Abstract
Dimethylsulfoniopropionate (DMSP) is an abundant marine organosulfur compound with roles in stress protection, chemotaxis, nutrient and sulfur cycling and climate regulation. Here we report the discovery of a bifunctional DMSP biosynthesis enzyme, DsyGD, in the transamination pathway of the rhizobacterium Gynuella sunshinyii and some filamentous cyanobacteria not previously known to produce DMSP. DsyGD produces DMSP through its N-terminal DsyG methylthiohydroxybutyrate S-methyltransferase and C-terminal DsyD dimethylsulfoniohydroxybutyrate decarboxylase domains. Phylogenetically distinct DsyG-like proteins, termed DSYE, with methylthiohydroxybutyrate S-methyltransferase activity were found in diverse and environmentally abundant algae, comprising a mix of low, high and previously unknown DMSP producers. Algae containing DSYE, particularly bloom-forming Pelagophyceae species, were globally more abundant DMSP producers than those with previously described DMSP synthesis genes. This work greatly increases the number and diversity of predicted DMSP-producing organisms and highlights the importance of Pelagophyceae and other DSYE-containing algae in global DMSP production and sulfur cycling.
Similar content being viewed by others
Main
Petagrams of dimethylsulfoniopropionate (DMSP) are made annually in Earth’s surface waters, with potentially much more in marine aphotic, sediment and coastal settings1,2,3,4,5. DMSP is an anti-stress compound6,7,8,9 produced to millimolar concentrations within diverse algae, corals, bacteria and some angiosperms10. When released into the environment, DMSP is also a major source of carbon and sulfur to marine microorganisms11 and of climate-cooling gases and/or signalling molecules11, including dimethyl sulfide (DMS) and methanethiol (MeSH), via DMSP catabolism12.
Recent work has categorized DMSP producers into low (<50 mM) and high (≥50 mM) accumulators13 and identified key genes encoding single-domain S-methyltransferase enzymes involved in, and that are robust indicators for, DMSP synthesis in diverse algae (DSYB and TpMMT) and bacteria (dsyB, mmtN and burB) (Fig. 1a)1,14,15,16,17,18. However, many known DMSP-producing algae19, bacteria14 and plants20,21,22,23 lack these DMSP synthesis genes and probably contain alternative DMSP synthesis enzymes. Thus, despite some recent attempts9,24,25,26, it is currently challenging to predict from omics data which organisms are important DMSP producers in environmental samples. In this Article, we elucidate and characterize the activity, biodiversity, potential role and environmental importance of previously unknown DMSP synthesis enzymes in cyanobacteria, other bacteria and eukaryotic algae and identify additional and important global DMSP producers.
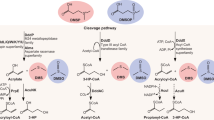
a, The ‘methylation’ pathway in some higher plants with the methionine (Met) S-methyltransferase (MMT) and bacteria containing MmtN or another methyltransferase (BurB) (left); the ‘transamination’ pathway in algae, bacteria and corals with DSYB/DsyB, DsyGD/DsyG, DSYE and/or TpMMT (middle); and the ‘decarboxylation’ pathway in Crypthecodinium cohnii (right). The pathways are named after their first reaction step (in larger font). AdoMet, S-adenosylmethionine; AdoHcy, S-adenosylhomocysteine; NADP, nicotine adenine dinucleotide phosphate; MAT, methionine aminotransferase; MR, MTOB reductase; MSM, MTHB S-methyltransferase; DDC, DMSHB decarboxylase; SMM, S-methylmethionine; MTOB, 4-methylthio-2-oxybutyrate; MTHB, 4-methylthio-2-hydroxybutyrate; DMSHB, 4-dimethylsulfonio-2-hydroxybutyrate; MTPA, 3-methylthiopropylamine; MMPA, 3-methylmercaptopropionate. The enzymes and domains identified here are coloured to match their corresponding genes in c. b, DMSP accumulation in G. sunshinyii incubated with DMSP synthesis intermediates (0.5 mM) or nothing added (NA, control). The results show the mean values of three independent biological replicates with error bars indicating standard deviations. The statistically significant differences compared with control conditions were determined using a two-sided Student’s t-test (**P = 0.0025 and ****P = 7.74 × 10−6). c, The genomic location of dsyGD/dsyG in DMSP-producing bacteria. The algal DSYE transcripts are included for size comparison. For Oscillatoria sp. SIO1A7, dsyGD is located at the start of the contig. MFS, major facilitator superfamily; tRNA, transfer RNA; ribonuclease BN, ribonuclease from Escherichia coli strain BN; ABC, ATP-binding cassette.
Results
G. sunshinyii makes DMSP by the transamination pathway
This study initially focused on G. sunshinyii, a rhizobacterium with anti-fungal activity isolated from the salt marsh plants Carex scabrifolia and Spartina alterniflora27,28. The S. alterniflora rhizosphere is rich in DMSP produced by this cordgrass29,30,31,32 and microbial DMSP cycling21,33,34,35. DMSP was also found in C. scabrifolia leaves, roots and rhizosphere samples (ranging from 5.51 ± 0.15 to 6.92 ± 0.13 nmol DMSP g−1; Supplementary Fig. 1). It was possible that these plants fed DMSP to G. sunshinyii in return for favourable bacterial traits and metabolites, for example, activity against fungal pathogens28,36,37,38. However, G. sunshinyii (strain YC6258 (ref. 27)) could not use DMSP as a sole carbon source nor liberate DMS or MeSH from DMSP, consistent with its genome lacking all known DMSP lyase genes39,40,41,42,43,44,45,46,47 and the DMSP demethylation gene dmdA48. Instead, G. sunshinyii produced DMSP when grown without added organosulfur compounds and at levels approximately threefold higher than the model DMSP-producing bacterium Labrenzia aggregata1 (101.11 ± 6.64 and 35.38 ± 3.94 pmol µg−1 of protein, respectively). DMSP synthesis in G. sunshinyii was investigated because its genome lacked all known DMSP synthesis genes.
Incubation of G. sunshinyii cells with DMSP synthesis intermediates from the transamination pathway49,50 (Fig. 1a), 4-methylthio-2-hydroxybutyrate (MTHB) and 4-dimethylsulfonio-2-hydroxybutyrate (DMSHB), significantly enhanced DMSP accumulation by 2- and 30-fold, respectively, whereas those from the methylation and decarboxylation pathways had no significant effects compared to controls with no added intermediates (Fig. 1b). DMSHB probably resulted in higher DMSP levels because it is specific to the transamination pathway for DMSP synthesis49, whereas MTHB is a substrate in competing pathways, for example, in methionine (Met) salvage51. Furthermore, G. sunshinyii cell extracts displayed in vitro MTHB S-methyltransferase (MSM) and DMSHB decarboxylase (DDC) activities (39.11 ± 0.21 and 9.23 ± 0.19 pmol DMSP per µg of protein per hour, respectively). These data implied that G. sunshinyii synthesized DMSP via the transamination pathway.
Identification of a bifunctional DMSP synthesis enzyme
A G. sunshinyii genomic library was constructed and screened for MSM activity in Rhizobium leguminosarum. One from 3,000 clones screened (termed pJDT0020) conferred MSM activity. Unlike dsyB/DSYB clones1,15, pJDT0020 conferred MSM activity in Escherichia coli (2.51 ± 0.12 pmol DMSP per µg of protein per hour), but intriguingly, also DDC activity (0.74 ± 0.08 pmol DMSP per µg of protein per hour), implying that G. sunshinyii contained a DMSP synthesis gene cluster. The ~30 kb insert in pJDT0020 contained a gene, termed dsyGD, adjacent to another predicted to encode a 4-methylthio-2-oxobutyrate (MTOB) reductase (Fig. 1a,c). DsyGD is a 494 amino acid protein with two domains (Supplementary Fig. 2). The N-terminal methyltransferase domain (Pfam PF08241.15, 76–175 amino acids), termed DsyG, had 31% amino acid identity to Thalassiosira pseudonana TpMMT16, was phylogenetically distinct and formed a separate clade from this and all other known S-methyltransferases involved in DMSP synthesis10 (Fig. 2). The DsyGD C-terminal domain (Pfam PF04115.15, 320–469 amino acids), termed DsyD, was similar to an ureidoglycolate lyase domain and was predicted to be a DMSHB decarboxylase (Supplementary Fig. 2).
The tree was constructed in MEGA X (ref. 87) using the sequences of previously characterized S-methyltransferases involved in DMSP synthesis (Supplementary Table 10) and others shown to be not functional1,14,15,16, including those from this study and homologues from MMETSP. Where proteins were multi-domain (labelled DsyGD), only the DsyG S-methyltransferase domain was analysed. Experimentally ratified (as functional) MSM or MmtN are marked with green ticks, while non-functional S-methyltransferases are labelled with a red cross. Eukaryotic (circles) and prokaryotic (triangles) proteins are coloured according to the taxonomy described in the key. The organisms containing both DSYE and DSYB are indicated with a rhombus. The proteins identified and discussed from previous studies are marked with purple branches. The predicted intracellular DMSP levels of the organisms13 are also indicated.
Cloned dsyGD conferred in vivo MSM (177.42 ± 3.23 pmol DMSHB per µg of protein per hour) and DDC activity (13.81 ± 0.97 pmol DMSP per µg of protein per hour) when expressed in E. coli and restored DMSP production in a L. aggregata LZB033 dsyB−mutant1, which does not produce DMSP (Table 1). Furthermore, purified DsyGD (Supplementary Fig. 3a) exhibited in vitro S-adenosylmethionine (AdoMet)-dependent MSM and DDC activity with an optimal temperature of 25 °C (Supplementary Fig. 4a) and pH of 7.0 for MSM activity (Supplementary Fig. 4b). Kinetic analysis of DsyGD showed it to have an approximate tenfold higher MSM (kcat/Km of 0.21 and 0.073 µM−1 s−1 for MTHB and AdoMet, respectively) than DDC catalytic efficiency (kcat/Km of 2.30 × 10−3 µM−1 s−1) (Supplementary Fig. 4c–e). Even with this lower DDC catalytic efficiency, G. sunshinyii still accumulated 23-fold higher DMSP than DMSHB under standard growth conditions (Fig. 3a).
a, G. sunshinyii DMSP and DMSHB accumulation measured by GC. b, dysG transcription from cultures grown under standard conditions (35 PSU MBM and 0.5 mM NH4Cl), low salt (5 PSU), high salt (50 PSU) or high nitrogen (10 mM NH4Cl). DMSP and DMSHB values in a represent the mean of three independent biological replicates with the error bars indicating the respective standard deviations. For a, statistically significant differences compared with control conditions were determined using a two-sided Student’s t-test (DMSP group, low salt: ***P = 0.0002; high salt: *P = 0.0409; high N: *** P = 0.0002. DMSHB group, low salt: **P = 0.0013 and ***P = 0.0004). For the RT–qPCR assays in b, the mean values of three technical replicates for each of three independent biological replicates are shown. The error bars indicate standard deviation. For b, statistically significant differences compared with control conditions were determined using a two-sided Student’s t-test (low salt: ****P = 1.31 × 10−6; high salt: *** P = 0.0009; high N: ****P = 3.219 × 10−5). c, Growth of wild type E. coli MC4100, the salt-sensitive E. coli otsA−mutant strain FF4169 (deficient in trehalose production) and FF4169 strains expressing cloned dsyGD was monitored in media containing 0.5 M NaCl alone or with 1 mM GB, DMSP or DMSP synthesis intermediates (MTHB and DMSHB). The arrows indicate the three strains that did not grow. The values shown represent the mean of three biological replicates with the error bars indicating the respective standard deviations. d, DMSP levels in selected cells after the 36 h incubation experiments shown in c. The mean values of three biological replicates are shown with the error bars indicating standard deviation. ND, not detected.
The individual G. sunshinyii DsyG and DsyD domains and the predicted MTOB reductase (MR) enzyme were either insoluble (for DsyG) and/or did not have the expected MSM, DDC or MR activities (Fig. 1a) when expressed in E. coli or as purified proteins (Supplementary Fig. 3b–d) under the conditions tested here. It is possible that these specific G. sunshinyii DsyG and DsyD domains evolved to require each other. Unfortunately, transformation and conjugation of plasmids into G. sunshinyii were not possible, preventing mutagenic and/or overexpression analysis of DsyGD in this host. Nevertheless, DsyGD is a bifunctional DMSP synthesis enzyme with two DMSP synthesis-specific and sequential enzyme activities in the transamination pathway50 and the only known enzyme with DDC activity.
DsyGD is confined to G. sunshinyii and some Oscillatoriales
Proteins with a high level of amino acid identity to GsDsyGD were not identified from any other sequenced microbial genomes or transcriptomes. However, proteins with MSM and DDC activity (Table 1) but only an ~46% amino acid identity to GsDsyGD (Supplementary Table 1) were encoded from metagenome-assembled genomes (MAGs) of two Oscillatoriales order cyanobacteria (Symploca sp. SIO3E6 and Oscillatoria sp. SIO1A7) (Figs. 1c and 2). Interestingly, a single-domain DsyG with MSM activity and an ~50% amino acid identity to this domain of GsDsyGD was also identified in Zarconia navalis LEGE 11467, an Oscillatoriales isolate from a subtidal epilithic marine sample52 (Figs. 1c and 2, Supplementary Table 1 and Supplementary Fig. 2). Unlike the truncated GsDsyG, ZnDsyG was expressed as a soluble protein in E. coli, explaining their differences in MSM activity (Supplementary Fig. 3d,e). Z. navalis lacked dsyD and accumulated 111- to 335-fold lower DMSP than DMSHB levels (0.34 ± 0.005 pmol DMSP per µg of protein versus 108.53 ± 8.06 pmol DMSHB per µg of protein in standard conditions; Supplementary Fig. 5a). These data support the hypotheses that the double domain GsDsyGD was responsible for the higher ratio of DMSP:DMSHB in G. sunshinyii than Z. navalis, that any Z. navalis enzyme(s) with DDC activity (currently unidentified) were not efficient or expressed at low levels and that DMSHB may have a more prominent role than DMSP in Z. navalis.
Unlike DsyG, a single-domain DsyD was not identified from any sequenced genomes, MAGs or transcriptomes. The most homologous proteins to the GsDsyD domain, from Prymnesium parvum Texoma1 and Alexandrium monilatum CCMP3105, contained only the PF04115.15 domain, showed 34% and 28% amino acid identity to the Oscillatoria sp. SIO1A7 DsyD domain and lacked DDC activity (Table 1, Supplementary Tables 1 and 2 and Supplementary Fig. 6). Thus, knowledge on the DDC step of the transamination pathway is still lacking. Note, the unidentified enzymes with DDC activity in DMSP-producing bacteria (such as L. aggregata), algae and non-DMSP producers (such as Rhizobium) are probably more widespread than DsyD1,15. DMSHB probably has more important physiological role(s) than DMSP in Z. navalis and potentially other organisms, inferring that DMSHB may be prominent in marine environments, and that a DDC enzyme is not always required in organisms with MSM activity.
After ZnDsyG, the next most homologous proteins to the GsDsyG domain, with an ~39% amino acid identity, were from a Planctomycetales bacterium MAG and the red alga Porphyra umbilicalis (Supplementary Table 1). These DsyG-like proteins either phylogenetically clustered more closely to the diatom MTHB S-methyltransferase (MSM) TpMMT than GsDsyG (P. umbilicalis) or were positioned in between TpMMT and GsDsyG (Planctomycetales bacterium) (Fig. 2). Note, the P. umbilicalis protein, like GsDsyGD, contained two domains, but its C-terminal domain belonged to the aspartate decarboxylase protein family (pfam02261), which seemed a good candidate DDC as a DsyD isoform enzyme. Despite this, both the recombinant Planctomycetales and the P. umbilicalis DsyG-like proteins lacked MSM and DDC activity (Table 1). There were also no proteins with high homology to DsyG or DsyD (>38% or 29% amino acid identity, respectively) predicted from the genomes and/or transcriptomes of eukaryotic algae. Overall, these data support dsyGD/dsyG as reliable indicators for DMSP/DMSHB synthesis in bacteria and filamentous cyanobacteria not previously suspected to produce these molecules. These data also highlight the need for careful functional analysis of DMSP synthesis genes and enzymes before predicting DMSP synthesis in organisms based on their presence. This is particularly relevant for TpMMT, which has only been characterized from T. pseudonana16.
Regulation of DMSP production in Gynuella and Zarconia
In G. sunshinyii, DMSP and DMSHB accumulation and GsdsyGD gene transcription were significantly upregulated by growth in media with increased salinity or decreased nitrogen levels, with DMSP and DMSHB either low or undetected under low salinity or high nitrogen conditions (Fig. 3a,b and Supplementary Fig. 7a,b). Note, G. sunshinyii accumulated nitrogenous glycine betaine (GB) as a probable major osmolyte, whose levels always far exceeded DMSP/DMSHB, except under low salinity conditions, where both GB and DMSP/DMSHB were undetected (Supplementary Fig. 7a,b). Z. navalis also accumulated more DMSHB (and ZndsyG transcripts) with increased salinity and showed reduced levels in high nitrogen conditions (Supplementary Fig. 5a,c). GB production was higher than DMSP/DMSHB (Supplementary Fig. 5b,d), indicating that this may also be a major osmolyte in Z. navalis. In contrast, DMSP accumulated to comparatively very low and constitutive levels in Z. navalis irrespective of the growth conditions (Supplementary Fig. 5a). These data are consistent with findings on other DMSP-producing organisms14,15,49, where DMSP and/or DMSHB potentially act as sulfur osmolytes, whose production over nitrogen-containing equivalents may be advantageous in sulfur-rich but nitrogen-sparse marine settings, and expression of any unknown Z. navalis DDC enzyme(s) either being very low and/or not regulated by salinity or nitrogen levels. Note, DMSHB and DMSP production also releases nitrogen from the transamination of Met (Fig. 1).
Further supporting the role of DMSP and DsyGD in osmoprotection, cloned GsdsyGD greatly enhanced the growth of an osmosensitive E. coli strain FF4169 (ref. 53) under increased salinity in the presence of MTHB (which has limited osmoprotective properties50) or, especially, DMSHB, compared with control strains lacking cloned dsyGD (Fig. 3c). This osmoprotection phenotype was probably due to DMSHB and/or DMSP produced from MTHB and DMSHB (5.49 ± 0.99 and 10.13 ± 0.63 pmol DMSP per µg protein per hour, respectively), since E. coli strain FF4169 lacking cloned GsdsyGD produced no DMSP from MTHB or DMSHB (Fig. 3d). Although this work was conducted in E. coli and not a marine organism, it demonstrates that cloned DMSP synthesis genes can confer osmoprotection, which may be of importance for biotechnological applications.
Identification of DSYE in diverse algae
Although no DsyGD proteins were predicted in eukaryotic algae, single-domain DsyG-like proteins were identified with <38% amino acid identity to GsDsyG from sequenced algal genomes (Fragilariopsis cylindrus CCMP1102 and Nitzschia inconspicua strain hildebrandi). Furthermore, 61 DsyG-like proteins were predicted from the 397 different marine eukaryotes in the Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP) database54 (Supplementary Table3). These algal proteins, termed DSYE (‘E’ for eukaryotes and DSY in upper case to denote their eukaryotic host), were phylogenetically distinct to cyano/bacterial DsyG and were themselves divided into five separate clades (termed DSYE clade A–E) (Fig. 2). Multiple representative DSYE proteins from the five clades were expressed in E. coli and all showed MSM activity (Table 1 and Fig. 2).
Clade A DSYE proteins were identified in Chloroarachniophyta, notably Bigelowiella natans, which are known to accumulate high levels of DMSP13 and Norrisiella spp., which are not previously known to produce DMSP (Table 1 and Fig. 2).
Clade B DSYE proteins were in diverse and highly abundant chlorophyte algae, including Tetraselmis sp.55, Pyraminonas sp.55, Bathycoccus sp.56 and Mantoniella sp.55 (which are known to accumulate low levels of DMSP); Micromonas sp. (which contain both high and low DMSP-producing representatives55,56) and Ostreococcus sp. (a widely distributed genus in Earth’s oceans57 not previously known to produce DMSP) (Fig. 2 and Supplementary Table 3). Tested Ostreococcus tauri cells contained DMSP (0.34 ± 0.003 nmol DMSP per µg of protein), consistent with members of this genus being DMSP producers (Supplementary Table 4).
Clade C DSYE proteins were mostly in pelagophyte algae, for example, Pelagococcus sp., such as P. subviridis CCMP1429, which had DSYE and DSYB15, and Pelagomonas spp., both thought to accumulate low levels of DMSP13,55,56 (Fig. 2). Pelagophyte algae were not thought to be globally important DMSP producers, and few had been studied for DMSP production, despite these picoeukaryotes often forming large blooms and being globally abundant58,59,60,61. Here, diverse axenic bloom-forming and sometimes toxin-producing pelagophytes58,59,60 Chrysocystis, Aureococcus, Pelagococcus, Chrysoreinhardia and Pelagomonas strains were shown to accumulate DMSP to intracellular concentrations ranging from 13.79 ± 0.46 to 233.81 ± 32.10 mM, (Supplementary Table 4 and Supplementary Fig. 8). Thus, pelagophytes, for example, Pelagomonas calceolata, one of the most abundant eukaryotic species in Earth’s oceans61, are potentially important global DMSP producers.
Haptophytes are generally thought to accumulate high DMSP levels and contain DSYB15,62. Pavlova spp. and Exanthemachysis spp. are exceptions that lack DSYB but contain a functional clade D DSYE (Fig. 2 and Supplementary Table 3). Most Pavlova spp. are high DMSP accumulators, but some, for example, P. lutheri, are considered low DMSP accumulators, as are all tested Exanthemachysis spp.13.
Clade E DSYE proteins were exclusively in diatoms, generally thought to accumulate low intracellular DMSP levels13,55. None of the diatoms with DSYE contained TpMMT, although some did also contain DSYB, for example, F. cylindrus CCMP1102 and Pseudonitzschia fraudulenta WWA7, while others, for example, N. inconspicua15, contained only DSYE (Fig. 2 and Supplementary Table 3).
Purified clade B and C DSYE from Ostreococcus prasinos BCC99000 and Chroomonas mesostigmatica CCMP1168 (Table 1 and Supplementary Fig. 9) showed in vitro AdoMet-dependent MSM activity with temperature and pH optima of 30 °C and 20 °C (Supplementary Fig. 10a,e) and 9.0 and 9.5 (Supplementary Fig. 10b,f), respectively. The C. mesostigmatica clade C DSYE was ~30-fold more efficient with MTHB (kcat/Km of 4.5 × 10−3 μM−1 s−1) than the O. prasinos clade B DSYE enzyme (kcat/Km of 0.15 × 10−3 μM−1 s−1) (Supplementary Fig. 10). Note, these DSYE enzymes were 40- and 1,400-fold, respectively, less efficient than GsDsyGD. Further work is required to establish whether DSYE catalytic efficiency and/or its expression levels are robust reporters of the DMSP levels that organisms accumulate.
Identification of DSYE, in addition to DSYB and TpMMT in algae and dsyGD, dsyG, dsyB and mmtN in diverse bacteria, has greatly expanded the ability to predict which organisms, particularly algae, can produce DMSP (Fig. 2 and Supplementary Table 3). With the inclusion of DSYE, 66% of the predicted 162 DMSP-producing eukaryotes13 within MMETSP expressed a known S-methyltransferase gene involved in DMSP synthesis, an increase from 44% when considering only DSYB and TpMMT (Supplementary Table 3). Most of the remaining candidate DMSP producers on MMETSP which lacked DSYE, DSYB or TpMMT had not been tested for DMSP production or were predicted to accumulate low DMSP levels (Supplementary Table 3). Outside of MMETSP data, there are still known DMSP-producing organisms which lack any of these S-methyltransferase genes, but their numbers are now reduced and are mainly confined to plants such as Spartina spp. and Melanthera biflora that utilize the methylation pathway for DMSP synthesis31,63, macroalgae, such as Ulva spp., and cyanobacteria such as Trichodesmium that accumulate low DMSP levels64.
Algae containing DSYE are abundant in Earth’s oceans
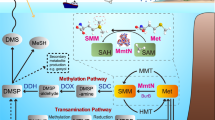
The Ocean Microbial Reference Gene Catalogue (OM-RGC_V2) metagenomic dataset65, generated from 0.22–3 µm fractionated samples and apportioned to bacterioplankton, was analysed for known DMSP synthesis genes. As previously described, dsyB and its transcripts were far more abundant than those for mmtN in Earth’s oceans, and these dsyB genes/transcripts were over twofold more abundant in the surface (SRF) and deep chlorophyll maximum (DCM) than in mesopelagic (MES) waters (Supplementary Fig. 11, Supplementary Fig. 12 and Supplementary Table 5). dsyGD/dsyG genes and transcripts were not detected in any OM-RGC_V2 dataset, consistent with this system being largely irrelevant to marine DMSP cycling. Alternatively, some bacteria, notably filamentous cyanobacteria, containing these genes, may have aggregated and not been captured by the bacterioplankton sampling methods. However, eukaryotic DSYE clade B genes and transcripts from chlorophyte algae (picoeukaryotes including Pyramimonas, Pterosperma, Ostreococcus, Micromonas and Tetraselmis), small enough to be in the bacterioplankton samples, were present in almost all stations, at approximately twofold lower levels than dsyB in SRF and DCM samples (Supplementary Fig. 12). Approximately 6% of the picoeukaryotes in these SRF and DCM samples contained DSYE. Consistent with the phototrophic lifestyle of their algal hosts, DSYE and its transcripts were barely detected in MES samples (Supplementary Table 5 and Supplementary Fig. 11). OM-RGC_V2 DSYE and dsyB genes and transcripts were most abundant in high-latitude polar samples, with a few exceptions. Notably, maximal dsyB abundance was seen in a mid-latitude DCM sample (Supplementary Fig. 12).
Within the eukaryotic Marine Atlas of Tara Ocean Unigenes (MATOU), algal DMSP synthesis genes and transcripts were also barely detected in data from MES but were much better represented in the SRF and DCM samples, consistent with their presence in phototrophs (Supplementary Table 6). Although DSYB genes, mostly from haptophytes and dinophytes, were detected in all stations, DSYE genes, predominantly from pelagophytes (clade C) and to a lesser extent, chlorophytes (clade B), were marginally and approximately twofold more abundant in the photic SRF and DCM samples, respectively (Supplementary Figs. 11 and 12 and Supplementary Table 5). The DSYB and DSYE genes showed similar biogeographical distribution patterns in MATOU stations, being concentrated in non-polar sites between −50° and 50° latitude (Supplementary Fig. 12). In contrast to the metagenomic data, DSYB transcripts were approximately twofold more abundant than those for DSYE in SRF and DCM samples from MATOU datasets (Supplementary Fig. 11 and Supplementary Table 5), and this may be a better indication of DMSP production than gene abundance. Diatom TpMMT and their transcripts were generally one to two orders of magnitude less abundant than those for algal DSYB or DSYE (Supplementary Fig. 11 and Supplementary Table 5). These data are consistent with previous reports of haptophytes and dinophytes15, and also now pelagophyte algae, being important global DMSP producers, with most diatoms having a less prominent role. Further model organism and environmental sampling work on diverse pelagophyte algae is required to explore their importance in global DMSP cycling, especially during blooms66, where they are likely to have a more considerable impact.
Discussion
DMSP is an abundant and ecologically important marine organosulfur compound. This study identifies the unusual DMSP synthesis genes dsyGD/dsyG in the rhizobacterium G. sunshinyii and filamentous cyanobacteria, not previously suspected to produce DMSP (Fig. 4), and provides evidence for DMSP and/or DMSHB being osmolytes in these bacteria. The origin and transfer of dsyG/dsyGD between organisms was interesting but difficult to address because these genes were rare in sequenced organisms and environmental samples.
a, Key DMSP synthesis and cleavage pathways are indicated with known algal and bacterial S-methyltransferases. b, The relative abundance of DMSP synthesis genes and transcripts in SRF, DCM and MES water layers from OM-RGC_V2 (0.22–3 µm size fraction) and MATOU (0.8–2,000 µm size fraction) datasets. c, Clades and taxonomy of DSYE sequences detected in MATOU datasets. The genes in OM-RGC_V2 and MATOU datasets were normalized to recA and β-actin genes, respectively. The size of the pie charts represents the gene relative abundance in the corresponding datasets. Note, no dsyG/dsyGD sequences were detected. CCN, cloud condensation nuclei. MetaG, metagenomes data; MetaT, metatranscriptomes data; RA, relative abundance. SMM, S-methylmethionine.
Functional genomics identified DSYE, forming a diverse family of eukaryotic MSM enzymes that were phylogenetically distinct from DsyG and other known enzymes with MSM activity. The five DSYE clades (A–E) comprised taxonomically distinct eukaryotic algae spanning low, medium and high DMSP accumulators, and algae not previously reported to produce DMSP (for example, O. tauri) and multiple pelagophyte algae. DSYE, with DSYB and TpMMT, serve as indicator genes of DMSP synthesis, and their combined presence in most known DMSP-producing algae with available transcriptomic and genomic data, allows more comprehensive predictions of key algal producers in marine environments with available multi-omics data.
A major unanswered question was whether the presence of a particular DMSP synthesis gene implies how much DMSP an organism accumulates. McParland et al. suggested that the presence of DSYB or TpMMT in algae was a reporter of high or low DMSP accumulation levels, respectively62. This appealing hypothesis was supported by a strong correlation between DSYB and high DMSP accumulators (Supplementary Table 3)13. However, the bacterial DsyB enzyme is as efficient as algal DSYB, despite bacteria generally accumulating low intracellular levels of DMSP13, and there are many examples of organisms with DSYB that also accumulate low intracellular DMSP levels (for example, F. cylindrus and Chrysochromulina tobin15). It is more difficult to infer the reverse correlation for TpMMT because this protein has only been studied in T. pseudonana16. However, all proteins with high homology to T. pseudonana TpMMT were from diatoms, predicted to accumulate low DMSP levels (Supplementary Table 3), supporting this notion. Considering DSYE was found in organisms predicted to be both low and high DMSP accumulators13, it would be difficult to predict an organism’s intracellular DMSP level based on DSYE occurrence (Supplementary Table 3), exemplified by the varied DMSP levels seen in pelagophyte algae with DSYE. Previous research has shown that gene transcript and protein levels were more robust indicators of an organism’s potential DMSP levels13, since these are guided by varying environmental conditions, for example, nitrogen and salinity levels, and govern DMSP synthesis potential, along with substrate availability. Finally, most studies only report the intracellular DMSP levels in producers, which is affected by both DMSP production and turnover, and DMSP production will be dependent on any variability in growth conditions. Therefore, although the different factors discussed here may give clues or indicate gross trends in DMSP production, prediction of a particular organism’s DMSP content is difficult in the absence of direct measurement.
The dsyGD/dsyG and DSYE genes were at different ends of the spectrum for their perceived importance in marine environments. Bacteria with dsyGD/dsyG were not detected in any TARA metagenomic or metatranscriptomic dataset, consistent with them having a negligible role in marine DMSP production. Furthermore, dsyGD/dsyG could not be detected in metagenomic data from Spartina rhizosphere samples in which G. sunshinyii was present67, suggesting that dsyGD may not even be universal in this species. In contrast, DSYE genes, particularly from pelagophyte and chlorophyte algae, were more abundant than DSYB (largely from haptophytes and dinophytes) and orders of magnitude more abundant than TpMMT from diatoms in Earth’s SRF waters. However, DSYE transcripts were approximately twofold less abundant than DSYB transcripts in these samples, which is probably a better reporter of DMSP production. Even with these reduced transcript levels, pelagophyte and chlorophyte algae with DSYE should be considered as potentially important marine DMSP producers, especially given that many of these algae form large blooms, are globally abundant66 and were shown here to accumulate medium to high levels of DMSP. Further work on these algae in the natural environment is vital because they have not received the same attention from DMSP biologists as, for example, haptophyte and dinophyte algae68,69.
Assuming that the known S-methyltransferase genes in microbial DMSP synthesis pathways were the major isoforms, which our analysis of algal transcriptomes implied, it was puzzling why these genes and their transcripts were not more abundant in marine systems. This is an especially relevant question considering the marine ubiquity of DMSP and DMSP catabolic genes, for example, dddP, predicted to be in 5.29% of SRF marine bacteria70. There are still many DMSP producers that lack known DMSP synthesis genes, for example, DMSP-producing plants, macroalgae such as Ulva spp., cyanobacteria such as Trichodesmium and Synechococcus and other bacteria, for example, Marinobacter sp.14, but these are not expected to be major DMSP producers on the same scale as haptophyte, dinophyte and now pelagophyte algae, for instance. It is possible that these phototrophs contain other unidentified isoform MSM enzymes or DMSP synthesis pathways with unknown enzymes. This was proposed for the dinophyte Crypthecodinium cohnii, which has multiple DSYB copies15 but is thought to utilize a Met decarboxylation pathway10,12, for which no genes or enzymes are known. Finally, it is also possible that the DMSP synthesis gene products are more abundant and active than their gene and transcript abundance implies. Further molecular work is required on model marine organisms to address these important questions, combined with more comprehensive environmental quantification of DMSP stocks, synthesis and catabolism rates and of DMSP biosynthetic enzyme abundance.
Methods
Strains, plasmids and culture conditions
Strains, plasmids and primers used in this study are listed in Supplementary Tables 7, 8 and 9. G. sunshinyii, L. aggregata dsyB− mutant strain and R. pomeroyi DSS-3 were grown in YTSS (yeast tryptone sea salts)71 or MBM minimal medium72 (10 mM succinate carbon source, 10 mM NH4Cl nitrogen source and 35 practical salinity units (PSU)) at 30 °C. Where indicated, MBM salinity and/or nitrogen content was adjusted by altering the amount of sea salts (Merck; S9883) or NH4Cl added, respectively. Z. navalis LEGE 11467 was grown in BG-11 medium73 supplemented with varying amounts of sea salts and NaNO3 at 22 °C under 12 h light (50 μmol photons per square metre per second)/12 h dark cycles with 170 rpm shaking. E. coli strains were grown in lysogeny broth (LB) or M9 minimal medium74 at 37 °C. R. leguminosarum J391 was grown in TY or Y medium at 28 °C (ref. 75) with 180 rpm shaking. All eukaryotic algae were grown in F/2 medium with 16 h light (50 μmol photons per square metre per second)/8 h dark cycles, as in Curson et al.15. Where necessary, algal medium was modified according to the requirements of the experimental conditions being tested. All liquid cultures were grown with shaking at 180–200 rpm, unless specified otherwise. Where necessary, antibiotics were added to media at the final concentrations specified as follows: ampicillin 100 µg ml−1, streptomycin 400 µg ml−1, kanamycin 20 µg ml−1, rifampicin 20 µg ml−1, tetracycline 10 µg ml−1 and gentamicin 20 µg ml−1 (or 80 µg ml−1 for L. aggregata dsyB− mutant with pLMB509).
Quantification of MeSH, DMS, DMSHB and DMSP
Gas chromatography (GC) was the primary method used to quantify DMSP and DMSHB. All GC assays involved measurement of either headspace MeSH, as described in Carrión et al.76 or of DMS (either produced directly or through alkaline lysis of DMSP and/or DMSHB), as described in Curson et al.15 for culture-dependent and protein work or as in Williams et al.14 for work on environmental samples. These assays were conducted using a flame photometric detector (Agilent 7890 A GC equipped with a 7693 autosampler) along with a capillary column (HPINNOWax 30 m × 0.320 mm, Agilent Technologies J&W Scientific). The detection limit for headspace DMS was 0.0067 µM DMSP and DMSHB in water and media respectively and 1 µM DMSP in methanol; MeSH was 27 µM in water/media.
DMSP content in C. scabrifolia
C. scabrifolia plants and rhizosphere soil were obtained in a saltern area in Shandong Province, China (120.745° E, 36.454° N). C. scabrifolia plants were carefully uprooted and placed into sterile plastic bags. The plant material was washed to remove sediment and separated into different tissue types (roots and leaves) using ethanol sterilized scissors or tweezers and assayed for DMSP. The C. scabrifolia rhizosphere was sampled, as in Williams et al.14. Briefly, 5 g roots were sampled, and rhizosphere was subjected to vortexing five times to collect the adhered soil. The samples were assayed for DMSP by GC as above and normalized to wet weight.
DMSP synthesis in G. sunshinyii
To infer the G. sunshinyii DMSP synthesis pathway, the cultures were incubated overnight in YTSS, adjusted to an OD600 of 0.3 and washed three times with 35 PSU MBM. The samples were then diluted 1:100 into 5 ml 35 PSU MBM with or without (control) 0.5 mM DMSP synthesis intermediates (l-Met (Merck, M9625), MTOB (Merck, K6000), MTHB (Merck, 55875), DMSHB, DMSP-amine, 3-methylthiopropylamine (Merck, 639095), methylmercaptopropionate (Tokyo Chemical Industry, M0811) and incubated for 24 h at 30 °C. DMSHB and DMSP-amine were synthesized, as in Curson et al.1. Apart from l-Met, all chiral DMSP intermediates were thought to be a 50:50 mixture of d- and l-forms.
To study DMSHB/DMSP accumulation in G. sunshinyii under varied environmental conditions, the cultures were grown under standard (35 PSU, 0.5 mM NH4Cl), low salinity (5 PSU, 0.5 mM NH4Cl), high salinity (50 PSU, 0.5 mM NH4Cl) and high nitrogen (35 PSU, 10 mM NH4Cl) conditions. G. sunshinyii was inoculated into 50 ml YTSS and incubated with shaking at 30 °C overnight. The cultures were then washed three times by centrifuging at 17,000g for 5 min and resuspending in 35 PSU MBM without nitrogen added. A total of 1 ml of washed cells was then inoculated into 10 ml MBM as described for the different conditions and incubated at 30 °C for 24 h. Three biological replicates were prepared for each condition, and DMSP amounts were normalized to protein concentrations determined using the Bradford method, as in Curson et al.1.
To quantify in vitro MSM and DDC activities in G. sunshinyii, 5 ml YTSS overnight cultures were collected by centrifugation at 17,000g for 5 min, washed three times with 1 ml 50 mM Tris–HCl buffer (pH 7.5) and then resuspended in 1 ml 50 mM Tris–HCl buffer. Subsequently, the cells were sonicated (3 × 10 s) on ice using a Markson GE50 Ultrasonic Processor set to an output of 70, then centrifuged at 17,000g for 5 min to pellet the debris. The resultant supernatants (cell-free extracts) were dialysed to remove any pre-existing metabolites, using dialysis tubing (3,500 Da molecular weight cut off; Spectrum Labs) in 2 l of dialysis buffer (20 mM HEPES, 150 mM NaCl, pH 7.5) at 4 °C overnight15. A total of 200 µl of cell-free extracts with nothing added (control) or with 1 mM MTHB plus 1 mM AdoMet (New England Biolabs, B9003S) or just 1 mM DMSHB were placed into GC vials and incubated at 30 °C for 30 min. After incubation, 100 µl 10 M NaOH was added to cell-free extracts and assayed for DMSHB and/or DMSP by GC, as above.
Prediction of G. sunshinyii DMSP synthesis and catabolic genes
The G. sunshinyii genome sequence and protein annotation data were downloaded from the National Center for Biotechnology Information (NCBI) (PRJNA233633) and searched for DMSP synthesis and catabolic proteins using local BLASTp and verified probe sequences (Supplementary Table 10) with an E-value threshold of ≤1 × 10−5, amino acid identity of ≥40% and coverage of ≥70%.
Screening of G. sunshinyii genomic library
A G. sunshinyii genomic library was constructed in the cosmid pLAFR3 (ref. 77), as described in Curson et al.45. Briefly, 2.5 µg of G. sunshinyii high-quality genomic DNA was partially digested with EcoRI, followed by ligation into 1.0 µg of pLAFR3 cosmid DNA that had been fully digested with EcoRI and dephosphorylated. Subsequently, 0.7 µg of ligated DNA was packaged into recombinant λ phage using Gigapack III XL packaging extracts (Agilent Technologies, 200209). The packaged DNA was then transfected into E. coli 803 to produce the G. sunshinyii genomic library. The library comprising 90,000 clones was transferred en masse into the heterologous host R. leguminosarum J391 by conjugation using an E. coli helper strain containing the plasmid pRK2013 (ref. 78). The transconjugants were inoculated into 200 µl Y medium containing 0.5 mM MTHB in 2 ml GC vials, incubated at 30 °C for 48 h and assayed for DMSHB and DMSP by GC analysis as above. The DMSHB and DMSP levels in the headspace were normalized to protein levels, as above. R. leguminosarum J391 with empty pLAFR3 cosmid and media only, with and without MTHB substrate, were used as controls. J391 has DDC activity, so any DMSHB produced through MSM activity would lead to DMSP production1.
Osmotolerance experiments in E. coli strains
E. coli strain MC4100 and FF4169 (otsA−)50,53 (Supplementary Table 7) were used to study osmotolerance conferred by cloned GsdsyGD. The GsdsyGD gene and its promoter region was synthesized and cloned in pUCm-T (by Sangon Biotech, Shanghai Co., Ltd.; Supplementary Table 8) to make pJDT0029 and transformed into E. coli FF4169. The E. coli strains MC4100, FF4169 and FF4169:pJDT0029 were grown in LB medium overnight (in triplicate). All starter cultures were adjusted to an OD600 of 0.3 and washed twice with M63 medium lacking NaCl and sulfur, followed by resuspension in 1 ml M63, as in Summers et al.50. The suspensions were diluted 1:100 in fresh M63 medium (22 mM d-glucose as carbon source and 1 mM MgSO4 as sulfur source) with high salinity (0.5 M NaCl) and DMSP, GB, MTHB or DMSHB at 1 mM final concentration. A total of 0.1 mM IPTG was added to induce expression of GsdsyGD from pJDT0029 in FF4169. The growth was monitored by measuring OD600 using a plate reader (Thermo Scientific, Multiskan GO) every 1 h until stationary phase.The DMSP production was confirmed by GC at the end of each experiment.
Identification of DsyGD, DsyG, DsyD and DSYE homologues
The prokaryotic GsDsyGD, GsDsyG and GsDsyD homologues in the NCBI were identified by BLASTp using an E-value cut-off of 1 × 10−55 and 38–50% amino acid identity (Supplementary Table 1). To identify eukaryotic DSYE and DsyD-like enzymes BLASTP searches (E-value of 1 × 10−55 and ≥70% coverage for GsDsyG and E-value of 1 × 10−5 for DsyD domains) were performed against the predicted proteomes from genomes on the NCBI and the 678 transcriptomes available at MMETSP54 (Supplementary Tables 1 and 2).
Growth of Z. navalis under different conditions
Z. navalis LEGE 11467 (ref. 52) was obtained from the Blue Biotechnology and Ecotoxicology Culture Collection (LEGE-CC) from CIIMAR in Portugal and grown with shaking at 22 °C in 50 ml BG-11 medium at 25 PSU (with 0.5 mM NaNO3 as the nitrogen source), unless otherwise stated, as described in Rippka et al.73. Note that Z. navalis grows as a floating mass or masses in liquid culture. The triplicate samples were then set up by introducing 100 mg of Z. navalis material into 25 ml BG-11 medium with different salinities or nitrate concentrations as follows: standard conditions (25 PSU, 0.5 mM NaNO3), low salinity (5 PSU, 0.5 mM NaNO3), high salinity (50 PSU, 0.5 mM NaNO3) and high nitrogen (25 PSU, 17.65 mM NaNO3). The samples were taken 14 days after inoculation by removing Z. navalis material with sterile forceps to 1.5 ml centrifuge tubes, and the wet weight of material (after removing any residual liquid by pipette) was recorded. The samples were stored at −80 °C until GC and/or nuclear magnetic resonance (NMR) analysis. DMSP, DMSHB or GB amounts were normalized to micrograms of protein (determined by Bradford assay as above).
Quantification of DMSP in Pelagophyceae algae
Cultures of Pelagophyceae algae (Supplementary Table 7) were incubated for 20 days at 22 °C under 16 h light (120 µmol photons per square metre per second)/8 h dark cycles. Subsequently, 4 ml of culture were centrifuged at 6,000g for 10 min, and the pellet was resuspended in 200 µl methanol. The samples were stored at −20 °C for 24 h to allow for extraction of cellular metabolites. The methanol extracts were transferred to GC vials and 100 µl 10 M NaOH was added. The vials were immediately sealed and incubated at 22 °C for 24 h in the dark before DMSP measurements by GC. All experiments were performed in triplicate. The cell numbers in the cultures were quantified using a using a CASY model TT cell counter (Sedna Scientific).
NMR analysis of DMSP, DMSHB and GB
NMR was used to confirm the presence of DMSP/DMSHB and GB in cyano/bacteria and algae and to estimate the concentration and relative levels of these osmolytes. G. sunshinyii, Z. navalis LEGE 11467 and Pelagophyceae algae cultures grown under the conditions described in their corresponding sections were spun down, and the cell pellets were resuspended in 800 μl of deuterium oxide (D2O, Merck; 113366). The samples were then transferred to 2 ml tubes containing 0.1–1.4 mm beads and homogenized using the FastPrep-24 5 G (FP5G, FastPrep system, MP Biomedicals) for three cycles of 40 s at 6.0 m s−1. The samples were centrifuged at 5,000g for 10 min at 4 °C. Subsequently, pyrazine (Sigma-Aldrich) was added at 1 mM final concentration to 500 µl supernatants as internal standard before NMR analysis. The NMR experiments were performed, as in Carrión et al.70, using a double echo excitation sculpting component for water suppression (Bruker library zgesgp) and 2 ms Sinc shaped pulses, 128 scans, relaxation delay of 1 s and acquisition delay of 2 s. All spectra were phased, base-corrected and calibrated for the pyrazine peak at 8.64 ppm. The chemical shift of the methyl groups of GB ((CH3)3N) was at 3.26 ppm (298 K). The methyl groups of DMSP and DMSHB ((CH3)2S) overlap at 2.91 ppm (298 K); therefore, it was not possible to distinguish them at low concentrations by NMR. Thus, the singlet at 2.91 ppm was taken as the sum of the DMSP and DMSHB concentrations (and refer to them as ‘DMSP/DMSHB’ thereinafter). The GB and DMSP/DMSHB concentrations were estimated by using the following equation:
where [A] is the molar concentration of the analyte, I is the absolute integral of either the analyte (A) or pyrazine (P), N is the number of nuclei corresponding to the peak (N = 4 for pyrazine, N = 9 for GB and N = 6 for DMSHB/DMSP) and [P] is the pyrazine molar concentration. These absolute concentrations were then multiplied by the dilution factor derived from manipulation of the initial culture to the NMR tube, divided by the correction factors derived from the calibration curves (2.96 for GB and 2.72 for DMSP/DMSHB) and normalized to cell volume or micrograms of protein. The calibration curves for GB and DMSP/DMSHB were performed using 0.2–1.6 mM standards and 1 mM pyrazine and plotted to obtain straight lines with R2 of 0.99, where the obtained slope was used as the correction factor. The detection limits for GB and DMSP/DMSHB were 10 and 15 µM, respectively. The DMSP/DMSHB concentrations in Z. narvalis samples were below the detection limit; therefore, only estimation of GB levels was possible in these samples.
RNA isolation and RT–qPCR assays
G. sunshinyii was cultured in triplicate under the conditions described in the ‘DMSP synthesis in G. sunshinyii’ section above. Z. navalis LEGE 11467 starter cultures were grown as in ‘Growth of Z. navalis under different conditions’ then inoculated to 50 ml BG-11 medium with 17.65 mM NaNO3 and different salinities for standard (25 PSU), low (5 PSU) and high salinity (50 PSU) and sampled after 14 days. The cell pellets were stored at −80 °C with RNAlater RNA stabilization reagent (Qiagen; 76104) before RNA extraction.
Total RNA from G. sunshinyii and Z. navalis LEGE 11467 cultures was extracted using a Direct-zol RNA Miniprep kit (Zymo Research; R2050) and reverse transcribed with a QuantiTect Reverse Transcription Kit (Qiagen; 205311) following the manufacturer’s instructions. Quantitiative polymerase chain reaction with reverse transcritption (RT–qPCR) assays were performed in triplicate with primers listed in Supplementary Table 9 on an AriaMx Real-Time PCR system (Agilent) using a QuantiTect SYBR Green PCR Kit (Qiagen; 204343) and the following cycling conditions: 95 °C for 3 min, 40 cycles of 95 °C for 20 s, 60 °C for 30 s and 72 °C for 30 s.
In vivo MSM, DDC and MR enzyme assays
Full-length G. sunshinyii dsyGD (including the dsyG methyltransferase and dsyD decarboxylase domains), the separate dsyG and dsyD domain genes and the putative reductase gene were PCR-amplified and cloned into pET-22b (Supplementary Tables 8 and 9). The individual GsdsyG and GsdsyD domain sequences were determined from their homology to Pfam domains (https://www.ebi.ac.uk/interpro/) and to the functional ZndsyG (Supplementary Fig. 2). An existing ATG start codon, corresponding to the penultimate codon of ZndsyG, was used to initiate the GsdsyD domain. For GsdsyG, a stop codon was introduced immediately before this ATG codon. The homologous dsyGD, dsyG, dsyD and DSYE genes were synthesized and cloned into pET-16b or pET-22b (by Sangon Biotech, Shanghai Co., Ltd.; Supplementary Table 8). All the clones were verified by sequencing and transformed into E. coli BL21 (DE3). The transformants were cultured in LB containing ampicillin at 37 °C to an OD600 of 0.8–1.0 and then incubated at 18 °C for 14 h with 0.1–0.4 mM IPTG for in vivo enzyme assays and protein purification work (see the ‘Protein purification’ section). These cells were either incubated with 0.5 mM MTHB or DMSHB and assayed for in vivo MSM or DDC activity (by GC, as above), respectively, or with nothing added for control experiments. Except for GsDsyG, all tested proteins overexpressed in E. coli were seen in the soluble fraction in SDS–polyacrylamide gel electrophoresis analysis (Supplementary Figs. 3 and 9).
To further study the in vivo MSM and DDC activity of dsyGD, dsyG, dsyD, DSYE and homologous genes, these were cloned into the wide host range taurine-inducible expression plasmid pLMB509 (ref. 79) (Supplementary Table 8). These plasmids were conjugated into the L. aggregata dsyB−mutant, which makes no DMSP and/or R. pomeroyi DSS-3 (for dsyD clones as it cannot produce DMSP from DMSHB1) using the helper plasmid pRK2013 (ref. 78), as described in Curson et al.1. For MSM and DDC activity assays, triplicate cultures were grown in YTSS at 30 °C for 24 h. The cultures were then adjusted to an OD600 of 0.3, washed three times with 35 PSU MBM and diluted 1:100 into 5 ml MBM medium with 5 mM taurine (Sigma-Aldrich, T0625). Where indicated, 0.5 mM MTHB or DMSHB was added as the substrate, and the samples were incubated at 30 °C for 24 h before the accumulation of DMSHB/DMSP was monitored by GC as above.
Protein purification
E. coli BL21 (DE3) cells overexpressing DsyGD, DsyG, DsyD, DSYE and the G. sunshinyii putative reductase (Supplementary Table 8 and Supplementary Figs. 3 and 9) were collected by centrifugation (20 min, 7,500g at 4 °C), washed and resuspended in 25 mM Tris–HCl (pH 8.0), 150 mM NaCl. Overexpressed recombinant proteins were purified by Ni2+–NTA (nitrilotriacetic acid) affinity chromatography (GE healthcare), followed by gel filtration on a Superdex200 column (Cytiva), as in Li et al.80. The purified proteins were flash frozen in liquid nitrogen and stored at −80 °C until required.
In vitro MSM, DDC and MR enzyme assays
Where appropriate, recombinant DsyGD, DsyD, DSYE, candidate MR and homologous proteins were assayed for MSM, DDC and MR activity, as in Curson et al.1.
For in vitro MSM activity, 0–1,000 μM MTHB, 10–1,000 μM AdoMet and 0.1 μM purified DsyGD/DsyG/DSYE were mixed in a total volume of 100 μl reaction buffer containing 100 mM Tris–HCl (pH 7.0) and incubated at 25 °C for 10 min in triplicate. A total of 15 μl of 20% HCl was added to stop the reactions. The reaction buffers with no enzymes were used as negative controls. MSM activity was determined by detecting S-adenosyl-homocysteine (AdoHcy) produced from AdoMet demethylation by HPLC, as described in Li et al.44.
For in vitro DDC activity, 0.5–3 mM DMSHB and 0.1 μM purified DsyGD or DsyD domain proteins were mixed in a total volume of 100 μl with reaction buffer (100 mM Tris–HCl (pH 7.0)), before incubation at 25 °C for 10 min in triplicate. A total of 15 μl of 20% HCl was added to stop the reaction. In vitro DDC activity of DsyGD and DsyD was monitored via the HPLC detection of acrylate produced from alkaline hydrolysis of the DMSP reaction product81,82.
To determine the optimal temperature of DsyGD and DSYE for MTHB, the reaction mixtures were incubated at 10–60 °C. The optimum pH values of purified enzymes on MTHB were examined at their optimal temperature using Britton–Robinson buffer at pH 4–11, as in Peng et al.83.
Kinetic parameters of DsyGD and DSYE for MTHB, AdoMet and DMSHB (for DsyGD) were determined by non-linear analysis based on the initial rates with 0–20,000 μM MTHB, 0–250 μM AdoMet or 500–3,000 μM DMSHB at the optimal temperature and pH, as described in Peng et al.83.
For in vitro MR activity, 1 mM MTOB and 0.25 mM NADPH were mixed in a total volume of 2 ml reaction buffer (10 mM Tris–HCl, pH 8.0) in triplicate and incubated at 30 °C. The reactions were initiated by the addition of 1 μM purified reductase enzyme and MR activity was monitored by NADPH reduction at 340 nm using a V550 ultraviolet–visible light spectrophotometre (Jasco) at 0, 15 and 180 min after enzyme addition. The reaction mixtures with no reductase enzyme were used as negative controls.
Distribution of DMSP synthesis genes in Tara Oceans datasets
To study the relative abundance and distribution of DMSP synthesis genes/transcripts in Tara Oceans OM-RGC_v2 and MATOU datasets65, a hidden Markov model profile of reported DMSP synthesis enzymes and experimentally ratified DsyGD, DsyG and DSYE proteins (Supplementary Table 10 and Supplementary Data 1) was created using HMMER tools (v.3.3, http://hmmer.janelia.org/)84. The hidden Markov model searches were performed on the online webserver Ocean Gene Atlas65 with default settings and an E-value of 1 × 10−30. The resultant sequences were further verified by BLASTp analysis. Only homologues with ≥40% amino acid identity and ≥70% coverage to ratified sequences were counted. In metagenomic samples, the relative abundance of eukaryotic DMSP synthesis genes was normalized to the relative abundance of ACTB, which encodes β-actin, except for DSYE, which was also normalized to recA. The relative abundance of prokaryotic DMSP synthesis genes was normalized to the relative abundance of recA85. In metatranscriptomic datasets, the relative abundance of DMSP synthesis transcripts is expressed as percentage of mapped reads. Finally, the biogeographic distribution of DMSP synthesis genes/transcripts was plotted with R (v. 4.0.3) using scatterpie and ggplot2 (ref. 86).
Relative abundance of dsyGD in terrestrial metagenomes
The relative abundance of dsyGD in metagenomic datasets of S. alterniflora, Rhizophora stylosa and mangrove sediment from the Chinese National Genomics Data Center GSA database (PRJCA002729) was analysed, as in Liu et al.85. Only homologues with ≥40% amino acid identity and ≥70% coverage to ratified sequences (Supplementary Table 10 and Supplementary Data 1) were counted.
Phylogenetic analysis of DMSP synthesis enzymes
All prokaryotic DsyB, MmtN, DsyGD, DsyG, DsyG-like (lacking MSM function) and DsyD sequences, and eukaryotic DSYB, TpMMT and DSYE sequences listed in Supplementary Table 10 were aligned in MAFFT version 7 (ref. 87) using default settings, then visually checked. The S-methyltransferase or decarboxylase domains sequences of these enzymes were used to construct maximum-likelihood phylogenetic trees using MEGA version X (ref. 88) (Fig. 2 and Supplementary Fig. 6). The maximum-likelihood phylogenetic trees were visualized and annotated using the Interactive Tree Of Life version 6.6 (ref. 89).
Statistical methods
All measurements of metabolites, for example, DMSP, DMSHB and DMS levels (in bacterial strains or enzyme assays) were based on the mean of three biological replicates per strain/condition tested, and the error bars indicate standard deviations. For RT–qPCR assays, the results shown represent the mean of three biological replicates and three technical replicates with their respective standard deviations. To identify statistically significant differences between standard and experimental conditions in Figs. 1b and 3a,b, Supplementary Fig. 5a,c and Supplementary Fig. 7a (P < 0.05), a two-sided independent Student’s t-test was applied to the data. For Supplementary Fig. 11a–d (P < 0.05), a Wilcoxon test was applied to the data.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Accession numbers of sequences from the NCBI and MMETSP analysed in this study are listed in Table 1 and Supplementary Tables 1 and 10. Source data are provided with this paper.
References
Curson, A. R. J. et al. Dimethylsulfoniopropionate biosynthesis in marine bacteria and identification of the key gene in this process. Nat. Microbiol. 2, 17009 (2017).
Gillian, M. & Gunter, O. K. Algal production of dimethyl sulfideand its atmospheric role. J. Phycol. 33, 889–896 (1997).
Ksionzek, K. B. et al. Dissolved organic sulfur in the ocean: biogeochemistry of a petagram inventory. Science 354, 456–459 (2016).
Vallina, S. M. & Simo, R. Strong relationship between DMS and the solar radiation dose over the global surface ocean. Science 315, 506–508 (2007).
Charlson, R. J., Lovelock, J. E., Andreae, M. O. & Warren, S. G. Oceanic phytoplankton atmospheric sulpher cloud albedo and climate. ISME J. 326, 655–661 (1987).
Yoch, D. C. Dimethylsulfoniopropionate: its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl. Environ. Microbiol. 68, 5804–5815 (2002).
Sunda, W., Kieber, D. J., Kiene, R. P. & Huntsman, S. An antioxidant function for DMSP and DMS in marine algae. Nature 418, 317–320 (2002).
Seymour, J. R., Simo, R., Ahmed, T. & Stocker, R. Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science 329, 342–345 (2010).
Zheng, Y. F. et al. Bacteria are important dimethylsulfoniopropionate producers in marine aphotic and high-pressure environments. Nat. Commun. 11, 4658 (2020).
Carrión, O. et al. Molecular discoveries in microbial DMSP synthesis. Adv. Microb. Physiol. 83, 59–116 (2023).
Nevitt, G. A. The neuroecology of dimethyl sulfide: a global-climate regulator turned marine infochemical. Integr. Comp. Biol. 51, 819–825 (2011).
Hopkins, F. E. et al. The biogeochemistry of marine dimethylsulfide. Nat. Rev. Earth Env. 4, 361–376 (2023).
McParland, E. L. et al. The role of differential DMSP production and community compositionin predicting variability of global surface DMSP concentrations. Limnol. Oceanogr. 64, 757–773 (2019).
Williams, B. T. et al. Bacteria are important dimethylsulfoniopropionate producers in coastal sediments. Nat. Microbiol. 4, 1815–1825 (2019).
Curson, A. R. J. et al. DSYB catalyses the key step of dimethylsulfoniopropionate biosynthesis in many phytoplankton. Nat. Microbiol. 3, 430–439 (2018).
Kageyama, H., Tanaka, Y., Shibata, A., Waditee-Sirisattha, R. & Takabe, T. Dimethylsulfoniopropionate biosynthesis in a diatom Thalassiosira pseudonana: identification of a gene encoding MTHB-methyltransferase. Arch. Biochem. Biophys. 645, 100–106 (2018).
Trottmann, F. et al. Sulfonium acids loaded onto an unusual thiotemplate assembly line construct the cyclopropanol warhead of a Burkholderia virulence factor. Angew. Chem. Int. Ed. 59, 13511–13515 (2020).
Liao, C. & Seebeck, F. P. In vitro reconstitution of bacterial DMSP biosynthesis. Angew. Chem. Int. Ed. 58, 3553–3556 (2019).
McParland, E. L. et al. Evidence for contrasting roles of dimethylsulfoniopropionate production in Emiliania huxleyi and Thalassiosira oceanica. New Phytol. 226, 396–409 (2020).
Paquet, L. et al. Accumulation of the compatible solute 3-dimethylsulfoniopropionate in sugarcane and its relatives, but not other gramineous crops. Funct. Plant Biol. 21, 37–48 (1994).
Otte, M. L. & Morris, J. T. Dimethylsulphoniopropionate (DMSP) in Spartina alterniflora Loisel. Aquat. Bot. 48, 239–259 (1994).
Hanson, A. D., Rivoal, J., Paquet, L. & Gage, D. A. Biosynthesis of 3-dimethylsulfoniopropionate in Wollastonia biflora (L.) DC. (Evidence that S-methylmethionine is an intermediate). Plant Physiol. 105, 103–110 (1994).
James, F., Paquet, L., Sparace, S. A., Gage, D. A. & Hanson, A. D. Evidence implicating dimethylsulfoniopropionaldehyde as an intermediate in dimethylsulfoniopropionate biosynthesis. Plant Physiol. 108, 1439–1448 (1995).
Sun, H. et al. Spatiotemporal distribution of bacterial dimethylsulfoniopropionate producing and catabolic genes in the Changjiang Estuary. Environ. Microbiol. 23, 7073–7092 (2021).
O’Brien, J. et al. Biogeographical and seasonal dynamics of the marine Roseobacter community and ecological links to DMSP-producing phytoplankton. ISME Commun. 2, 16 (2022).
O’Brien, J. et al. The microbiological drivers of temporally dynamic dimethylsulfoniopropionate cycling processes in Australian coastal shelf waters. Front. Microbiol. 13, 894026 (2022).
Chung, E. J., Park, J. A., Jeon, C. O. & Chung, Y. R. Gynuella sunshinyii gen. nov., sp. nov., an antifungal rhizobacterium isolated from a halophyte, Carex scabrifolia Steud. Int. J. Syst. Evol. Microbiol. 65, 1038–1043 (2015).
Mavrodi, O. V. et al. Rhizosphere microbial communities of Spartina alterniflora and Juncus roemerianus from restored and natural tidal marshes on Deer Island, Mississippi. Front. Microbiol. 9, 03049 (2018).
Dacey, J. W. H., King, G. M. & Wakeham, S. G. Factors controlling emission of dimethylsulfide from salt marshes. Nature 330, 643–645 (1987).
Steudler, P. A. & Peterson, B. J. Contribution of gaseous sulphur from salt marshes to the global sulphur cycle. Nature 311, 455–457 (1984).
Kocsis, M. G. et al. Dimethylsulfoniopropionate biosynthesis in Spartina alterniflora1. Evidence that S-methylmethionine and dimethylsulfoniopropylamine are intermediates. Plant Physiol. 117, 273–281 (1998).
Challenger, F. & Simpson, M. I. A precursor of the dimethyl sulphide evolved by Polysiphonia fastigiata; dimethyl-beta-propiothetine (dimethyl-beta-carboxyethylsulphonium hydroxide) and its salts. Biochem. J. 41, 1591–1597 (1947).
Rolando, J. L., Kolton, M., Song, T. & Kostka, J. E. The core root microbiome of Spartina alterniflora is predominated by sulfur-oxidizing and sulfate-reducing bacteria in Georgia salt marshes. Microbiome 10, 37 (2022).
McNeil, S. D., Nuccio, M. L. & Hanson, A. D. Betaines and related osmoprotectants. Targets for metabolic engineering of stress resistance. Plant Physiol. 120, 945–950 (1999).
Kiehn, W. M. & Morris, J. T. Variability in dimethylsulfoniopropionate (DMSP) concentrations in Spartina alterniflora and the effect on Littoraria irrorata. Mar. Ecol. Prog. Ser. 406, 47–55 (2010).
Ueoka, R. et al. Genome mining of oxidation modules in trans-acyltransferase polyketide synthases reveals a culturable source for lobatamides. Angew. Chem. Int. Ed. 59, 7761–7765 (2020).
Wang, Y., Qin, Z., Fan, L. & Zhao, L. Structure-function analysis of Gynuella sunshinyii chitosanase uncovers the mechanism of substrate binding in GH family 46 members. Int. J. Biol. Macromol. 165, 2038–2048 (2020).
Helfrich, E. J. N. et al. Evolution of combinatorial diversity in trans-acyltransferase polyketide synthase assembly lines across bacteria. Nat. Commun. 12, 1422 (2021).
Todd, J. D. et al. The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some Ascomycete fungi. Environ Microbiol 11, 1376–1385 (2009).
Todd, J. D. et al. DddQ, a novel, cupin-containing, dimethylsulfoniopropionate lyase in marine Roseobacters and in uncultured marine bacteria. Environ. Microbiol. 13, 427–438 (2011).
Todd, J. D., Kirkwood, M., Newton-Payne, S. & Johnston, A. W. B. DddW, a third DMSP lyase in a model Roseobacter marine bacterium, Ruegeria pomeroyi DSS-3. ISME J. 6, 223–226 (2012).
Curson, A. R. et al. DddY, a periplasmic dimethylsulfoniopropionate lyase found in taxonomically diverse species of Proteobacteria. ISME J. 5, 1191–1200 (2011).
Curson, A. R. et al. Identification of genes for dimethyl sulfide production in bacteria in the gut of Atlantic herring (Clupea harengus). ISME J. 4, 144–146 (2010).
Li, C. Y. et al. A novel ATP dependent dimethylsulfoniopropionate lyase in bacteria that releases dimethyl sulfide and acryloyl-CoA. eLife 10, e64045 (2021).
Curson, A. R. J., Rogers, R., Todd, J. D., Brearley, C. A. & Johnston, A. W. B. Molecular genetic analysis of a dimethylsulfoniopropionate lyase that liberates the climate-changing gas dimethylsulfide in several marine alpha-proteobacteria and Rhodobacter sphaeroides. Environ. Microbiol. 10, 757–767 (2008).
Uria, A. et al. Identification of the algal dimethyl sulfide-releasing enzyme: a missing link in the marine sulfur cycle. Science 348, 1466–1469 (2015).
Todd, J. D. et al. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science 315, 666–669 (2007).
Erinn, C. H. et al. Bacterial taxa that limit sulfur flux from the ocean. Science 314, 649–651 (2006).
Gage, D. A. et al. A new route for synthesis of dimethylsulphoniopropionate in marine algae. Nature 387, 891–894 (1997).
Summers, P. S. et al. Identification and stereospecificity of the first three enzymes of 3-dimethylsulfoniopropionate biosynthesis in a chlorophyte algal. Plant Physiol. 116, 369–378 (1998).
Miyazaki, J. H. & Yang, S. F. Metabolism of 5-methylthioribose to methionine. Plant Physiol. 84, 277–281 (1987).
Hentschke, G. S. et al. Zarconia navalis gen. nov., sp. nov., Romeriopsis navalis gen. nov., sp. nov. and Romeriopsis marina sp. nov., isolated from inter- and subtidal environments from northern Portugal. Int. J. Syst. Evol. Microbiol. 72, 005552 (2022).
Giaever, H. M., Styrvold, O. B., Kassen, I. & Strøm, A. R. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J. Bacteriol. 170, 2841–2849 (1988).
Keeling, P. J. et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12, e1001889 (2014).
Keller, M. D. Dimethyl sulfide production and marine phytoplankton: the importance of species composition and cell size. Biol. Oceanogr. 6, 375–382 (1989).
Corn, M., Belviso, S., Partensky, F., Simon, N. & Christaki, U. in Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds (eds Kiene, R. P. et al.), Ch. 17, 191–201 (Springer, 1996).
Demir-Hilton, E. et al. Global distribution patterns of distinct clades of the photosynthetic picoeukaryote Ostreococcus. ISME J. 5, 1095–1107 (2011).
Schaffelke, B. et al. Blooms of Chrysocystis fragilis on the Great Barrier Reef. Coral Reefs 23, 514 (2004).
Gobler, C. J. et al. Niche of harmful alga Aureococcus anophagefferens revealed through ecogenomics. Proc. Natl Acad. Sci. USA 108, 4352–4357 (2011).
Gobler, C. J. & Sunda, W. G. Ecosystem disruptive algal blooms of the brown tide species, Aureococcus anophagefferens and Aureoumbra lagunensis. Harmful Algae 14, 36–45 (2012).
Guerin, N. et al. Genomic adaptation of the picoeukaryote Pelagomonas calceolata to iron-poor oceans revealed by a chromosome-scale genome sequence. Commun. Biol. 5, 983 (2022).
McParland, E. L. et al. DMSP synthesis genes distinguish two types of DMSP producer phenotypes. Environ. Microbiol. 23, 1656–1669 (2021).
Kocsis, M. G. & Hanson, A. D. Biochemical evidence for two novel enzymes in the biosynthesis of 3-dimethylsulfoniopropionate in Spartina alterniflora. Plant Physiol. 123, 1153–1161 (2000).
Keller, M. D., Bellows, W. K. & Guillard, R. R. L. in Biogenic Sulfur in the Environment (eds Saltzman, E. S. and Cooper, W. J.), Ch. 11, 167–182 (American Chemical Society, 1989).
Villar, E. et al. The Ocean Gene Atlas: exploring the biogeography of plankton genes online. Nucleic Acids Res. 46, 289–295 (2018).
Gobler, C. J. et al. The central role of selenium in the biochemistry and ecology of the harmful pelagophyte, Aureococcus anophagefferens. ISME J. 7, 1333–1343 (2013).
Mo, S. et al. Integration of microbial transformation mechanism of polyphosphate accumulation and sulfur cycle in subtropical marine mangrove ecosystems with Spartina alterniflora invasion. Microb. Ecol. 85, 478–494 (2023).
Van Alstyne, K. L., Koellermeier, L. & Nelson, T. A. Spatial variation in dimethylsulfoniopropionate (DMSP) production in Ulva lactuca (Chlorophyta) from the Northeast Pacific. Marine Biol. 150, 1127–1135 (2006).
Kessler, R. W. et al. Macroalgal–bacterial interactions: role of dimethylsulfoniopropionate in microbial gardening by Ulva (Chlorophyta). Mol Ecol. 27, 1808–1819 (2018).
Carrión, O. et al. DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms. Nat. Microbiol. 8, 2326–2337 (2023).
González, J. M., Whitman, W. B., Hodson, R. E. & Moran, M. A. Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture. Appl. Environ. Microbiol. 62, 4433–4440 (1996).
Baumann, P. & Baumann, L. in The Prokaryotes: A Handbook on Habitats, Isolation and Identification of Bacteria (eds Starr, M. P., Stolp, H., Trüper, H. G., Balows, A. & Schlegel, H. G.) 1302–1331 (Springer, 1981).
Rippka, R. Isolation and purification of cyanobacteria. Methods Enzymol. 167, 3–27 (1988).
Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular Cloning, A Laboratory Manual 2nd edn Vol. 3 (Cold Spring Harbor Laboortory Press, 1989).
Beringer, J. E. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84, 188–198 (1974).
Carrión, O. et al. A novel pathway producing dimethylsulphide in bacteria is widespread in soil environments. Nat. Commun. 6, 6579 (2015).
Staskawicz, B., Dahlbeck, D., Keen, N. & Napoli, C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169, 5789–5794 (1987).
Figurski, D. H. & Helimshi, H. D. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl Acad. Sci. USA 76, 1648–1652 (1979).
Tett, A. J., Rudder, S. J., Bourdes, A., Karunakaran, R. & Poole, P. S. Regulatable vectors for environmental gene expression in Alphaproteobacteria. Appl. Environ. Microb. 78, 7137–7140 (2012).
Li, C. Y. et al. Mechanistic insights into the key marine dimethylsulfoniopropionate synthesis enzyme DsyB/DSYB. mLife 1, 114–130 (2022).
Kuek, F. W. I. et al. DMSP production by coral-associated bacteria. Front. Mar. Sci. 9, 869574 (2022).
Peng, M. et al. Structure–function analysis indicates that an active-site water molecule participates in dimethylsulfoniopropionate cleavage by DddK. Appl. Environ. Microbiol. 85, e03127–18 (2019).
Peng, M. et al. Insights into methionine S-methylation in diverse organisms. Nat. Commun. 13, 2947 (2022).
Wheeler, T. J. & Eddy, S. R. nhmmer: DNA homology search with profile HMMs. Bioinformatics 29, 2487–2489 (2013).
Liu, J. et al. Oceanospirillales containing the DMSP lyase DddD are key utilisers of carbon from DMSP in coastal seawater. Microbiome 10, 110 (2022).
R Core Team. R: a language and environment for statistical computing. (R Foundation for Statistical Computing, 2018); https://www.R-project.org/
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, 293–296 (2021).
Acknowledgements
We acknowledge Y.R. Chung from Gyeongsang National University for kindly providing the G. sunshinyii strain and Y. Zheng and J. Liu for experimental advice. We also thank G. Yang from the Ocean University of China for help with the sampling and identification of Carex scabrifolia. Axenic O. tauri was kindly provided by V. Jackson and A. Monier at the University of Exeter. This work was funded by the National Natural Science Foundation of China (92251303 and 32370118, PI (Principal Investigator): X.-H.Z.), the Marine S&T Fund of Shandong Province for Laoshan Laboratory (2022QNLM030004-3, Co-PI: X.-H.Z.), the Fundamental Research Funds for the Central Universities (202172002, PI: X.-H.Z.), the European Union’s Horizon 2020 Research and Innovation programme under grant agreement no. 952374 (PI: P.N.L.) and the Fundação para a Ciência e a Tecnologia, Portugal, through the strategic funding grant UIDP/04423/2020 and UIDB/04423/2020 (PI: P.N.L.), Biotechnology and Biological Sciences Research Council, United Kingdom (BB/X005968, PI: J.D.T.), Natural Environmental Research Council, United Kingdom (NE/P012671, NE/S001352, NE/X000990, PI: J.D.T.) and NE/X014428 with D.L.S. and the Leverhulme trust (RPG-2020-413, PI: J.D.T.). We acknowledge funding support from the BBSRC Norwich Research Park Doctoral Training Partnership programme (BB/2244848, K.S.W.) and (BB/M011216/1, L.H.), UKRI for funding via a Future Leaders Fellowship (MR/T044020/1, M.W.), the National Natural Science Foundation of China (32330001, PI: Y.-Z.Z.), Shandong Provincial Natural Science Foundation (ZR2022QD003, J.L.) and the Taishan Scholars Program of Shandong Province, China (tsqn202306092, PI: C.-Y.L.).
Author information
Authors and Affiliations
Contributions
J.D.T. and X.-H.Z. conceived and designed all experiments, analysed the data and wrote the paper. J.W. wrote the paper, designed all of the experiments and performed or contributed to all of the experiments, analysed all the data and prepared figures and tables. A.R.J.C. performed the following experiments: dsyGD cloning for protein expression, GC assays of DMSP and DMSHB production by Z. navalis under different conditions and by G. sunshinyii with different synthesis intermediates and under different growth conditions, as well as provided advice on genomic library construction. S.Z. performed G. sunshinyii RNA isolation and dsyG RT–qPCR. J.L. performed G. sunshinyii RT–qPCR and performed critical revision of the pictures. A.R.V., P.N.L., A.R.J.C. and O.C. performed Z. navalis LEGE 11467 growth experiments. A.R.J.C. and O.C. wrote the paper. K.S.W. performed the G. sunshinyii and Z. navalis LEGE 11467 protein assays for normalizing DMSP production. P.P.L.R. performed the phytoplankton growth experiments and DMSP production assays. S.M. and M.W. performed the NMR detection and analysis. L.H. performed the O. tauri DMSP production measurements. X.-Y.Z. performed the Z. navalis LEGE 11467 dsyG RT–qPCR. C.-Y.L. and Y.-Z.Z. performed protein purification and activity assays. X.-D.W. and M.Z. performed the purified DsyGD, DsyG, DsyD protein and activity assay experiments. Q.-Y.D. and Y.W. performed protein purification and enzymatic assays. D.J.L.-S. performed critical revision of the manuscript. All authors edited and approved the manuscript. We acknowledge the Chinese Scholarship Council (CSC) who funded the visitations and/or PhD’s of JW, SZ, XYZ in JDT’s lab at UEA.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Frederik De Boever and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1, 4 and 7–10 and Figs. 1–12.
Supplementary Data 1
Amino acid sequences of proteins tested for functionality in this study.
Supplementary Table 1
Supplementary Tables 2, 3, 5 and 6.
Supplementary Data 5
Source data for Supplementary Figs. 1.
Supplementary Data 5
Source data for Supplementary Figs. 5.
Supplementary Data 5
Source data for Supplementary Figs. 7.
Supplementary Data 5
Source data for Supplementary Figs. 8.
Supplementary Data 5
Source data for Supplementary Figs. 11.
Source data
Source Data Fig. 1
DMSP production data from three independent biological replicates are displayed in Fig. 1.
Source Data Fig. 3
Data from three independent biological replicates in experiments are shown in Fig. 3.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, J., Curson, A.R.J., Zhou, S. et al. Alternative dimethylsulfoniopropionate biosynthesis enzymes in diverse and abundant microorganisms. Nat Microbiol 9, 1979–1992 (2024). https://doi.org/10.1038/s41564-024-01715-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-024-01715-9
- Springer Nature Limited