Abstract
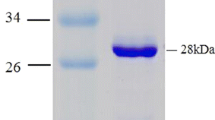
4-Chlorobenzoate:CoA ligase, the first enzyme in the pathwayfor 4-chlorobenzoate dissimilation, has been partially purifiedfrom Arthrobacter sp. strain TM-1, by sequential ammoniumsulphate precipitation and chromatography on DEAE-Sepharoseand Sephacryl S-200. The enzyme, a homodimer of subunitmolecular mass approximately 56 kD, is dependent onMg2+-ATP and coenzyme A, and produces 4-chlorobenzoyl CoA and AMP. Besides Mg2+, Mn2+, Co2+, Fe2+ and Zn2+ are also stimulatory, but not Ca2+. Maximal activity is exhibited at pH 7.0 and 25 °C. The ligase demonstrates broad specificity towards otherhalobenzoates, with 4-chlorobenzoate as best substrate.The apparent Michaelis constants (Km) of the enzymefor 4-chlorobenzoate, CoA and ATP were determined as3.5, 30 and 238 μM respectively. 4-ChlorobenzoylCoA dehalogenase, the second enzyme, has been purified tohomogeneity by sequential column chromatography onhydroxyapatite, DEAE-Sepharose and Sephacryl S-200. It isa homotetramer of 33 kD subunits with an isoelectric pointof 6.4. At pH 7.5 and 30 °C, Km andkcat for 4-CBCoA are 9 μM and1 s-1 respectively. The optimum pH is 7.5, andmaximal enzymic activity occurs at 45 °C. Theproperties of this enzyme are compared with those ofthe 4-chlorobenzoyl CoA dehalogenases from Arthrobactersp. strain 4-CB1 and Pseudomonas sp. strain CBS-3, whichdiffer variously in their N-terminal amino acid sequences, optimalpH values, pI values and/or temperatures of maximal activity.
Similar content being viewed by others
References
Ahmed M & Focht DD (1973) Degradation of polychlorinated biphenyls by two species of Achromobacter. Can. J.Microbiol. 19: 47–52
Ajithkumar PV & Kunhi AAM (2000) Pathways for 3-chloro-and 4-chlorobenzoate degradation in Pseudomonas aeruginosa 3mT. Biodegradation 11: 247–261
Bradford, MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254
Chae JC & Kim CK (1997) Dechlorination of 4-chlorobenzoate by Pseudomonas sp. DJ-12. Kor. J. Microbiol. 35: 290–294
Chae JC, Kim Y, Min KH, Kim YC & Kim CK (1999) Cloning and sequencing of the fcbB gene encoding 4-chlorobenzoatecoenzyme A dehalogenase from Pseudomonas sp. DJ-12. Mol. Cells 9: 225–229
Chang K, Liang P, Beck W, Scholten JD & Dunaway-Mariano D (1992) Isolation and characterisation of the three polypeptide components of 4-chlorobenzoate dehalogenase from Pseudomonas sp. strain CBS-3. Biochemistry 31: 5605–5610
Crooks GP & Copley SD (1994) Purification and characterisation of 4-chlorobenzoyl CoA dehalogenase from Arthrobacter sp. strain 4-CB1. Biochemistry 33: 11645–11649
Evans CG, Herbert D & Tempest DW (1970) The continuous cultivation of microorganisms. II. In: Construction of a Chemostat. Methods in Microbiology, Vol. 2 (pp 278–324). Academic Press, London
Gartemann K-H, Fiedler J, Schmitz A, Zellermann E-M & Eichenlaub R (1998) GenBank Accession Number: AF042490
Hartmann J, Reineke W & Knackmus, H-J (1979) Metabolism of 3-chloro-, 4-chloro-and 3,5-dichlorobenzoate by a pseudomonad. Appl. Environ. Microbiol. 37: 421–428
Hützinger O & Veerkamp W (1981) Xenobiotic chemicals with pollution potential. In: Microbial Degradation of Xenobiotics and Recalcitrant Compounds. FEMS Symp. 12: 3–45
Klages U & Lingens F (1979) Degradation of 4-chlorobenzoic acid by a Nocardia sp. FEMS Microbiol. Lett. 6: 201–203
Klages U & Lingens F (1980) Degradation of 4-chlorobenzoic acid by a Pseudomonas sp. Zbl. Bakt. Hyg. I. Abt. Orig. C1: 215–223
Laemmli, UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685
Löffler F & Müller R (1991) Identification of 4-chlorobenzoylcoenzyme A as intermediate in the dehalogenation catalyzed by 4-chlorobenzoate dehalogenase from Pseudomonas sp. CBS-3. FEBS Lett. 290: 224-226
Löffler F, Müller R & Lingens F (1991) Dehalogenation of 4-chlorobenzoate by 4-chlorobenzoate dehalogenase from Pseudomonas sp. strain CBS3: an ATP/coenzyme A dependent reaction. Biochem. Biophys. Res. Comm. 176: 1106–1111
Löffler F, Müller R & Lingens F (1992) Purification and properties of 4-halobenzoate-coenzyme A ligase from Pseudomonas sp. CBS3. Biol. Chem. Hoppe-Seyler 373: 1001–1007
Löffler F, Lingens F & Müller R (1995) Dehalogenation of 4-chlorobenzoate: Characterisation of 4-chlorobenzoyl coenzyme A dehalogenase from Pseudomonas sp. CBS3]. Biodegradation 6: 203–212.
Marks, TS, Smith, ARW & Quirk, AV (1984a) Degradation of 4-chlorobenzoic acid by Arthrobacter sp. strain TM-1. Appl. Environ. Microbiol. 48: 1020–1025
Marks TS, Wait R, Smith ARW & Quir, AV (1984b) The origin of oxygen incorporated during the dehalogenation/hydroxylation of 4-chlorobenzoate by an Arthrobacter sp. strain TM-1. Biochem. Biophys. Res. Commun. 124: 669–674
Merkel SM, Eberhard AE, Gibson J & Harwood CS (1989) Involvement of coenzyme A thioesters in anaerobic metabolism of 4-hydroxybenzoate by Rhodopseudomonas palustris. J. Bacteriol. 171: 1–7
Müller R, Thiele J, Klages U & Lingens F (1984) Incorporation of [18O]-water into 4-hydroxybenzoic acid in the reaction of 4-chlorobenzoate dehalogenase from Pseudomonas sp. Strain CBS3. Biochem. Biophys. Res. Commun. 124: 178–182
Müller R, Oltmanns RH & Lingens F (1988) Enzymatic dehalogenation of 4-chlorobenzoate by extracts from Arthrobacter sp. strain SU DSM 20407. Biol. Chem. Hoppe-Seyler 369: 567–571
Peyton TO (1984) Biological disposal of hazardous waste. Enzym. Micro. Tech. 6: 146–154
Quinn JA, McKay DB & Entsch B (2001) Analysis of the pobA and pobR genes controlling expression of p-hydroxybenzoate hydroxylase in Azotobacter chroococcum. Gene 264: 77–85 (GenBank Accession Number: AF019891)
Reinecke W, Jeenes DJ, Williams PA & Knackmuss H-J (1982) TOL plasmid pWWO in constructed halobenzoate-degrading Pseudomonas strains: prevention of meta-pathway. J. Bacteriol. 150: 195–201
Reinecke W & Knackmuss H-J (1979) Construction of haloaromatics-utilizing bacteria. Nature 277: 385–386
Reinecke W & Knackmuss H-J (1980) Hybrid pathway for chlorobenzoate metabolism in Pseudomonas sp. B13 derivatives. J. Bacteriol. 142: 467–473
Schmitz A, Gartemann K-H, Fiedler J, Grund E & Eichenlaub R (1992) Cloning and sequence analysis of genes for dehalogenation of 4-chlorobenzoate from Arthrobacter sp. strain SU. Appl. Environ. Microbiol. 58: 4068–4071
Seibold B, Matthes M, Eppink MH, Lingens F, van Berkel WJ & Müller R (1996) 4-Hydroxybenzoate hydroxylase from Pseudomonas sp. CBS3. Purification, characterisation, gene cloning, sequence analysis and assignment of structural features determining the coenzyme specificity. Eur. J. Biochem. 239: 469–478 (GenBank Accession Number: X74827)
Sheets TJ, Smith JW & Kaufman DD (1968) Persistence of benzoic acids and phenylacetic acids in soils. Weed Science 16: 217–222
Shimao M, Onishi S, Mizumori S, Kato N & Sakazawa C (1989) Degradation of 4-chlorobenzoate by facultatively alkalophilic Arthrobacter sp. strain SB8. Appl. Environ. Microbiol. 55: 478–482
Smith RJ (1991) The purification of an aryl dehalogenase from Arthrobacter strain TM-1. Ph.D. thesis. Thames Polytechnic, London
van den Tweel WJJ, ter Burg WJJ, Kok JB & de Bont JAM (1986) Bioformation of 4-hydroxybenzoate from 4-chlorobenzoate by Alcaligenes denitrificans NTB-1. Appl. Microbiol. Biotech. 25: 289–294
Zaitsev GM & Karasevich YN (1981a) Utilisation of 4-chlorobenzoic acid in Arthrobacter globiformis. Microbiology (Russia) 50: 23–27
Zaitsev GM & Karasevich YN (1981b) Preparative metabolism of 4-chlorobenzoic acid in Arthrobacter globiformis. Microbiology (Russia) 50: 287–291
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Zhou, L., Marks, T.S., Poh, R.P.C. et al. The purification and characterisation of 4-chlorobenzoate:CoA ligase and 4-chlorobenzoyl CoA dehalogenase from Arthrobacter sp. strain TM-1. Biodegradation 15, 97–109 (2004). https://doi.org/10.1023/B:BIOD.0000015614.94615.34
Issue Date:
DOI: https://doi.org/10.1023/B:BIOD.0000015614.94615.34