Abstract
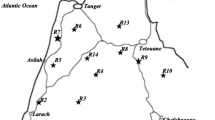
Genetic variation within and among several Sorghum populations from different agroecological zones in Malawi were investigated using random amplified polymorphic markers (RAPDs). DNA samples from individual plants were analyzed using 35 oligonucleotides of random sequence. Twenty five of these primers allowed amplifications of random polymorphic (RAPD) loci. Overall, 52% of the scored loci were polymorphic. Every accession was genetically distinct. The analysis of molecular variance revealed that the within-region (among accessions) variations accounted for 96.43% of the total molecular variance. Observed variations in allelic frequency was not related to agroecological differences. The degree of band sharing was used to evaluate genetic distance between accessions and to construct a phylogenetic tree. Further analysis revealed that the sorghum accessions analyzed were genetically close despite considerable phenotypic diversity within and among them. It is suggested that all the sorghum landraces currently available in Malawi should be conserved both ex situ and in situ to maintain the current level of genetic diversity.
Similar content being viewed by others
References
Ahnert D., Lee M., Austin D.F., Livini C., Openshaw S.J., Smith J.S.C. et al. 1996. Genetic diversity among elite sorghum inbred lines assessed with DNA markers and pedigree information. Crop Sci. 36: 1385–1392.
Aldrich P.R. and Doebley J. 1992. Restriction fragment variation in the nuclear and chloroplast genomes of cultivated and wild Sorghum bicolor. Theor. Appl. Genet. 85: 293–302.
Anonymous 1996. Lost crops of Africa: grains, National Academy species of Sciences. National Academy Press, USA. vol 1.
Armstrong J.S., Gibbs A.J., Peakall R. and Weiller G. 1994. The RAPDistance Package. Http://life.anu.edu.au /molecular/software/rapd.html.
Avise J.C. 1994. Molecular markers, natural history and evolution. Chapman and Hall, New York.
Brown A.M., Hopkins M., Mitchell S.E., Senior M.L., Wang T.Y., Duncan R.R. et al. 1996. Multiple methods for the identification of polymorphic simple sequence repeats (SSRs) in sorghum [Sorghum bicolor (L.) Moench]. Theor. Appl. Genet. 93: 190–198.
Xu J.X., Cui F.W., Magill C.W., Schertz K.F. and Hart G.E. 1995. RFLP-;based assay of Sorghum bicolor (L.) Moench genetic diversity. heor. Appl. Genet. 90: 787–796.
Crow J.F. 1986. Basic concepts in population, quantitative and evolutionary genetics.W. H. Freeman and Company, New York.
Dahlberg J.A., Hash T., Kresovich S., Maunder A.B. and Gilbert M. 1997. Sorghum and pearl millet genetic resources utilisation Proceedings of the International Conference on Genetic Improvement of Sorghum & Millet. SICNA, Lubbock, TX, pp. 42–54.
Deu M., Gonzalez-;De-;Leon D., Glaszmann J.C., Degremont I., Chantereau J., Lanaud C. et al. 1994. RFLP diversity in cultivated sorghum in relation to racial differentiation. Theor. Appl. Genet. 88: 838–844.
Doggett H. 1988. Sorghum. Longman, London, UK.
Duncan R.R., Bramel-;Cox P.J. and Miller F.R. 1991. Contributions of introduced sorghum germplasm to hybrid development in the USA. In: Shands H.L. and Weisner L.E. (eds), Use of plant unintroduction in cultivar development. CSSA Spec. Publ. 17. CSSA, Madison, WI, pp. 69–102.
Eberhart S.A., Bramel-;Cox P.J. and Prasada Rao K.E. 1997. Prerequired serving genetic resources. In: Proceedings of the International Conference on Genetic Improvement of Sorghum and Millet. SICNA, Lubbock, TX, pp. 25–41.
Hartl D.L. 1988. A primer of population genetics. 2nd edn. Sinauer Assoc. Inc., Sunderland, M.A.
Excoffier L., Smouse P.E. and Quattro J.M. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction sites. Genetics 131: 479–491.
Hedrick P.W. 1986. Genetic polymorphism in heterogeneous environments: A decade later. Ann. Rev. Ecol. Syst. 17: 535–566.
Loveless M.D. and Hamrick J.L. 1984. Ecological determinants of genetic structure in plant populations. Annu. Rev. Ecol. Syst. 15: 65–95.
Maniatis T., Fitsch E.F. and Sambrook J. 1989. Molecular Cloning: A laboratory Manual. 2nd edn. Cold Spring Harbor Laboratory, Long Island, NY. vol I–III.
Menkir A., Goldsbrough P. and Ejeta G. 1997. RAPD based assessment of genetic diversity in cultivated races of sorghum. Crop Sci. 37: 564–569.
Morden W.C., Doebley J. and Schertz K.F. 1989. Allozyme variation in old world races of Sorghum bicolor (Poaceae). Am. J. Bot. 76: 245–255.
Nkongolo K.K. 1999a. RAPD variations among pure and hybrid populations of Picea mariana, P. rubens andP. glauca, and cytogenetic stability of Picea hybrids: identification of speciesof specific RAPD markers. Pl. Syst. Evol. 215: 229–239.
Nkongolo K.K. 1999b. RAPD and cytological analyses of Picea spp. from different provenances: genomic relationships among taxa. Hereditas 130: 137–144.
Ollitrault P. 1987. Evaluation genetique des sorghos cultives (Sor-;ghum bicolor L. Moench) par l'analyse conjointe des diversities enzymatiques et morphophysiolgiques, Universite de Paris.
Saitou N. and Nei M. 1987. The neigbor-;joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425.
Tao Y., Manners J.M., Ludlow M.M. and Henzell R.F. 1993. DNA polymorphism in grain sorghum (Sorghum bicolor (L.) Moench). Theor. Appl. Genet. 86: 679–688.
Teshome A., Fahrig L., Torrance J.K., Lambert J.D., Arnason T.J. and Baum B.R. 1999. Maintenance of Sorghum (Sorghum bicolor, Poaceae) landrace diversity by farmers selection in Ethiopia. Econ. Bot. 53: 79–88.
Vierling R.A., Xiang Z., Joshi C.P., Gilbert M.L. and Nguyen H.T. 1994. Genetic diversity among elite sorghum lines revealed by restriction fragment length polymorphism and random amplified polymorphic DNAs. Theor. Appl. Genet. 87: 816–820.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Nkongolo, K., Nsapato, L. Genetic diversity in Sorghum bicolor(L.) Moench accessions from different ecogeographical regions in Malawi assessed with RAPDs. Genetic Resources and Crop Evolution 50, 149–156 (2003). https://doi.org/10.1023/A:1022996211164
Issue Date:
DOI: https://doi.org/10.1023/A:1022996211164