Abstract
Copy number variations (CNVs) are commonly reported in STRC, the causal gene for DFNB16. Various techniques are used clinically for CNV detection, and droplet digital PCR (ddPCR) provides highly precise absolute quantification of DNA copy number. We aimed to validate the feasibility and efficiency of ddPCR in combination with long-range PCR (LR-PCR) in identifying CNVs and mutations in STRC. Additionally, we determined the frequency of CNVs and mutations in STRC in Japanese patients with mild-to-moderate hearing loss. We evaluated 84 unrelated Japanese patients with mild-to-moderate bilateral idiopathic or autosomal recessive nonsyndromic sensorineural hearing loss. The ratio of STRC copy number to the copy number of the internal control RPP30 ranged from 0.949 to 1.009 (0.989 ± 0.017) in 77 patients; it ranged from 0.484 to 0.538 (0.509 ± 0.024) in five patients and was 0.000 in two patients, indicating heterozygous and homozygous deletions, respectively. The copy number deletion prevalence rates were 7.7% and 0.9% in the patients and healthy controls, respectively. In combination with LR-PCR, ddPCR revealed that at least three patients (3.6%) had STRC-related hearing loss. Detecting STRC CNVs by ddPCR was rapid, precise, and cost-effective and facilitated the identification of STRC CNVs.
Similar content being viewed by others
Introduction
Copy number variations (CNVs), defined as the presence of variable copy numbers of specific DNA segments, can be associated with a wide range of disorders. The most recent study has estimated that CNVs constitute 4.8–9.5% of the human genome and comprise a greater fraction of nucleotides than single nucleotide variants (SNVs). Specifically, in bilateral sensorineural hearing loss (SNHL), CNVs were identified in ~20% of all deafness-causing genes and found in ~15% of patients undergoing genetic testing1,2,3. Until recently, real-time quantitative PCR (qPCR) assays and microarray hybridization were the two main methods used to determine CNVs in the genome. Conversely, droplet digital PCR (ddPCR) allows for high-precision absolute quantification of DNA copy number. During ddPCR, each sample is partitioned into several tens of thousands of nanoliter-sized droplets prior to amplification. PCR amplification proceeds to the reaction endpoint, which is followed by the determination of whether every droplet in a well is positive or negative for both the target and the reference. The fluorescence intensity in each droplet is measured, and those above the threshold are counted as positive, thus providing a digital yes/no result. ddPCR overcomes several limitations of qPCR and microarray techniques for CNV analysis.
STRC (MIM: 606440) encodes the extracellular structural protein stereocilin, which is necessary for proper hair cell function and is the causal gene for DFNB164,5. Mutations in STRC, commonly reported as CNVs, were described mainly in association with bilateral mild-to-moderate hearing loss6,7. With ~1–1.6% of individuals in the general population carrying a heterozygous STRC deletion8,9,10, the estimated incidence of STRC-associated hearing loss is 1 in 16,0009. Numerous studies suggest that DFNB16 constitutes a significant proportion of the genetically heterogeneous SNHL, and STRC is increasingly recognized as one of the most significant contributors to autosomal recessive nonsyndromic SNHL.
STRC contains 29 exons encompassing approximately 19 kb and is tandemly duplicated with the coding sequence of the second copy, the pseudo-STRC (pSTRC) gene4. The first half of the functional STRC copy is 100% identical to pSTRC. Over the entire locus, STRC and pSTRC are 98.9% homologous across exons and introns7. Therefore, pathogenic SNVs identified by targeted sequences across the entire STRC coding region should always be differentiated from those derived from the amplification products of pSTRC. Conventional direct PCR and short-read massively parallel sequencing (MPS) are unable to provide consistently reliable data on STRC. In fact, some variants previously reported as pathogenic likely reflect codetection with pseudogenes rather than causal mutations in the STRC coding sequence4. In this study, we present rapid, cost-effective, and precise methods for the diagnosis of STRC-related hearing loss by ddPCR and long-range PCR (LR-PCR).
Methods
Patients
We analyzed archival DNA samples from 84 unrelated Japanese patients (51 females and 33 males; age range, 5–77 years; mean ± standard deviation, 40.02 ± 18.5 years) with mild-to-moderate SNHL who visited the Department of Otolaryngology at Tokyo Medical and Dental University Medical Hospital between April 2000 and September 2010 and who were diagnosed with idiopathic or autosomal recessive nonsyndromic bilateral SNHL11. None of the patients were from consanguineous families. A total of 107 Japanese individuals with normal hearing were included as controls. Based on Sanger sequencing results, we previously excluded pathogenic mutations in the coding sequences of GJB2 and Mt-155511. This study was approved by the research ethics committee of Tokyo Medical and Dental University.
Screening of CNVs with ddPCR
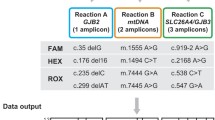
DNA integrity and quality were verified by a NanoDrop spectrophotometer and agarose gel electrophoresis. The ratio of absorbances at 260 nm and 280 nm for each DNA sample was ≥1.8, and all samples were diluted at a concentration of 10 ng/μl by pure water. CNVs were detected by ddPCR with standard protocols using the QX100TM ddPCR system (Bio-Rad, Hercules, CA). Briefly, the master mix for ddPCR included 1 × ddPCR supermix for probes (no dUTP; Bio-Rad), 1.0 μM primer, and 0.25 μM probe (Integrated DNA Technologies, Skokie, IL, USA), together with 20 ng sample DNA. Primers and a probe were designed for intron 23 of STRC (Fig. 1, Table 1). This region was chosen as one of the few regions of the STRC genomic locus with base pairs that differ from those of pSTRC and therefore allows for unique probe sequences. To define the extent of STRC CNVs, additional primers and probes recognizing approximately 10-kbp upstream and 39-kbp downstream sequences of STRC were also designed (Fig. 1, Table 1). Primers and a probe were also designed for RPP30 (Fig. 1, Table 1) as the internal control. A black hole quencher was used in combination with the fluorescent dye reporters FAM and HEX. The samples were thoroughly mixed and transferred to a DG8 cartridge for a QX100TM/QX200 droplet generator (Bio-Rad). Droplet generation oil for probes (Bio-Rad) was then added to the cartridge, which was placed into the QX200 Droplet GeneratorTM (Bio-Rad). After droplet generation, the droplets were carefully transferred to a twin-tec semi-skirted 96-well PCR plate (Eppendorf AG, Hamburg, Germany), and the plate was sealed for 5 s at 180 °C using a PX1 PCR plate sealer (Bio-Rad). Subsequent amplification was performed in a C1000 Touch thermal cycler (Bio-Rad) with a ramp rate of 2 °C/sec. First, the enzyme was activated at 95 °C for 10 min, followed by 40 cycles of denaturation at 94 °C for 30 s and 55 °C for 1 min. The enzyme was deactivated at 98 °C (10 min), and the reaction was held at 4 °C. Droplets were read using a QX200 droplet reader TM (Bio-Rad), and the ddPCR data were analyzed using the Quantasoft software program version 1.7.4 (Bio-Rad). Manual thresholds were applied for both STRC and RPP30.
LR-PCR-based STRC sequencing
To exclude pseudogene sequences, two LR-PCR products were generated for the subsequent nested PCR. Sequences of primers that specifically amplify STRC but not pSTRC were obtained from the literature7,12. LR-PCR was performed with the Qiagen Long-Range PCR Kit (Qiagen, Hilden, Germany) using cycling profiles. Amplification products were loaded and run on a 0.6% agarose gel, and the fragments of appropriate DNA sizes were extracted from the gel using NucleoSpin® gel and a PCR clean-up kit (Macherey-Nagel, Dueren, Germany). Nested PCRs and sequencing continued after LR-PCR products for a portion of STRC exon 16, which were used for verification of amplification from only the STRC sequence. PCR products were purified using a QIAquick PCR Purification Kit (Qiagen) and were directly sequenced using the Applied Biosystems Prism BigDye Terminator Cycle Sequencing Ready Kit and an ABI Prism 3130xl Genetic Analyzer.
Results
STRC CNVs and SNVs
First, we performed ddPCR to identify STRC CNVs in 84 patients with SNHL and 107 healthy controls. Among the patients with SNHL, the STRC/RPP30 copy number ratio ranged from 0.949 to 1.009 (0.989 ± 0.017) in 77 patients and from 0.484 to 0.538 (0.509 ± 0.024) in five patients; the ratio was 0.000 in two patients. The ratios of 0.509 and 0.000 likely indicated heterozygous and homozygous deletions, respectively (Fig. 2). Accordingly, seven of the 84 patients carried at least one STRC copy loss, which was detected in only one of the 107 healthy controls as a heterozygous deletion (Table 2). The samples from those individuals with STRC deletions showed the same deletion pattern by both the 10-kbp upstream and 39-kbp downstream probes. The prevalence of copy number deletion among the patients with mild-to-moderate hearing loss was significantly higher than that among the healthy controls (7.7% vs. 0.9%, p < 0.05, Student’s t test).
All STRC/RPP30 copy number ratios are arranged in descending order and classified into groups centered at approximately 1.00, 0.50, or 0.00. Each group reflects preservation (no deletion), loss of one copy, or loss of two copies of STRC. The error bars for each sample represent the 95% confidence intervals using Poisson statistics
Next, we performed LR-PCR-based STRC sequencing in five patients carrying the heterozygous STRC deletion. LR-PCR-based sequencing successfully detected SNVs without pSTRC contamination, which was observed in conventional direct sequencing (Fig. 3). LR-PCR-based sequencing revealed six types of SNVs in the exons and the untranslated regions of STRC in the opposite allele (Table 3). Among these, the c.4057 C > T mutation detected in case 3 was previously reported as a pathogenic SNV resulting in a stop codon. The pathogenicity of c.−28 C > G remained uncertain, while the remaining four SNVs were synonymous substitutions. Overall, three cases (cases 1, 2, and 3) among the study cohort were considered to exhibit STRC-related hearing loss.
Clinical and audiologic findings
Clinical findings and audiograms of the patients with at least one STRC copy loss are summarized in Table 3 and Fig. 4, respectively. The ascertained onset of hearing loss varied from the first to the sixth decade of life, and five patients complained of hearing loss progression. The severity of hearing loss ranged from mild to moderate, and the audiograms showed symmetrical flat or high frequency SNHL. Episodes of vertigo were reported by two of the seven patients. None of the patients exhibited overt family histories and were recognized as solitary cases.
The vertical axis represents the hearing level, and the horizontal axis represents the frequency. Circles and crosses in audiograms represent the right and left ears, respectively. A typical normal audiogram shows hearing thresholds of both ears above the 20 dB line for all frequencies. All audiograms of patients show mild-to-moderate, horizontal, or high frequency hearing loss
Discussion
We evaluated the CNVs and SNVs in Japanese patients with mild-to-moderate SNHL using ddPCR in combination with LR-PCR. The prevalence of copy number deletion was 7.7% among the patients. In addition to the homozygous deletion of STRC in two patients, LR-PCR-based STRC sequencing detected six distinct SNVs in the exons and the untranslated regions of STRC in patients with heterozygous STRC deletions. One of the SNVs was previously reported as pathogenic13. Therefore, at least three patients (3.6%) were considered to exhibit hearing loss associated with STRC dysfunction. All patients were recognized as sporadic cases.
Recent studies indicate that approximately two-thirds of the entire human genome is composed of repeats14, and 4.8%–9.5% of the human genome can be classified as CNVs15. Among these, large repeats, such as whole gene repeats, are generally detected and identified by fluorescent in situ hybridization, array comparative genomic hybridization, single nucleotide polymorphism or oligo arrays, and paired-end mapping. Recently, MPS has also been used to detect CNVs with high genomic resolution. Indeed, MPS is used in the clinic and can efficiently detect CNVs in most Japanese patients with hearing loss2,10. These techniques are suitable for searching the whole genome at once but require a considerable amount of work, time and cost. Conversely, ddPCR is insufficient for global identification; however, it provides rapid and easy completion at less than $10 per sample, and the total assay time is 2.5 h, illustrating its efficacy in determining the copy number of specific DNA areas16. Additionally, the results are highly reproducible, with minimal variability among DNA samples that does not require strict sample preparation and that is accurate and acceptable for homogenous and heterogeneous statuses. Considering the relatively high prevalence of STRC deletion in the Japanese population and challenges in diagnosing inheritable causes of hearing loss, screening with ddPCR is a potential approach before evaluation of patients by MPS.
As a causal gene for mild-to-moderate idiopathic SNHL, STRC is garnering increasing attention. Our results reconfirm that CNVs in STRC are a common phenomenon in mild-to-moderate SNHL, found in more than 7% of the patients with a carrier rate of less than 1% in healthy Japanese controls, which is consistent with previous reports2,10. However, no mutations in the exons of the translated regions of the opposite allele were detected in four of the five patients with heterozygous gene deletion. It is possible that these patients might harbor mutations in genes other than STRC, such as GJB2 and Mt-1555. Alternatively, the significantly high prevalence of CNVs in the patients compared to the healthy controls suggests undetectable mutations that might be present in the intronic, promoter, or enhancer regions of STRC. Further studies are warranted to verify this hypothesis.
Numerous pathogenic SNVs across the entire STRC coding sequence have been reported thus far. However, direct PCR or MPS cannot distinguish whether such variants originate from STRC or pSTRC. Indeed, the c.4057 C > T, Q1353X variant is coincident with the reference sequence for exon 20 from the pSTRC17. Therefore, novel nonsense variants that are detected in patients with hearing loss should be verified to ensure that they are a pathogenic variant in the STRC coding sequence and not a pseudogene contamination. We recommend LR-PCR-based sequencing as an excellent technique that might be necessary for comprehensive screening for STRC-related hearing loss.
In conclusion, STRC copy number deletion was detected in approximately 1% of the Japanese healthy controls, whereas STRC was the causal gene in more than 3% of patients with mild-to-moderate hearing loss in Japan. The detection of CNVs using ddPCR is a rapid, precise, and cost-effective method for relatively prevalent deafness genes, such as STRC. The combination of ddPCR and LR-PCR-based sequencing is a reliable approach to detect patients with STRC-associated hearing loss.
References
Shearer, A. E. et al. Copy number variants are a common cause of non-syndromic hearing loss. Genome Med. 6, 37 (2014).
Nishio, S. Y., Moteki, H. & Usami, S. I. Simple and efficient germline copy number variant visualization method for the Ion AmpliSeq custom panel. Mol. Genet. Genom. Med. 6, 678–686 (2018).
Sloan-Heggen, C. M. et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum. Genet. 135, 441–450 (2016).
Verpy, E. et al. Mutations in a new gene encoding a protein of the hair bundle cause non-syndromic deafness at the DFNB16 locus. Nat. Genet. 29, 345–349 (2001).
Verpy, E. et al. Stereocilin connects outer hair cell stereocilia to one another and to the tectorial membrane. J. Comp. Neurol. 519, 194–210 (2011).
Moteki, H. et al. Detection and confirmation of deafness-causing copy number variations in the STRC gene by massively parallel sequencing and comparative genomic hybridization. Ann. Otol. Rhinol. Laryngol. 125, 918–923 (2016).
Francey, L. J. et al. Genome-wide SNP genotyping identifies the Stereocilin (STRC) gene as a major contributor to pediatric bilateral sensorineural hearing impairment. Am. J. Med. Genet. A. 158A, 298–308 (2012).
Hoppman, N. et al. Genetic testing for hearing loss in the United States should include deletion/duplication analysis for the deafness/infertility locus at 15q15.3. Mol. Cytogenet. 6, 19 (2013).
Knijnenburg, J. et al. A homozygous deletion of a normal variation locus in a patient with hearing loss from non-consanguineous parents. J. Med. Genet. 46, 412–417 (2009).
Yokota, Y. et al. Frequency and clinical features of hearing loss caused by STRC deletions. Sci. Rep. 9, 4408 (2019).
Ito, T., Noguchi, Y., Yashima, T., Ohno, K. & Kitamura, K. Hereditary hearing loss and deafness genes in Japan. J. Med. Dent. Sci. 57, 1–10 (2010).
Vona, B. et al. DFNB16 is a frequent cause of congenital hearing impairment: implementation of STRC mutation analysis in routine diagnostics. Clin. Genet. 87, 49–55 (2015).
Momeni, P. et al. Mutations in a new gene, encoding a zinc-finger protein, cause tricho-rhino-phalangeal syndrome type I. Nat. Genet. 24, 71–74 (2000).
de Koning, A. P., Gu, W., Castoe, T. A., Batzer, M. A. & Pollock, D. D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 7, e1002384 (2011).
Zarrei, M., MacDonald, J. R., Merico, D. & Scherer, S. W. A copy number variation map of the human genome. Nat. Rev. Genet. 16, 172–183 (2015).
Okada, R. et al. The quantitative analysis of the human papillomavirus DNA load in submandibular gland lesions with droplet digital polymerase chain reaction. Acta Otolaryngol. 139, 105–110 (2019).
Mandelker, D. et al. Comprehensive diagnostic testing for stereocilin: an approach for analyzing medically important genes with high homology. J. Mol. Diagn. 16, 639–647 (2014).
Acknowledgements
This work was supported by JSPS KAKENHI Grant Numbers [JP17K11314], [JP17K11315], [JP17K11316], and [JP16K11174].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ito, T., Kawashima, Y., Fujikawa, T. et al. Rapid screening of copy number variations in STRC by droplet digital PCR in patients with mild-to-moderate hearing loss. Hum Genome Var 6, 41 (2019). https://doi.org/10.1038/s41439-019-0075-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41439-019-0075-5
- Springer Nature Limited
This article is cited by
-
Frequency of the STRC-CATSPER2 deletion in STRC-associated hearing loss patients
Scientific Reports (2022)
-
Robust and accurate estimation of paralog-specific copy number for duplicated genes using whole-genome sequencing
Nature Communications (2022)
-
Detecting copy number variation in next generation sequencing data from diagnostic gene panels
BMC Medical Genomics (2021)
-
Measurement of gene amplifications related to drug resistance in Plasmodium falciparum using droplet digital PCR
Malaria Journal (2021)
-
Detection of non-targeted transgenes by whole-genome resequencing for gene-doping control
Gene Therapy (2021)