Abstract
There is a significant effort to explore the potential pharmacological activity of natural products derived from plant extracts. The goal is to generate new compounds that could be used for therapeutic purposes. Eugenia punicifolia is a shrub that grows widely all over the world. As a natural cure for diabetes, fever, flu and inflammation, leaves of this medicinal plant are often used in decoction or aqueous infusion. The goal of the study was to record the molecular docking, antioxidant, chemical composition, and alpha-glucosidase inhibition of E. punicifolia. A number of parameters were evaluated in order to confirm its efficacy, including Total Phenolic Content (TPC), TFC (Total Flavonoid Content), FRAP (ferric reducing antioxidant power), DPPH radical scavenging activity, inhibition of α-glucosidase, chemical composition, and molecular docking. The leaf ethanolic exhibited robust antioxidant activity with respect to radical scavenging activity, TPC (Total phenolic content), reducing power, TFC (Total Flavonoid Content), and alpha-glucosidase inhibitory activity at concentrations of 26.7 µg/mL, 18.9 mmol/g, 11.9 mg GAE/g, 13.5 mg QE/g, and 18.9 µg/mL, respectively. In the ethanolic leaf extract, Z-8-Methyl-9-tetradecenoic acid (28.6%) was the main component. The five compounds (− 121.208 kcal/mol, − 120.465 kcal/mol, − 105.572 kcal/mol, − 104.497 kcal/mol, and − 102.256 kcal/mol) were selected as the top lead compounds, respectively. The lead compounds that were found had drug-like properties and were suitable for oral administration. In addition, it was noted that they exhibited non-toxic features and favorable ADME-Tox characteristics, except for compound 6, which displayed toxicity. The leaves exhibited potential applications in the food and pharmaceutical sectors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
As a therapy strategy, the use of alternative therapies is becoming more frequent [1]. In this context, the use of medicinal plants in treating different diseases is significant [2]. Most parts of the world practice folk medicine very much. Plants and plant extracts are used by more than 80% of people in Asia and Africa to treat a variety of illnesses [2]. Because of the special chemical compounds they contain, which have a known physiological effect on both humans and animals, plants have medicinal relevance [3]. Secondary metabolites produced by different plant components are widely used in many aspects of daily human life [4]. Both in their unrefined and refined forms, these substances are acknowledged for their broad biological value [5]. The concept of computer-aided drug design (CADD) has gained significant support as a means of producing or discovering novel, more effective small molecules faster and at a lower cost [6]. The concept of computer-aided drug design (CADD) has gained significant support as a means of producing or discovering novel, more effective small molecules faster and at a lower cost [6]. Molecular docking virtual screening is widely used in structure-based drug design (SBDD) to predict the binding orientation and affinities of two molecules when they form a stable complex [6]. The shrub E. punicifolia is widely found around the world. This medicinal plant's leaves are frequently used as a natural remedy for diabetes, fever, flu, and inflammation in decoctions or aqueous infusions. In the treatment of infections and wounds, it is also administered in alcoholic infusion [7]. Eugenia punicifolia has clearly been important historically and currently, but not much has been done to document its biological function or chemical composition. The study sought to document E. punicifolia molecular docking, antioxidants, chemical composition, and inhibition of alpha-glucosidase.
2 Materials and methods
2.1 Sample collection
The plant specimens were recognized by a certified botanist from Ahmadu Bello University (ABU) in Zaria, Kaduna State, Nigeria. Eugenia punicifolia was found and is kept in the herbarium of the department (Voucher ABU1684). The species name was confirmed using the World Flora Online (WFO) at https://www.worldfloraonline.org/.
2.2 Sample extraction
With the grinding machine, the dry materials were crushed to a solid powder. Powdered plant samples weighed 100 g. The plant components were extracted into ethanol, hexane, and aqueous forms using the Soxhlet process [5]. The byproduct of the extraction was filtered through Whatman no. 2 filter paper [5]. Using an evaporation apparatus (E-Z-2-Elite), the crude extract was made from hexane, ethanol, and aqueous plant leaves. The solvent pressure was adjusted to 300, 20, and 72 for the ethanol, hexane, and aqueous extracts, and the vacuum was set to 40ºC for each [5].
2.3 DPPH radical scavenging activity
About 100µL of leaves and roots have been mixed with the DPPH solution in methanol at a concentration of 0.004% wv (extracted using ethanol, hexane, and aqueous) at different concentrations ranging from 1.56 to 100 μg/mL, respectively. The mixture was assessed for DPPH reduction at a wavelength of 517 nm following a 30-min storage time at 37 °C in a dark environment. The scavenging activity percentage were determined by comparing it to the control, which consisted of 100 L of methanol and 200 L of DPPH. The subsequent equation was employed to compute the radical scavenging activity.
C1 represents the sample absorbance, while Co represents the control absorbance. The IC50 values, which indicate the concentration of the extract required to eliminate 50% of the DPPH radical, were determined by plotting the percentage of scavenging against different extract concentrations of 1.56 to 100 μg/mL, respectively [8,9,10].
2.4 Antioxidant activity by (FRAP) assay
The FRAP antioxidant assay was determined using G-Biosciences’ FRAP assay kit according to the manufacturer’s instructions.
2.5 Total phenolic content (TPC)
The Folin Ciocalteu reagent was employed to quantify the overall phenolic content (TPC). Simply put, 150 µl of Folin-Ciocalteu reagent (10%) were mixed with 30 µl of extract (1 mg/mL). After 4 min, a further 120 μL of 7.5% Na2CO3 was added. Following a period of 45 min in a dark environment, the resulting mixture was assessed for its optical density at a wavelength of 760 nm [9]. The quantification of the results was done in milligrams of Gallic Acid Equivalent (GAE) per gram of dry weight (DW) using the gallic acid standard curve equation, y = 0.0038x + 0.0673, with an R2 value of 0.9964 (Fig. 1) [9].
2.6 Total flavonoid content (TFC)
To perform the estimation, methanol was mixed with an equivalent volume of a predetermined quantity of the sample. Subsequently, a weighed combination of diluted aluminum chloride, potassium acetate, and 1.4 mL of distilled water was introduced into the aforementioned mixture, which was sealed in aluminum foil and left undisturbed for a duration of thirty minutes to avoid light exposure [11, 12]. The measurement was obtained at a wavelength of 415 nm. Quercetin was employed as the flavonoid standard [12]. The results were expressed as mg of Quercetin Equivalent (QE) per g of dry weight (DW) [12] (Fig. 2).
2.7 α-glucosidase inhibition assay
The reaction was done by mixing of plant extracts 10 μL from different plant sections with 50 μL of 0.1 M phosphate buffer at a pH of 7.0. Afterwards, a 25µL solution of alpha-glucosidase with a concentration of 0.2 U/mL in buffer was added to the mixture. The samples were incubated at 37 °C for 10 min. Following that, a volume of 25 μL of a 0.5 mM solution of (pNPG) substrate was added to the process. Subsequently, the combination was incubated at 37 °C for an additional 30 min. The reaction was stopped by adding 100 μL of a sodium carbonate solution. Quercetin was used as a positive control [13]. The absorbance was measured at 410 nm. The inhibition % was determined by:
Cab represents absorbance of the control, while Sab represents the sample absorbance.
2.8 Gas chromatography mass spectrometry (GC–MS) conditions
The experiment was performed utilizing an Agilent GC–MS. The chemical mixtures were separated utilizing an HP-5MS column measuring 30 m in length and 0.25 mm in diameter. The separation procedure consisted of initially heating the column to a temperature of 60 °C for a duration of 10 min, and thereafter raising the temperature to 230 °C over a period of 1 min, with a heating rate of 3 °C per minute. The column was thereafter maintained at a 230 °C for a duration of 1 min. The injector's helium flow rate was 1 mL/min, at a temperature of 245 °C. The ion source and analyzer for 70 eV MS were maintained at a temperature of 260 °C [14].
2.9 Molecular docking methodology
The search for the stable conformations of the studied compounds was achieved using the Density Functional DFT (B3LYP) and the 6–31G* basis set and saved in pdb format [15]. The alpha-glucosidase 3D structure (pdb entry:3top) was collected from the online RCSB protein data bank. The protein was prepared with the Discovery Studio software. Here, ligand groups, water molecules, and heteroatoms attached to the target protein were removed and saved in pdb format. The investigated compounds (ligands) were docked into the binding site of the receptor with Molegro Virtual Docker (MVD) software [16]. Furthermore, the visualisation of protein–ligand interactions was carried out using the Discovery Studio software [17].
2.10 Pharmacokinetics properties estimation
The Pharmacokinetics Properties Estimation was conducted at http://www.SwissADME.ch (Daina et al. 2017). Similarly the The “Adsorption, Distribution, Metabolism, Excretion and Toxicity properties of the compounds was estimated on the pkCSM”website at https://biosig.lab.uq.edu.au/pkcsm [18].
2.11 Frontier molecular orbitals Dft method
Density functional theory (DFT) is a computational method used in physics, chemistry, and materials science to study the electronic structure of many-body systems, such as atoms, molecules, and condensed phases. It primarily focuses on the ground state and can provide insights into the nuclear structure as well [19, 20]. Two fundamental ideas in quantum chemistry are the HOMO and LUMO, or lowest unoccupied molecular orbitals. The lowest empty orbital is known as the LUMO, while the highest energy orbital is known as the HOMO, where electrons are present. The HOMO–LUMO gap is a measure of molecular stability that is defined as the energy difference between the HOMO and LUMO [21].
2.12 Molecular dynamics
The stability assessment of a promising molecule in conjunction with MMP-2 receptors, identified by the Protein Data Bank entry entry:3top, was conducted through molecular dynamics simulations using the force field (OPLS3e) within the 2020–3 Desmond software by Schrodinger LLC [22, 23]. A spacing of 10 Å was maintained among the protein atoms, and the boundaries of the orthorhombic periodic box that contained the solvent, while the ligand–protein complex was immersed therein. The SPC model, which stands for single-point charge water, was used as the solvent model. The Desmond System Builder panel helped achieve charge neutralization by adding Na + and Cl − counterions, and the concentration of NaCl salt was changed to 0.15 M to match physiological circumstances [24]. Subsequently, the solvated system underwent relaxation and minimization utilizing the default OPLS3e force field parameters in Desmond. The model ran in the isothermal isobaric ensemble (NPT) mode, with the Nose–Hoover chain thermostat controlling the temperature to 300 K and the Martyna-Tobias-Klein barostat the atmospheric pressure to 1.0315 Bar [25]. The estimation of binding free energy was performed using the MM/PBSA approach from the beginning and end of the MD simulation trajectory.
2.13 Statistical analysis
Statistical Analysis System (SAS) version 9.4 was used to analyzed the data. Completed Randomized Design (CRD) was used, with three replications. At the p ≤ 0.05 level, significant changes in the means were identified by comparing the results using ANOVA with a post hoc Duncan’s Multiple Range Test (DMRT) [5].
3 Results and discussion
3.1 Extraction yield
The yield exhibited substantial variation based on the extraction solvents employed (Table 1). It is possible that the high yield seen in the aqueous extracts (0.12%) is owing to the solvent's capacity to extract more compounds (Table 1). These findings differ from other studies, which assert that ethanol has a higher capacity to extract from plant material [26]. The yield of the extract varied due to four extraction parameters: extraction temperature, extraction duration, and solvent concentration [27]. The main factors influencing the yield extract variance across medicinal plant sections are the extraction technique, solvent type, chemical type, and metabolite polarity [5].
3.2 Biological activity
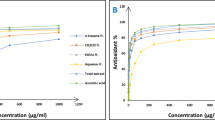
A single approach is incapable of fully quantifying all antioxidants and radicals present in complex matrices. The two most commonly employed forms of in vitro tests are basically two [28]. One type of test involves the use of hydrogen atoms to assess an individual's ability to remove free radicals [29]. The DPPH method has been widely employed by researchers to assess the antioxidant ability of specific plant extracts. The procedure is reliable and capable of transforming a DPPH free radical found in an antioxidant plant extract with an abundance of electrons into a non-radical state [30,31,32]. The other type of complex reduces its complexity by transferring a single electron, for example, in the case of a metal or a free radical [9]. The scavenging properties of DPPH were assessed from the leaves and root of E. punicifolia using ethanol, aqueous, and hexane extracts (Table 2). The ethanol leaf extract had the most potent scavenging effect, as demonstrated by its IC50 value of 26. 7 ± 1.4 µg/mL (Table 2). The leaves' aqueous extract exhibited the least scavenging action, with an IC50 value of 71.2 ± 2.5 µg/mL (Table 2). The highest ability to convert Fe3+ to Fe2+ was observed in ethanol leaves extracts, with 18.9 mmol/g (Table 2). The significant amount of phenolic (11.9 ± 0.9 mg GAE/g) and flavonoid (13.5 ± 0.9 mg QE/g) compounds in the leaf extract enabled the scavenging effect and ferric reduction capability (Table 2). Phenolic and flavonoid compounds are important types of molecules that operate as primary antioxidants and have the ability to interact with radicals such as hydroxyl, superoxide, and lipid peroxy [32]. The activity of different plant sections is significantly influenced by the nature of the chemicals involved, the polarity of the metabolites, and the kind and character of the extraction solvents. The plant's antioxidant activity was evaluated by the TPC, TFC, FRAP and DPPH tests. The reason for this is that no single technique can provide a reliable assessment of a substance's antioxidant capacity. Instead, many antioxidant compounds may function in vivo through different mechanisms [5]. Due to the complexity of phytochemicals, it is crucial to evaluate the antioxidant activity of plant materials using many methodologies [32]. The ethanol and aqueous extracts of the Leaves exhibited α-glucoside inhibitory activity of 18.9 and 45.6 µg/mL, respectively (Table 2). The IC50 values were not detected in the extracts of leaf hexane, root ethanol, and root hexane, as shown in Table 2. Alpha glucosidase is the main enzyme responsible for carrying out the last stage of carbohydrate digestion. The investigations corroborate the assertions made by the traditional herbalist that E. punicifolia has the potential to alleviate diabetes. The inhibitory impact of α-glucoside in the ethanol leaf extract is attributed to the extract's strong antioxidant content (Table 2).
3.3 Chemical composition
Leaf ethanol extract was analyzed with GC–MS, a hybrid analytical technique that combines the separation characteristics of gas–liquid chromatography with the detection capabilities of mass spectrometry. This approach was used to determine the existence of compounds in the extract, as it has shown high activity. The data analysis revealed that Z-8-Methyl-9-tetradecenoic acid was identified as the primary chemical, with the largest proportion of 28.6, as indicated in Table 3. The composition of the extract is significantly influenced by the polarity of the metabolites, the presence of chemicals, the extraction solvent, and the extraction procedure. The presence of compounds in an extract is affected by several factors, including temperature, geographical distribution, plant species, freshness, drying duration, and extraction technique [14]. The subsequent chemicals underwent molecular docking analysis to assess their individual contributions to the biological activity of the entire plant extract. An essential aspect in the rational design of small molecules and the comprehension of underlying biological processes is the evaluation of binding behavior [6].
3.4 Molecular docking
The significant antioxidants found in the ethanolic extract along with their prominent ability to scavenge free radicals and alpha glucosidase motivate us to conduct more in-depth analysis of the identified compounds present in the extract as potential anti-diabetic agent through in silico simulation. The binding interaction among the studied compounds and the binding site of alpha-glucosidase (pdb entry:3top) was studied through molecular docking to investigate whether the studied compounds will have favourable binding interactions with the alpha-glucosidase. They were found to possess favourable interactions with the receptor within the range of energy total values of − 41.4218 to − 121.208 kcal/mol as shown in Table 4. of − 105.572 kcal/mol, respectively (Table 4).
Based on the molecular docking investigation, the best five lead compounds identified are: molecule 14 (13-Docosenamide) possesses the highest energy total value of − 121.208 kcal/mol, followed by molecule 10 (15-Tetracosenoic acid) with an energy total value of − 120.465 kcal/mol, then by molecule 6 (Adenosine) with energy total value of − 105.572 kcal/mol, then by molecule 16 (Z-8-Methyl-9-tetradecenoic acid) with energy total value of − 104.497 kcal/mol, and lastly by molecule 9 (9,12-Octadecadien-1-ol) with energy total value − 102.256 kcal/mol, respectively as shown in Table 5.
From Table 5, 13-Docosenamide interacted with the binding site of alpha-glucosidase via hydrogen bond interaction with TYR1010 amino acid residue with a distance of 2.19 in Å unit and hydrophobic bond interactions with VAL1008, LYS1089, LYS1099, LEU1082 & TYR1010 groups of amino acid respectively (Fig. 3).
15-Tetracosenoic acid is the second-best lead compound as shown in Table 5 associated with the binding site of alpha-glucosidase via hydrogen bond interaction with GLN1109 amino acids with a distance of 2.43 a in Å unit. It further interacted with LYS1088, LYS1129, LYS1129, TYR1130 & TRP1648 group of amino acids via hydrophobic interactions, respectively (Fig. 4).
As also shown on Table 5, Adenosine was seen to have associated with the binding site of alpha-glucosidase via hydrogen bond interaction with SER1100 (1.83 Å) (2.73 Å) (2.76 Å), ILE1087(2.16 Å), PRO1078(2.84 Å), GLN1081(2.35 Å), GLU1079(1.85 Å), SER1006(1.68 Å) & VAL1008 (3.77 Å) group of amino acids (Fig. 5). Not only that, it interacted with VAL1008, LYS1099 & LYS1099 groups of amino acids via hydrophobic interactions, respectively.
Figure 6 have interactions with ARG1098(2.26 Å), ARG1098(2.68 Å), & PRO1220(2.78 Å) group of amino acids using hydrogen bond and with TYR1010, VAL1008, LEU1082, VAL1008, LYS1089, LYS1099 (2) & TYR1010 (2) classes of amino acids using hydrophobic interactions, as respectively shown in Table 3.
The last in the category which is 9,12-Octadecadien-1-ol (Fig. 7) also observed hydrogen bond interaction with LYS1089(1.83 Å), SER1004(2.23 Å), & LYS1089(2.44 Å) classes of amino acids and hydrophobic interactions with LYS1089 & LYS1089 classes of amino acids, as noticed in Table 5.
3.5 Drug-likeness features estimation of the reported five lead compounds
The drug-likeness of the reported best five lead compounds was estimated as follows: Lipinski's rule of 5 (Mw ≤ 500, hydrogen bond donors ≤ 5, hydrogen bond acceptors ≤ 10, Calculated MLog p ≤ 5) filtering criteria. which states that any small molecule that fails two of these stated conditions will have a problem related to oral bioavailability. All the reported compounds have a molecular weight ranging from 240.38 to 366.62 Da as presented in Table 4, which is within the accepted range for the Lipinski's rule of filtering conditions. They all have their number of hydrogen bond acceptors between 1 and 7 as clearly shown in Table 6, which is also within the range accepted. They further have their number of hydrogen bond donors between 1 and 4 as observed in Table 6 and is within the accepted range. Their Calculated MLog p value was between − 2.32 and 5.89 as shown in Table 4. Three compounds (6, 9, and 16) out of the 5 reported compounds were found to have zero (0) violation of the Lipinski's rule of 5, while two (2) molecules (10 and 14) were observed to have one (1) violation of the Lipinski's rule of 5 (in which their calculated Mlog p value has passed the threshold value set for this criteria). They are all orally bioavailable with an oral bioavailability score of 0.55 and 0.85, respectively. Furthermore, they will all be easily synthesized as indicated by their synthetic accessibility score of 3, respectively.
3.6 ADME-Tox feature estimation of the reported five lead compounds
The ADME-Tox features of the reported best five lead compounds were estimated to ascertain their pharmacokinetics properties as to how the body will respond to their administration. The estimated features are presented in Table 7. They were all reported to have intestinal absorption (human) between 54.036 and 93.138%, this is beyond the minimum threshold value set at 30% for the estimation of this property. They can be inferred to have a good intestinal absorption value since they have passed the minimum threshold value set. Their permeability to the skin is between the range of − 2.703–− 2.818 as clearly observed in Table 7. This is a good indication that they can permeate the skin. The threshold value set for the permeability of the blood–brain barrier (BBB) and central nervous system (CNS) is > 0.3 to < − 1 and > − 2 to < − 3, respectively. Based on this value, they are all ascertained to be within the range set. As such, they can also permeate both the BBB and CNS (except for compound 6 with CNS value of − 3.918), respectively. A good small molecule drug is anticipated to serve as a substrate and inhibitor of cytochrome a (CYP3A4). Three compounds were seen to be substrate to cytochrome a (CYP3A4) and the remaining two (that is, compounds 6 and 16) were not substrate to cytochrome a (CYP3A4), respectively. Additionally, none of them were observed to be an inhibitor to cytochrome a (CYP3A4). They can be regarded as drugs looking at other properties. Their total excretion clearance values are in the range of 0.737–2.09, this is an indication that they can be excreted from the body when the need arises. Finally, all of them are never toxic to the body except for compound 6 as demonstrated by their ames toxicity [33,34,35]. As such, they can be regarded as good alpha-glucosidase inhibitors for the treatment of diabetes mellitus.
3.7 Frontier molecular orbitals Dft estimation
The frontier molecular orbitals are the highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO), which are interconnected with the chemical reactivity and stability of a particular compound. The energy HOMO is linked to the electron-donor orbital, whereas the energy LUMO is connected to the electron-acceptor orbital [36]. The orbital energy gap aids to depict the kinetic stability and chemical reactivity of the compounds. The frontier molecular orbitals govern the path in which a compound relates to other species [37]. It was detected that compound 14 has the lowest reactivity with a bigger energy gap of 7.24 eV compared to compound 6 which is the most reactive with the smallest energy band gap of 5.42 eV (Table 8), respectively.
3.8 Molecular dynamics
Analyzing and verifying the stability of the expected ligand binding mechanism is greatly assisted by molecular dynamics (MD) simulations. To ensure a stable binding under dynamic conditions, our work used a 100 ns MD simulation to evaluate the mechanical stability of the promising molecule within its complexes. Based on characteristics such as root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF) and radius of gyration (Rg) of the protein backbone, MD simulation trajectories were meticulously analyzed to comprehend the many pathways followed by the system. By employing MM-GBSA-based methodologies, a thorough assessment of the stability and interactions within the ligand–protein complexes was accomplished in these analyses. Each parameter examined during the trajectory has its values presented in supplementary Table 1 as (average maximum and minimum).
3.8.1 Analyse RMSD
A protein's structural change from its initial configuration to its final disposition was evaluated using the RMSD (Root Mean Square Deviation). It is possible to evaluate the stability of a protein in respect to its conformation using simulation data. Less variation means more stability, and a lower RMSD indicates that the complex is more stable [38]. Figure 8(a) shows the RMSD graph of the Cα atoms in the protein. It shows that the ligand–protein complex maintained a balanced range of 0.005 Å to 0.170 Å over the 100 ns MD simulation. The fact that this range is less than 3 Å indicates that the combination was stable during the protein simulation, which is an acceptable outcome.
3.8.2 RMSF analysis
Analysis of amino acid fluctuation in the simulated protein provides valuable insights into how they contribute to maintaining the dynamic states of the protein–ligand complex [39]. Consequently, the fluctuation of each specific amino acid was evaluated, and the results are depicted in Fig. 8(b). Throughout the simulation, the RMSF (Root Mean Square Fluctuation) pattern of the amino acids bound to the ligand exhibited no significant or excessive fluctuations. It is noteworthy that the C-terminal amino acids displayed higher RMSF values owing to their intrinsic floating or flexible nature. The average RMSF serve as an indicator of amino acid stability during the MD simulation. For the ligand-3top complex, the average RMSF was determined to be 0.104 Å, as indicated in Supplementary Table 1. Generally, there was minimal fluctuation within the binding site region of the studied complex. These findings suggest that all amino acids in the protein maintained a consistent and strong binding to the ligand in the dynamic state.
3.8.3 Analysis of the radius of gyration
The amount of compactness and folding properties of protein structures and protein–ligand complexes can be evaluated using the radius of gyration (Rg) method [38]. Rg analysis provides insight into the protein–ligand complex's stiffness and compactness by estimating the atomic distribution relative to the mass center. In addition, smaller Rg gap values support unfolding and high values support folding of proteins, respectively. The computed Rg for each protein bound conformation to the simulated chemical is shown in Fig. 8(c). It looked like the complex was really compact and tight throughout the MD simulation. Under dynamic conditions, protein folding was not possible in any one system that showed significant divergence. A parameter's overall deviation can be seen as the difference between its maximum and minimum Rg values. The range of the Rg values was from 1.73316 to 1.7973 Å. It is worth mentioning that the observed system is both more compact and thermodynamically stable, as indicated by the minor difference between the maximum and lowest RGyr.
3.9 Solvent accessible surface (SASA) results
SASA can be used as an indicator of the stability of protein–ligand complexes because changes in the solvent accessible surface area can reflect conformational changes in the protein that can influence ligand binding. For example, a decrease in SASA may indicate a tightening of the protein structure around the ligand, which can be interpreted as an increase in complex stability. The results of the SASA analysis were shown in Fig. 8(d). Interestingly, the SASA values show stability around 115 nm2 from 17 ns until the end of simulation.
3.9.1 Analysis of binding free energy
When evaluating potential drugs, the binding free energy is still the most important parameter to consider, especially because lower Gbind values suggest that the complex is more stable. The binding free energy was determined at the beginning (0 ns) and end (100 ns) of the simulation using the MM/GBSA method, which confirmed the ligand's binding affinity with the protein website. As shown in Supplementary Table 2, the binding free energy of the ligand decreased from -16.1918 kJ/mol at the start of the simulation to − 19.3445 kJ/mol at its completion. It is worth mentioning that the decrease in binding free energy throughout the simulation suggests that interactions in the dynamic environment have become more stable. The main factors that comprise the binding free energy are the Coulombic energy (ΔGCoulomb), the Van der Waals energy (EvdW), and the lipophilicity energy. It appears from these results that hydrophobic contacts, Coulombic electrostatic interactions, and Van der Waals forces are very important in this intricate system. The effects of packing energy (ΔGBind packaging) and hydrogen bonding energy (ΔG H-bond) are not as noticeable. Polar solvation energy (ΔGSolvGB) and covalent energy, both of which are positive, indicate that ∆Gbind is not favorably affected.
4 Conclusion
This paper is the first of its kind to offer a comprehensive description of the molecular docking prediction, chemical composition, and biological activity. Strong antioxidant activity was demonstrated by the ethanol leaf extract in terms of alpha-glucosidase inhibitory activity, total phenolic compounds, reducing power, and radical scavenging activities. Based on the molecular docking tool we virtually screened, some compounds from the ethanol leaf the extract of E. punicifolia serve as potential alpha-glucosidase inhibitors. Five compounds (14: − 121.208 kcal/mol, 10: − 120.465 kcal/mol, 6: − 105.572 kcal/mol, 16: − 104.497 kcal/mol and 9: − 102.256 kcal/mol) were identified as the best lead compound, respectively. The identified lead compounds were seen to be drug-like compounds and orally bioavailable. Furthermore, they were also observed to be none-toxic with good ADME-Tox properties except for compound 6 that was toxic. Based on their computed frontier molecular orbitals, compound 6 was seen to be the most reactive and compound 14 as the less reactive, respectively. Therefore, the lead compounds except for 6 can serve as potential alpha-glucosidase inhibitors when subjected to a clinic trials.
Data availability
The manuscript has no associated data.
References
Abdulrahman MD, Zakariya AM, Hama HA, Hamad SW, Al-Rawi SS, Bradosty SW, Ibrahim AH (2022) Ethnopharmacology, biological evaluation, and chemical composition of Ziziphus spina-christi (L.) Desf.: a review. Adv Pharmacol Pharm Sci 2022:1–36
Caruntu S, Ciceu A, Olah NK, Don I, Hermenean A, Cotoraci C (2020) Thuja occidentalis L. (Cupressaceae): ethnobotany, phytochemistry and biological activity. Molecules 25(22):5416
Khan A, More K, Mali M, Deore SV, Patil M (2023) Phytochemical screening and Gas chromatography-mass spectrometry analysis on Ischaemumpilosum (Kleinex Willd.). Plant Sci Today 10(4):88–96
Usman M, Abdulrahman MD, Hamad S, Hama HA, Lema AA (2022) Antioxidants, Anti-inflammation, anti-hyperglycemia and chemical evaluation of the whole plant extracts of Anisopus mannii NE Br. Zanco J Pure Appl Sci 34(5):114–122
Abdulrahman MD, Hasan Nudin N, Khandaker MM, Ali AM, Mat N (2019) In vitro biological investigations on Syzygium polyanthum cultivars. Int J Agric Biol 22(6):1399–1406
Abdulrahman MD, Hamad W (2022) Traditional methods for treatment and management of measles in Northern Nigeria: medicinal plants and their molecular docking. Ethnobot Res Appl 23:1–18
Basting RT, Nishijima CM, Lopes JA, Santos RC, Périco LL, Laufer S, Bauer S, Costa MF, Santos LC, Rocha LR (2014) Antinociceptive, anti-inflammatory and gastroprotective effects of a hydroalcoholic extract from the leaves of Eugenia punicifolia (Kunth) DC. in rodents. J Ethnopharmacol 157:257–267
Abdulrahman M (2021) Antioxidant, alpha glucosidase and antibacterial evaluation of Syzygium mytifolium (Roxb.) Walp. Plant Sci Today 8(2):410–415
Yunusa AK, Abdullahi N, Rilwan A, Abdulkadir AR, Dandago MA (2018) DPPH radical scavenging activity and total phenolic content of rambutan (Nephelium lappaceum) peel and seed. Ann Food Sci Technol 19 (4)
Behera B, Satapathy KB (2023) Studies on in vitro evaluation of antibacterial and antioxidant activity of Hydrilla verticillata (Lf) Royle. Plant Sci Today 10(4):66–71
Yunusa AK, Dandago MA, Abdullahi N, Rilwan A, Barde A (2018) Total phenolic content and antioxidant capacity of different parts of cucumber (L.). Acta Univ Cibiniensis Ser E: Food Technol 22(2):13–20
Yunusa AK, Nouruddeen ZuB, Adam SA, Usman M, Dogara AM (2023) Effects of roasting conditions on sensory attributes, polyphenolic content and DPPH radical scavenging activity of peanut (Arachis hypogaea). Croat J Food Sci Technol 15(2):130–140
Yunusa AK, Rashid ZM, Mat N, Bakar CAA, Ali AM (2018) Chemicals and bioactivity discrimination of syconia of seven varieties of Ficus deltoidea Jack via ATR-IR spectroscopic-based metabolomics. Pharmacog J 10(6s):s147
Dogara AM (2023) Chemical composition of Corymbia citriodora. Nusant Biosci. https://doi.org/10.13057/nusbiosci/n150205
Ibrahim MT, Tahir SM, Umar AB, Abdulfatai U (2020) Theoretical investigation and design of some indole derivatives as potent β-glucuronidase inhibitors. Bull Nat Res Centre 44(1):1–11
Abdullahi M, Uzairu A, Shallangwa GA, Mamza PA, Ibrahim MT, Chandra A, Goel VK (2023) Molecular modelling studies of substituted indole derivatives as novel influenza a virus inhibitors. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2023.2280735
Ibrahim MT, Uzairu A (2022) 2D-QSAR, molecular docking, drug-likeness, and ADMET/pharmacokinetic predictions of some non-small cell lung cancer therapeutic agents. J Taibah Univ Med Sci 18(2):295–309
Pires DE, Blundell TL, Ascher DB (2015) pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem 58(9):4066–4072
Walter K, Pierre H (1964) Inhomogeneous electron gas. Phys Rev 136:B864
Hohenberg P, Kohn W (1964) Inhomogeneous electron gas. Phys Rev 136(3B):B864
Abdullahi M, Uzairu A, Shallangwa GA, Mamza PA, Ibrahim MT, Chandra A, Goel VK (2023) In-silico molecular modelling studies of some camphor imine based compounds as anti-influenza A (H1N1) pdm09 virus agents. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2023.2280735
Schrödinger L (2021) Schrödinger Release 2022–3: LigPrep. Schrödinger Inc, New York, NY, USA
Tabti K, Ahmad I, Zafar I, Sbai A, Maghat H, Bouachrine M, Lakhlifi T (2023) Profiling the structural determinants of pyrrolidine derivative as gelatinases (MMP-2 and MMP-9) inhibitors using in silico approaches. Comput Biol Chem 104:107855
Elbouhi M, Ouabane M, Tabti K, Badaoui H, Abdessadak O, El Alaouy MA, Elkamel K, Lakhlifi T, Sbai A, Ajana MA (2024) Computational evaluation of 1, 2, 3-triazole-based VEGFR-2 inhibitors: anti-angiogenesis potential and pharmacokinetic assessment. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2023.2301686
Tabti K, Sbai A, Maghat H, Lakhlifi T, Bouachrine M (2023) Computational exploration of the structural requirements of triazole derivatives as colchicine binding site inhibitors. ChemistrySelect 8(26):e202301707
Alrashidi M, Derawi D, Salimon J, Yusoff MF (2022) The effects of different extraction solvents on the yield and antioxidant properties of Nigella sativa oil from Saudi Arabia. J Taibah Univ Sci 16(1):330–336
Ngamkhae N, Monthakantirat O, Chulikhit Y, Boonyarat C, Maneenet J, Khamphukdee C, Kwankhao P, Pitiporn S, Daodee S (2022) Optimization of extraction method for Kleeb Bua Daeng formula and comparison between ultrasound-assisted and microwave-assisted extraction. J Appl Res Med Aromat Plants 28:100369
Tan ND, Tuyen VTX, Khang TN (2023) Determination of the content of major chemical components and bioactive compounds and antioxidant ability of Hong Quan (Flacourtia jangomas (Lour.) Raeusch) fruit cultivated in An Giang Vietnam. Plant Sci Today 10(4):39–43
Aftab A, Yousaf Z, Rashid M, Younas A, Yasin H, Riaz N, Mansoor Q (2023) Vegetative part of Nigella sativa L. potential antineoplastic sources against Hep2 and MCF7 human cancer cell lines. J Taibah Univ Sci 17(1):2161294
Gulcin İ, Alwasel SH (2023) DPPH radical scavenging assay. Processes 11(8):2248
He Y, Lin Y, Li Q, Gu Y (2020) The contribution ratio of various characteristic tea compounds in antioxidant capacity by DPPH assay. J Food Biochem 44(7):e13270
Shehu S, Abdurahman EM, Danmalam UH, Mohammed A, Barau AI (2022) Evaluation of antioxidant activity of Thesium viride hill (Santalaceae). The Niger J Pharm. https://doi.org/10.51412/psnnjp.2022.30
Kraithong S, Teerapattarakan N, Balasubramanian B, Issara U (2022) Bioactive compounds in tea: effect of imbalanced intake on digestive enzymes activity, cytochrome inhibition and drug interaction. S Afr J Bot 150:58–68
Li H, Sheng Y, Li W, Yuan L (2022) Recent advances in molecular fluorescent probes for CYP450 sensing and imaging. Chemosensors 10(8):304
Ahmed A, Saeed A, Ejaz SA, Aziz M, Hashmi MZ, Channar PA, Abbas Q, Raza H, Shafiq Z, El-Seedi HR (2022) Novel adamantyl clubbed iminothiazolidinones as promising elastase inhibitors: design, synthesis, molecular docking ADMET and DFT studies. RSC Adv 12(19):11974–11991
Khalid N, Qayyum A, Bilal M, Al-Fuqaha A, Qadir J (2023) Privacy-preserving artificial intelligence in healthcare: techniques and applications. Comput Biol Med 158:106848
Charanya C, Sampathkrishnan S, Kumutha R, Praveena J, Bhaskaran A, Prabakaran A, Balamurugan N (2023) Synthesis, quantum computational analysis and molecular docking of 3-(2-hydroxyphenyl)-1-phenyl propanone: a combined experimental and theoretical analysis. Polycycl Aromat Compd 43(10):8903–8932
Abdessadak O, Tabti K, Alaqarbeh M, Elmchichi L, Koubi Y, Sbai A, Ajana MA, Lakhlifi T, Bouachrine M (2023) Computational studies for development of triazole-pyrimidines as inhibitor of α-tubulin receptor. ChemistrySelect 8(46):e202301992
Hassan F, Jamil F, Hussain A, Ali HM, Janjua MM, Khushnood S, Farhan M, Altaf K, Said Z, Li C (2022) Recent advancements in latent heat phase change materials and their applications for thermal energy storage and buildings: a state of the art review. Sustain Energy Technol Assess 49:101646
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by [Abdulrahman Mahmoud Dogara], [Muhammad Tukur Ibrahim] [Aisha Abdullahi Mahmud] [Musa Daniel Danladi] [Abubakar Abdullahi Lema] [Muhammad Usman] and [Abdulsalam Salisu Tahir], Molecular dynamic simulation by [Kamal TABTI] The first draft of the manuscript was written by [Abdulrahman Mahmoud Dogara and Muhammad Tukur Ibrahim] and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.”
Corresponding author
Ethics declarations
Conflict of interest
The authors declares no conflict of interest.
Ethical approval
Not applicable.
Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dogara, A.M., Ibrahim, M.T., Mahmud, A.A. et al. Biological activity, chemical composition, and molecular docking of Eugenia punicifolia (Kunth) DC. J.Umm Al-Qura Univ. Appll. Sci. (2024). https://doi.org/10.1007/s43994-024-00160-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s43994-024-00160-y