxifen metabolism and survival outcomes, whole blood and serum samples and clinical information and follow-up regarding pre- and post-menopausal female patients encompassed in the CYPTAM study were used. Concisely, from February 2008 till December 2010, a total of 667 patients were enrolled in this study, which comprises research from 25 hospitals from Belgium and The Netherlands. The primary objective was to associate CYP2D6 predicted phenotypes and endoxifen serum concentration to breast cancer recurrence32. In this study, female individuals diagnosed with early-breast cancer and starting 20 mg QD tamoxifen as adjuvant endocrine therapy, were eligible to participate in this observational study. Also, patients were allowed to participate if they were receiving tamoxifen for no longer than twelve months. Exclusion criteria were pregnancy, breast-feeding and earlier malignancy, with the exception of adequately treated in-situ cervix carcinoma and basal-cell carcinoma. The study protocol was approved by the Institutional Review Board of the Leiden University Medical Center (The Netherlands) in accordance with the Declaration of Helsinki (Tokyo 2004) and registered in the Netherlands Trial Registry (NTR1509). All encompassed female individuals gave written informed consent. For this pharmacogenetic study, the CYPTAM population was analysed, which is described in more detail elsewhere14,34.
Quantification analysis and genotyping
Serum and whole blood specimens were collected for quantification analysis of tamoxifen and its metabolites and genotyping, respectively. Samples were retrieved after at least two-month of tamoxifen therapy in order to assure steady-state concentrations. Also, a minimum of twelve hours after the last intake was required for steady state trough concentrations.
Quantification of tamoxifen and its metabolites concentrations were performed by high-performance liquid chromatography-tandem mass spectrometry (HPLC–MS/MS)35. CYP2D6 Genotyping was performed with Amplichip CYP450 test (Roche Diagnostic, Indianapolis, USA). In accordance with their CYP2D6 genotypes, all individuals were classified in predicted phenotypes, as defined by Schroth et al. More comprehensive description about CYP2D6 predicted phenotypes is outlined elsewhere. In the same manner, CYP2C19 genotyping was performed with Amplichip CYP450 test (Roche Diagnostic, Indianapolis, USA) for CYP2C19*2 and TaqMan7500 (Applied Biosystems, Nieuwerkerk a.d. IJssel, The Netherlands) SNP Genotyping Assays for CYP2C19*17.
Due to the low allele frequency of other CYP2C19 genotypes in the Caucasian population, no other genotypes were assessed in this study. For instance, CYP2C19*3 variant has a reported frequency of occurrence of 0.04%, while it has an allele frequency of 5–11% in Asian population groups36,37. Therefore, only the two most common of CYP2C19 variants among Caucasians, CYP2C19*2 and CYP2C19*17, were investigated.
Statistical analysis
To evaluate the role of CYP2C19 genotypes on tamoxifen metabolism, concentrations and metabolic ratios (MRs) of tamoxifen, endoxifen, NDM-tamoxifen and 4-hydroxy-tamoxifen were used. In this case, MRs were considered as concentration of substrate divided by metabolite concentration. To assess differences between groups, analysis of variance (ANOVA) test were carried out. Also, multiple linear regression analyses were performed to investigate the contributions of these CYP2C19 genotypes to the total explained variability of MRs and concentrations of tamoxifen, endoxifen, NDM-tamoxifen and 4-hydroxy-tamoxifen.
For the second objective, Cox regression was carried to analyse whether RFS varied across all groups [Hazard Ratio: HR; 95% Confidence Interval (CI)]. When in the univariable analysis, a p value below 0.1 was obtained, this covariate was adopted in the multivariable analysis. In addition, the following covariates were fitted in the multivariate analysis due to their known clinical relevance: tumor and nodal stage, histological classification and grade, and Her2 receptor status and menopausal status. Since the variable menopausal status was not available, a surrogate variable based on age at enrolment was used. Premenopausal and postmenopausal patients had an age at enrolment of ≤ 45 years and ≥ 45 years, respectively.
At the same time, we conducted an exploratory examination by performing classification tree analyses in order to determine the levels of interactions by polymorphisms (CYP2D6 and CYP2C19 genotypes) on the effect of endoxifen concentrations. More detailed information about how these type of analyses are performed is described elsewhere38. All statistical analyses were performed with IBM SPSS for Windows, Version 23.0. Statistical significance was accepted for p values below 0.05.
Results
Allele frequencies and distributions: CYP2C19 genotypes
The genotype distributions of CYP2C19 variants are described in Table 2. In this study, both genotypes were found to be in consistency with Hardy–Weinberg equilibrium (CYP2C19*2: χ2 = 0.518, p value = 0.472; CYP2C19*17: χ2 = 0.135, p value = 0.713). Also, a summary of the overall tamoxifen activity groups depending on CYP2C19 variants and CYP2D6 genotypes is shown in Table 2. Of note, CYP2D6 ultra-rapid metabolizers (n = 5) were included in the high activity group, independently of the CYP2C19 genotype. As illustration, we also divided the CYPTAM patients according to the previously proposed overall tamoxifen activity groups based on CYP2C19 and CYP2D6 genotypes by Schroth and colleagues and later reproduced by Damkier and colleagues26. An overview of these groups (low, intermediate and high) is listed as Supplementary Table 2.
Study population
Baseline characteristics of the CYPTAM study by CYP2C19 genotypes (CYP2C19*1/*1; CYP2C19*1/*2; CYP2C19*2/*2; CYP2C19*1/*17; CYP2C19*17/*17; CYP2C19*2/*17) is presented in Table 3. At baseline, no statistically significant differences were found regarding tumor stage, histological classification and grade, progesterone status, type of surgery and axillar surgery, adjuvant chemotherapy and radiotherapy and trastuzumab treatment. Similarly, a second overview of the baseline demographics by CYP2C19*2 and CYP2C19*17 separately, is shown as Supplementary Table 3. At enrolment, all groups were found to be comparable since no statistical differences were observed (Supplementary Table 3). Also, an overview of the demographics at enrolment by the proposed overall tamoxifen activity groups based on CYP2D6 and CYP2C19 genotypes is listed as Supplementary Table 4. In the same manner, all groups were similar at baseline, with the exception of nodal stage (p value: 0.038).
Associations of tamoxifen and its metabolites concentrations and MRs with CYP2C19 genotypes
No differences in mean concentrations and MRs of tamoxifen, endoxifen, 4-hydroxy-tamoxifen and NDM-tamoxifen were observed when all CYP2C19 genotypes were compared (CYP2C19*1/*1; CYP2C19*1/*2; CYP2C19*2/*2; CYP2C19*1/*17; CYP2C19*17/*17; CYP2C19*2/*17). In Fig. 2, mean concentrations of tamoxifen, NDM-tamoxifen, 4-hydroyx-tamoxifen and endoxifen by CYP2C19 genotypes are presented. In the same way, assessing each polymorphism individually (CYP2C19*1/*1 and CYP2C19*1/*2 versus CYP2C19*2/*2; CYP2C19*1/*1 and CYP2C19*1/*17 versus CYP2C19*17/*17), also reached the same outcomes of no differences in mean concentrations and MRs of tamoxifen and its metabolites. In Supplementary Table 4, mean concentrations and MRs by CYP2C19 genotypes are listed. In contrast, statistically significant differences in mean concentrations and MRs were found when the overall tamoxifen activity groups (high, intermediate and low) were compared (Supplementary Table 5), with the exception of tamoxifen concentrations. In the same line, analysing the proposed tamoxifen activity groups by Schroth and colleagues and later reproduced by Damkier et al.26, yielded statistical differences in mean concentrations and MRs (Supplementary Table 6).
To study the additional effect of CYP2C19 genotypes to the explained variance of tamoxifen and its metabolites concentrations and MRs, these variants were fitted in a multiple regression model in which previously CYP2D6 genotypes and concentrations of tamoxifen and its metabolites were already assessed14. When both CYP2C19 genotypes were introduced in the model, the explained variability of the concentration levels of tamoxifen and its metabolites barely differed. In Supplementary Table 7, a summary of CYP2C19 variants covariate analysis is presented.
Breast cancer recurrence and CYP2C19 genotypes
In this study, no differences in terms of HR were observed when comparing all the CYP2C19 genotypes (CYP2C19*1/*1; CYP2C19*1/*2; CYP2C19*2/*2; CYP2C19*1/*17; CYP2C19*17/*17; CYP2C19*2/*17) (Table 4). In the same line, a log-rank test showed no associations (p value: 0.898) across the CYP2C19 genotypes (Fig. 3). At the same time, evaluating each variants separately did not modify these outcomes (Table 4). Similarly, we did not find any type of association when analysing the proposed tamoxifen activity based on CYP2D6 and CYP2C19 genotypes. In Table 4, the uni- and multi-variable Cox regression results are listed. In the same manner, analysing the association between the tamoxifen activity groups, as described by Schroth and Damkier26, with RFS, did not modify these outcomes (Supplementary Table 8).
Classification tree analyses
As an exploratory analysis, we conducted different Classification Tree Analyses (CTA) to evaluate the levels of interactions between CYP2D6 predicted phenotypes and CYP2C19 genotypes on endoxifen concentrations. The first CTA was performed with the only focus on the CYP2D6 predicted phenotypes and endoxifen concentrations. In this CTA, patients were subdivided in only one level of the CTA with three different groups of CYP2D6 phenotypes that statistically different (EM/UM verus hetEM versus IM/PM; p value < 0.001) (Fig. 4). In contrast, adding CYP2C19 genotypes to this CTA, did not allowed to achieve another level in the CTA.
Discussion
In this study, we assessed the effect of CYP2C19 genotypes on tamoxifen metabolism and efficacy in an extensive cohort of Caucasian breast cancer patients receiving tamoxifen as adjuvant endocrine therapy. In our study, we failed to find any differences in mean concentrations and MRs of tamoxifen, endoxifen, 4-hydroxy-tamoxifen and NDMA-tamoxifen when comparing CYP2C19 genotypes (CYP2C19*1/*1; CYP2C19*1/*2; CYP2C19*2/*2; CYP2C19*1/*17; CYP2C19*17/*17; CYP2C19*2/*17). Additionally, the same outcomes were obtained when each variant was analysed separately. Interestingly, an analysis accounting for CYP2D6 and CYP2C19 genotypes, in which the overall tamoxifen enzymatic activity was categorized as high, intermediate and low activity, resulted in statistically significant differences in mean concentrations of endoxifen, NDM-tamoxifen, and 4-hydroxy-tamoxifen and their corresponding MRs. In contrast, tamoxifen mean concentrations were comparable across all the groups. Similarly, we did not find an association between CYP2C19 genotypes and RFS. In the same manner, dividing patients in low, intermediate or high activity based on their CYP2D6 and CYP2C19 genotypes did not show a survival difference.
Tamoxifen has a complex metabolic pathway and many different enzymes are implicated in its activation into endoxifen. Yet, the most relevant enzyme of tamoxifen metabolism is CYP2D6, but it only partially contributes to explaining the inter-variability in endoxifen concentrations between patients. Therefore, many studies have been conducted to find other potentials sources which could clarify the high variability in concentration levels and response to therapy between patients, such as CYP2C19 genotypes.
According to Schroth and colleagues, the CYP2C19*17/*17 with its higher functioning genotypes has been correlated with improved clinical outcome. In theory, tamoxifen may be more easily metabolized into its active metabolites, e.g. endoxifen, mainly due to the higher enzymatic activity among CYP2C19*17 carriers19. Consequently, a higher exposure to the anti-estrogenic activity of tamoxifen and its metabolites could be expected, which potentially may clarify why CYP2C19*17/*17 patients may have an increased survival outcome. However, we also evaluated differences in mean concentrations of tamoxifen and its metabolites, and no differences by CYP2C19 genotypes were observed. Also, categorizing patients according to their tamoxifen enzymatic activity did not yield any type of associations in our study, nor using the previously proposed groups by Schroth and Damkier26. This hypothesis of higher exposure to anti-estrogenic activity due to higher concentration levels of tamoxifen active metabolites was not observed in our study.
Likewise, for the CYP2C19*2 allele, Van Schaik and colleagues27, Beelen et al.28 and Ruiter and colleagues29, found improved survival outcomes in the metastatic setting and in the adjuvant scenario. In this case, the decreased enzymatic activity of CYP2C19*2 may probably lead to a lower exposure to antiestrogenic activity of tamoxifen and its metabolites, due to the potentially lower concentration levels, and therefore, a worsened clinical outcome. Nevertheless, all of these studies reported improved survival outcomes. A potential explanation for this increased clinical outcome among CYP2C19*2 carriers may be due to the increased transformation from endoxifen into norendoxifen, which has a dual antiendocrine mechanism of action20. However, we did not find a statistically significant variations in mean concentration levels or MRs. In this case, our results are again in agreement with Damkier and colleagues26, still the main advantage of our study might rely on the use of concentration levels.
Following the approach of Schroth, we also created a new combined variable accounting for CYP2D6 and CYP2C19 genotypes. However, our classification slightly varied from the one of Schroth and colleagues, since we used the actual CYP2C19 genotypes. Although the creation of this activity groups is complicated, the use of the real CYP2C19 genotypes, instead of the simple allele, has a relevant advantage. For instance, a CYP2C19*2/*17 individual could be wrongly classified in the intermediate activity group for the analysis of CYP2C19*2 variant .In contrast, the same patient, would be categorized in the high activity group if the CYP2C19*17 variant was studied. While this interesting difference on enzymatic activity might be critical for the creation of this type of activity groups, the remaining question would be the interpretation of this difference for the clinical practice. Although differences in the stratification could potentially also affect the obtained results, a second analysis following the classification of Schroth did also not show any type of association: no differences in mean concentrations or MRs or clinical outcomes. In this case, we questioned the utility of this type of groups due to the limited role of CYP2C19 genotypes on tamoxifen metabolism.
To evaluate the rationale after this variable, we conducted a CTA. Interestingly, we failed to find any improvement in the prediction of endoxifen concentrations when CYP2C19 genotypes were fitted in the corresponding models, whereas only when CYP2D6 predicted phenotypes were introduced, significant differences were observed. Our interpretation is that the use of CYP2C19 genotypes only in order to predict endoxifen concentrations, might lack of usefulness in the clinical setting, and that CYP2D6 genotypes might have the most relevant role in the prediction of endoxifen concentrations. Due to differences in mean concentrations and metabolic ratios when using this type of groups (based on CYP2D6 and CYP2C19 genotypes), we hypothesize that CYP2C19 genotypes might help to compensate the formation of endoxifen and 4-hydroxy-tamoxifen in the case of low CYP2D6 enzymatic activity. Yet, this minor effect did not translate in better clinical outcomes.
A limitation of our study might be our sample size of 667 patients compared to the cohort of 2102 female patients of the ITPC. However, we believe that our study was sufficiently powered to replicate the results of Damkier and colleagues26, with the main advantage of the use of concentrations and MRs.
Another possible limitation in our study might the potential effect of CYP2C19 phenoconversion in cancer patients. While an acquired loss of activity in CYP2C19 has been described in the literature in cancer patients39, the real impact of this discrepancy between CYP2C19 pheno- and genotype in breast cancer patients receiving adjuvant tamoxifen is unclear. However, we believe that the effect of CYP2C19 phenoconversion in tamoxifen metabolism might be small. In our opinion, since CYP2C19 genotypes barely contributes to explaining the inter-patient variability of tamoxifen and its metabolites concentrations and MRs, small differences in concentrations or MRs due to a CYP2C19 phenoconversion might be unnoticed.
To conclude, we have shown that CYP2C19 polymorphisms have no or little impact on concentration levels and MRs of tamoxifen, endoxifen, 4-hydroxy-tamoxifen and NDM-tamoxifen, or clinical outcomes in breast cancer patients. Therefore, CYP2C19 genotypes might not be clinically decisive for patients with early-breast cancer treated with adjuvant tamoxifen.
References
WHO Media Centre: Cancer fact sheet (2018).
Torre, L. A., Siegel, R. L., Ward, E. M. & Jemal, A. Global cancer incidence and mortality rates and trends–an update. Cancer Epidemiol. Biomark. Prev. 25, 16–27 (2016).
Burstein, H. J. et al. Adjuvant endocrine therapy for women with hormone receptor-positive breast cancer: American society of clinical oncology clinical practice guideline focused update. J. Clin. Oncol. 32, 2255–2269 (2014).
Senkus, E. et al. Primary breast cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 26(Suppl 5), v8-30 (2015).
Early Breast Cancer Trialists’ Collaborative, G. et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet 378, 771–784 (2011).
Schiavon, G. & Smith, I. E. Endocrine therapy for advanced/metastatic breast cancer. Hematol. Oncol. Clin. N. Am. 27, 715–736 (2013).
de Vries Schultink, A. H., Zwart, W., Linn, S. C., Beijnen, J. H. & Huitema, A. D. Effects of pharmacogenetics on the pharmacokinetics and pharmacodynamics of tamoxifen. Clin. Pharmacokinet. 54, 797–810 (2015).
Klein, D. J. et al. PharmGKB summary: tamoxifen pathway, pharmacokinetics. Pharmacogenet. Genom. 23, 643–647 (2013).
Brauch, H., Murdter, T. E., Eichelbaum, M. & Schwab, M. Pharmacogenomics of tamoxifen therapy. Clin. Chem. 55, 1770–1782 (2009).
Stearns, V. et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J. Natl. Cancer Inst. 95, 1758–1764 (2003).
Lim, Y. C., Desta, Z., Flockhart, D. A. & Skaar, T. C. Endoxifen (4-hydroxy-N-desmethyl-tamoxifen) has anti-estrogenic effects in breast cancer cells with potency similar to 4-hydroxy-tamoxifen. Cancer Chemother. Pharmacol. 55, 471–478 (2005).
Goetz, M. P. et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J. Clin. Oncol. 23, 9312–9318 (2005).
Rae, J. M. et al. CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J Natl Cancer Inst 104, 452–460 (2012).
Sanchez Spitman, A. B. et al. Effect of CYP3A4*22, CYP3A5*3, and CYP3A combined genotypes on tamoxifen metabolism. Eur. J. Clin. Pharmacol. 73, 1589–1598 (2017).
Murdter, T. E. et al. Activity levels of tamoxifen metabolites at the estrogen receptor and the impact of genetic polymorphisms of phase I and II enzymes on their concentration levels in plasma. Clin. Pharmacol. Ther. 89, 708–717 (2011).
Binkhorst, L., Mathijssen, R. H., Jager, A. & Van, G. T. Individualization of tamoxifen therapy: much more than just CYP2D6 genotyping. Cancer Treat. Rev. 41, 289–299 (2015).
Pharmacogene Variation Consortium. CYP2C19 variation. Vol. 2018 (2018).
Lim, J. S. et al. Impact of CYP2D6, CYP3A5, CYP2C9 and CYP2C19 polymorphisms on tamoxifen pharmacokinetics in Asian breast cancer patients. Br. J. Clin. Pharmacol. 71, 737–750 (2011).
Gjerde, J. et al. Associations between tamoxifen, estrogens, and FSH serum levels during steady state tamoxifen treatment of postmenopausal women with breast cancer. BMC Cancer 10, 313 (2010).
Lim, J. S. et al. Association of CYP2C19*2 and associated haplotypes with lower norendoxifen concentrations in tamoxifen-treated Asian breast cancer patients. Br. J. Clin. Pharmacol. 81, 1142–1152 (2016).
Lu, W. J. et al. The tamoxifen metabolite norendoxifen is a potent and selective inhibitor of aromatase (CYP19) and a potential lead compound for novel therapeutic agents. Breast Cancer Res. Treat. 133, 99–109 (2012).
Lv, W., Liu, J., Lu, D., Flockhart, D. A. & Cushman, M. Synthesis of mixed (E, Z)-, (E)-, and (Z)-norendoxifen with dual aromatase inhibitory and estrogen receptor modulatory activities. J. Med. Chem. 56, 4611–4618 (2013).
Schroth, W. et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J. Clin. Oncol. 25, 5187–5193 (2007).
Bai, L. et al. Association of CYP2C19 polymorphisms with survival of breast cancer patients using tamoxifen: results of a meta- analysis. Asian Pac. J. Cancer Prev. 15, 8331–8335 (2014).
Moyer, A. M. et al. SULT1A1, CYP2C19 and disease-free survival in early breast cancer patients receiving tamoxifen. Pharmacogenomics 12, 1535–1543 (2011).
Damkier, P. et al. CYP2C19*2 and CYP2C19*17 variants and effect of tamoxifen on breast cancer recurrence: analysis of the International tamoxifen pharmacogenomics consortium dataset. Sci. Rep. 7, 7727 (2017).
van Schaik, R. H. et al. The CYP2C19*2 genotype predicts tamoxifen treatment outcome in advanced breast cancer patients. Pharmacogenomics 12, 1137–1146 (2011).
Beelen, K. et al. CYP2C19 2 predicts substantial tamoxifen benefit in postmenopausal breast cancer patients randomized between adjuvant tamoxifen and no systemic treatment. Breast Cancer Res. Treat. 139, 649–655 (2013).
Ruiter, R. et al. CYP2C19*2 polymorphism is associated with increased survival in breast cancer patients using tamoxifen. Pharmacogenomics 11, 1367–1375 (2010).
Balian, J. D. et al. The hydroxylation of omeprazole correlates with S-mephenytoin metabolism: a population study. Clin. Pharmacol. Ther. 57, 662–669 (1995).
Herrlin, K. et al. Slow chloroguanide metabolism in Tanzanians compared with white subjects and Asian subjects confirms a decreased CYP2C19 activity in relation to genotype. Clin. Pharmacol. Ther. 68, 189–198 (2000).
Sanchez-Spitman, A. et al. Tamoxifen pharmacogenetics and metabolism: results from the prospective CYPTAM study. J. Clin. Oncol. 37, 636–646 (2019).
Fernandez-Santander, A. et al. Relationship between genotypes Sult1a2 and Cyp2d6 and tamoxifen metabolism in breast cancer patients. PLoS ONE 8, e70183 (2013).
Sanchez-Spitman, A. B. et al. Genetic polymorphisms of 3’-untranslated region of SULT1A1 and their impact on tamoxifen metabolism and efficacy. Breast Cancer Res. Treat. 172, 401–411 (2018).
Teunissen, S. F. et al. Development and validation of a quantitative assay for the determination of tamoxifen and its five main phase I metabolites in human serum using liquid chromatography coupled with tandem mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 879, 1677–1685 (2011).
Tamminga, W. J. et al. The prevalence of CYP2D6 and CYP2C19 genotypes in a population of healthy Dutch volunteers. Eur. J. Clin. Pharmacol. 57, 717–722 (2001).
Sugimoto, K., Uno, T., Yamazaki, H. & Tateishi, T. Limited frequency of the CYP2C19*17 allele and its minor role in a Japanese population. Br. J. Clin. Pharmacol. 65, 437–439 (2008).
Pander, J., Wessels, J. A., Mathijssen, R. H., Gelderblom, H. & Guchelaar, H. J. Pharmacogenetics of tomorrow: the 1 + 1 = 3 principle. Pharmacogenomics 11, 1011–1017 (2010).
Klomp, S. D., Manson, M. L., Guchelaar, H. J. & Swen, J. J. Phenoconversion of cytochrome P450 metabolism: a systematic review. J. Clin. Med. 9, 2890 (2020).
Acknowledgements
We thank all of the patients who participated in the CYPTAM study. We also thank Roche for providing the Amplichip P450 test, Integraal Kankercentrum Nederland for data management, and ZOLEON for its grant.
Author information
Authors and Affiliations
Contributions
Conception and design: V.O.D., H.G. and H.J.G. Administrative support: A.B.S.S., V.O.D.; Collection and assembly of data: A.B.S.S., V.O.D., D.J.A.R.M., J.J.S.; Data analysis and interpretations: A.B.S.S., J.J.S., V.O.D., D.J.A.R.M., H.G. and H.J.G. Manuscript writing: A.B.S.S., J.J.S., V.O.D., D.J.A.R.M., H.G. and H.J.G.; Final approval of manuscript: A.B.S.S., J.J.S., V.O.D., D.J.A.R.M., H.G. and H.J.G.; Accountable for all aspects of the work: A.B.S.S., J.J.S., V.O.D., D.J.A.R.M., H.G. and H.J.G.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sanchez-Spitman, A.B., Swen, J.J., Dezentjé, V.O. et al. Effect of CYP2C19 genotypes on tamoxifen metabolism and early-breast cancer relapse. Sci Rep 11, 415 (2021). https://doi.org/10.1038/s41598-020-79972-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-79972-x
- Springer Nature Limited
This article is cited by
-
Tamoxifen pharmacokinetics and pharmacodynamics in older patients with non-metastatic breast cancer
Breast Cancer Research and Treatment (2023)
-
Preliminary results using a kit to measure tamoxifen and metabolites concentrations in capillary blood samples from women with breast cancer
Scientific Reports (2022)
-
Tamoxifen and oxidative stress: an overlooked connection
Discover Oncology (2021)