Abstract
Background
β-glucuronidase enzyme is mostly found in plants and animals. It plays a vital role in detoxification of reactive metabolites that are interrelated to several illnesses and the growth of colon cancer. It speeds up the breaking down of β-glucuronosyl-O-bonds. Lack of β-glucuronidase enzyme leads to Sly syndrome in humans, and overexpression of this enzyme leads to many diseases. Therefore, it becomes necessary to mediate the effect of this enzyme.
Result
Theoretical investigation via QSAR modeling on 30 indole derivatives was performed to build a model which could be used to predict the activity of the indole derivatives. QSAR was carried out using multi-linear regression (MLR) method utilizing genetic function approximation (GFA) to develop the QSAR models. A very high predictive QSAR model was reported based on its statistical fitness with good internal and external validation parameters: R2trng = 0.954942, Qcv2 = 0.925462, R2test = 0.855393, and LOF = 0.042924. Molecular docking on the 30 indole derivatives was also performed to screen and identify the lead compound that would be used as template for designing new indole compounds. The docking investigation reveals that ligand 10 binds very tight in the binding pocket of β-glucuronidase enzyme with binding energy of − 9.5 kcal/mol. The ligand (10) was chosen as a template for designing new β-glucuronidase inhibitors. The four design compounds were found to be better than the template and the standard drug (D-saccharic acid 1, 4-lactone) with binding energies of − 9.6, − 9.7, − 9.8, and − 9.9 kcal/mol.
Conclusion
A very high predictive QSAR model with good internal and external validation parameters: R2trng = 0.954942, Qcv2 = 0.925462, R2test = 0.855393, and LOF = 0.042924, was built and reported in this study. Molecular docking investigation reveals that the most potent compound among all the data set was compound 10 with binding energy of − 9.5 kcal/mole. It bound to the binding pocket of β-glucuronidase enzyme via hydrophobic, electrostatic, and hydrogen bond, and it was retained as template for designing new indole compounds. The design compound with serial number ID 4 was identified to have the highest binding energy of − 9.9 kcal/mole among the designed compounds. It bound to the binding site of the β-glucuronidase enzyme via halogen, hydrophobic, electrostatic, and hydrogen bond. The design compounds were discovered to be better than the template used in the design and the standard drug.
Similar content being viewed by others
Background
Indole is a heterocyclic aromatic organic compound consisting of a six-membered benzene ring fused to a five-membered pyrrole ring (Bouchikhi et al. 2008) with numerous physiological activities which include anti-inflammatory, anticancer, antiulcer, antimalarial, antioxidant, antibiotic, antimicrobial, and anticonvulsant (Patil and Dandagvhal 2016). Indole is broadly disseminated in the real environment and can be generated by different species of bacteria (Lee and Lee 2010). The synthesis of indole has been performed utilizing different starting materials in the literature of organic chemistry (Chadha and Silakari 2017).
β-Glucuronidase enzyme is mostly found in plant in the Escherichia, anaerobic bacteroides, and Clostridia. β-Glucuronidase enzyme is also found in human fluids, serum, blood cells, gastric juice, bile, urine, spleen, and it can also be found in organs like the kidney, liver, lung, and muscles (Ali et al. 2016; Gloux et al. 2011). It plays a vital role in detoxification of reactive metabolites that are interrelated to several illnesses and the growth of colon cancer (De Moreno de LeBlanc and Perdigón 2005). β-Glucuronidase enzyme speeds up the breaking down of β-glucuronosyl-O-bonds. Lack of β-glucuronidase enzyme leads to Sly syndrome in animals (humans) that is related to the high level of glycosaminoglycans in cells, and overexpression of this enzyme leads to many diseases which include urinary tract infections, active pyelonephritis, acute renal necrosis, and some pathological conditions including epilepsy, cancer, AIDS, renal diseases, neoplasm of the breast, larynx, bladder, and testes, and inflammation in joints and hepatic. Therefore, it becomes necessary to mediate the effect of this enzyme (Salar et al. 2015; Sharma et al. 2014; Taha et al. 2015).
Ibrahim et al. performed quantitative structure-activity relationship (QSAR) modeling, molecular docking, and pharmacokinetic study on 30 N-Arylidenequinoline-3-carbohydrazides analogs utilizing multi-linear regression analysis adopting genetic function algorithm method. Semi empirical method using PM6 basis set was used for complete geometry optimization of the data set. The molecular docking study showed that the most active chemical in the data set was better than the standard β-glucuronidase inhibitor both in terms of binding scores and the amino acid residues that interacted with the drug and β-glucuronidase enzyme. The pharmacokinetic studies indicated that none of the chemicals violated any of the condition set by the Lipinski’s Rule of five which confirm the bioavailability of these chemicals (Ibrahim et al. 2020a).
Theoretical modeling is a unique area of interest in computer-aided drug design which provides computational software and tools that are utilized in the design and development of novel compounds of medicinal benefit (Jorgensen 2004). Quantitative structure-activity relationship (QSAR) is a computational modeling technique used to study the connection between the experimental activities with different physicochemical properties associated with the structures of a particular molecule (Ibrahim et al. 2020b; Ojha Lokendra et al. 2013). Molecular docking is also a computational modeling technique used to predict the binding energy of intermolecular complexes based on their 3D structures when one bind to the other (Abdullahi et al. 2020; Kitchen et al. 2004). This study is aimed at carrying out QSAR and docking simulation on indole derivatives against β-glucuronidase enzyme and design new β-glucuronidase inhibitors using structure-based drug design.
Methods
QSAR modeling methodology
Data retrieving, activity normalization, and structure drawing
Thirty set of indole derivatives and their corresponding β-glucuronidase inhibitory activities (IC50) were retrieved from the work of (Baharudin et al. 2017) for the purpose of this study. After the data were collected from the literature, the inhibitory activities (IC50) of these compounds were transformed to their corresponding negative logarithm (pIC50) using Eq. 1. The Chemdraw software Ultra version 12.0 was employed for drawing of the structures of all the data set prior to optimization as shown in Fig. 1 (Ibrahim et al. 2019). Figure 1 shows the 2D general structure of the studied compounds.
Searching for optimum geometry and descriptor computations
In order to search the structure of all the data set at global minima on potential energy surface (PES), semi-empirical method using PM6 basis set was used for the searching of the most optimum geometry of all the data set on the PES (Amin and Gayen 2016). For the computation of physicochemical parameters (descriptors), Pharmaceutical Data Exploration Laboratory (PaDEL) descriptor tool kit was used to generate all the descriptors used in this research (Yap 2011).
Dataset pretreatment and division
Before dataset division, the data were pre-treated using data pre-treatment software retrieved from drug theoretic and cheminformatics (DTC) laboratory to remove redundant values (Ambure et al. 2015). Data division software was further employed to separate the data into model building set (70%) and validation set (30%) based on the Kennard-Stone algorithm also gotten from DTC laboratory (Kennard and Stone 1969).
Model generation and validation
The model building set was subjected for further analysis (regression analysis) adopting genetic function algorithm (GFA) method in material studio software to generate the models.
The equation for the regression analysis is shown in Eq. (2).
where Y is the pIC50, “B’s” are coefficients for the corresponding “Z’s” which are the descriptors, and the last variable “C” is the regression constant (Ibrahim et al. 2020c).
After model development, the model must be subjected to validation assessments, in order to ascertain the model’s quality, reliability, and predictive ability. The most widely used methods for the assessments of QSAR models were the internal validations, external validations, and applicability domain. The square correlation coefficients (R2) of both the training and test sets are used alongside with (Tropsha 2015) cross-validation coefficient (Qcv2) as the assessment parameters. Cross-validation is the statistical theory of separating of data into subsets such that the analysis is first performed on a single subset, while the other subset(s) are retained for subsequent use to confirm and validate the first analysis (Jalali-Heravi and Kyani 2004) and are defined as:
where Yprd and Yobs are the predicted and observed activity (pIC50) respectively of the validation set, and Ymntrng is the mean activity value of the model building set.
The multi-collinearity between the descriptors can be detected using their variation inflation factors (VIF), which can be calculated as follows:
where R2 is the correlation coefficient of the model. If VIF is equal to 1, it means there is no inter-correlation between the descriptors; if it is with the range of 1–5, the model is acceptable; and if greater than 10, the model is bad and therefore rejected and must be rechecked (Beheshti et al. 2016).
In order to confirm the predictability, strength, reliability, and accuracy of the QSAR model developed and also identify influential and outlier compounds, the mechanistic space of the model must be exploited (applicability domain) (Tropsha et al. 2003). In exploiting the applicability domain of the studied model, leverage approach was employed and is given as hi:
where yi is the model building compound matrix I, Y is n × k descriptor matrix of the model building set compounds, and YT is the transpose matrix Y used to develop the model. As a prediction tool, the thresh-hold value (h*) which is the limit for Y values and is given as:
where q is the number of descriptors in the model, and z is the number of model building compounds. For any QSAR model to be considered as valid and used, it must pass both the internal and external validations as recommended by Veerasamy et al. (Veerasamy et al. 2011) (see Table 1).
Docking methodology
Docking studies was conducted on personal Dell computer system, with Intel ® Core™ i7, 8GB of RAM using the Auto dock vina of Pyrex virtual screening software, UCSF Chimera version 1.10.2, and Discovery Studio Visualizer software.
Preparation of ligands and the enzyme β-glucuronidase
Before docking commences, the optimum geometry optimized using Spartan’14 was saved as protein data bank file format (ligand preparations) (Abdulfatai et al. 2017). Figure 2a presents the 3D structure of the prepared of the most potent ligand.
3D crystal structure of human β-glucuronidase was retrieved from protein data bank (with PDB ID 1bhg). Discovery Studio Visualizer was used to prepare the human β-glucuronidase enzyme for the docking studies; in the course of the preparation, chain B, heteroatoms, and co-ligands were deleted from the dimer structure and saved as protein data bank file format (Veerasamy et al. 2011). Figure 2b shows the prepared structure of the human β-glucuronidase.
Docking simulation
Vina of Pyrex virtual screening software was used to dock the ligands with human β-glucuronidase (Trott and Olson 2010). Vina of Pyrex virtual screening software has a short-coming of separating the receptor and the ligand after performing the docking. As a result of that, the UCSF Chimera 1.10.2 software was utilized to re-build the complexes (ligand-receptor) for further investigation. Discovery Studio Visualizer was used to visualize the ligand-receptor to show the interactions.
Design
Structure-based drug design also called direct drug design was used to design new β-glucuronidase inhibitors. Ligand 10 was chosen as the template as it has the highest binding energy of − 9.5 kcal/mol. Structural modification was carried out on the C-4 of the indole moiety by adding halo-substituted phenyl ring. Table 2 shows the structures of the design compounds

Results
The results of the QSAR modeling is given below:
Model 1
pIC50 = − 0.250653769 * AATS4i − 0.007819193 * ATSC2m + 0.001714704 * ATSC8v + 0.949838372 * SpMAD_Dzs − 0.050437194 * SpMax_Dt + 36.58388647.
R2trng = 0.954942, R2adj = 0.93885, Qcv2 = 0.925462, Ntrng = 20, R2test = 0.855393, Ntest = 10, R2 − Q2 = 0.016092, and LOF = 0.042924.
Results of docking studies
The results of the docking studies are shown below in Table 6 and Fig. 5.
Results of the design compounds
The result of the newly designed β-glucuronidase inhibitors is given in Table 7 and Fig. 6 a and b.
Discussion
QSAR modeling results
Three QSAR models were built; out of these built models, the best reported model was based on its statistical fitness with R2 value of 0.954942, R2adj value of 0.93885, Qcv2 value of 0.925462, and the R2pred value of 0.855393 and also lowest Friedman’s Lack of fit value of 0.042924. On taking the comparison of the statistical parameters of the reported model with those reported by Veerasamy et al. (Veerasamy et al. 2011) shown in Table 1, it can be seen clearly that the statistical parameters of the reported model were all greater than the minimum recommended values which confirmed the reliability of the model. Table 3 presents the symbols, descriptions, and the classes of descriptors that appear in the reported model. The physicochemical parameters responsible for the inhibitory activities of the β-glucuronidase inhibitors were identified to be AATS4i, ATSC2m, ATSC8v, SpMAD_Dzs and SpMax_Dt. Among these descriptors as seen in the model, SpMAD_Dzs has the highest coefficient of + 0.949838372 which gave the highest contribution in the model followed by ATSC8v. ATSC2m gave the lowest contribution in the model as it has the lowest coefficient of − 0.007819193. Either increase or decrease in these physicochemical parameters, the inhibitory activities of these compounds may increase or decrease.
Predicted activities of both the model building set and that of the validation set were plotted against the actual activities (Fig. 3a) in order to affirm the strength of the reported model. The tool used in this case is R2 value of both the plot and that of the internal validation. The strength of the reported model was confirmed by the corroboration of R2 value of the plot (0.9963) and that of the internal validation (0.954942).
Every good QSAR model is expected to be free from systematic error. In order to determine whether the selected model is free from systematic error, predicted activity was plotted against standardized residuals (Fig. 3b). The random propagation of the standardized residuals on either side of zero on the plot confirmed that the reported model is free from systematic error.
The observed activities were seen to have good correlations with the predicted activities. Table 4 represents the structures, IC50, pIC50, predicted pIC50, and residual values of the data set. The low residual values observed in the table further confirmed the stability and reliability of the reported model.
The correlation analysis of descriptors in the model building set of the reported model in Table 5 shows the importance of descriptors in the model. From the table, no correlation exists between the descriptors that appear in the reported model. The computed VIF values for all descriptors were obtained to be less than 5 (Table 5) indicating that the reported model was statistically significant and the descriptors were orthogonal (no multi-collinearity between the descriptors).
To confirm the quality of the reported model, it is subjected to applicability domain as shown in Fig. 4 (the plot of leverage value calculated for all the data set vs. standardized residuals or Williams plot), which permit a graphical identification of both the outliers and influential compounds in the chosen model (Beheshti et al. 2016). It can be seen from the plot that all the compounds of the model building set and the validation set were in the square area (mechanistic space). Also, there were no outliers or influential compounds with standard residuals > 3d for both model building set and the validation set. This confirmed the quality of the reported model which means the model can make good prediction of activities of new compounds.
Results of docking analysis
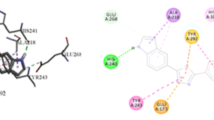
Docking analysis on the four highly potent indole derivatives and the standard drug was performed to investigate the nature of interactions between them and their target receptor (β-glucuronidase). Table 6 gives the results of the docking analysis. It was seen from the docking results that ligand 10 being the most active compound has the highest binding free energy of − 9.5 kcal/mol. The most active ligand bound to the active site of β-glucuronidase enzyme by hydrophobic, electrostatic, and hydrogen bond. Furthermore, it bound via hydrophobic with TYR29, LEU300, PRO323, MET393, CYS396, ARG436, and LEU322 amino acid residues and bound through electrostatic with HIS440 amino acid residue. More so, it bound via hydrogen bond with TYR29 (1.93247 Å), HIS440 (2.3299 Å), and GLU390 (3.68037 Å) amino acid residues. Looking at ligand 2 having the docking scores of − 8.8 kcal/mol, it forms hydrophobic interactions with TYR29, PRO148, LEU322, PRO323, MET393, ARG436, and ARG436; electrostatic interactions with CYS396; and hydrogen bond interactions with ASN439 (2.20367 Å), HIS440 (2.8928 Å), and LEU28 (3.55365 Å) amino acid residues. The standard drug with docking score of − 5.7 kcal/mol formed only hydrogen bond with ASN502 (2.42729 Å) and GLN524 (3.44886 Å) amino acid residues of β-glucuronidase enzyme. On comparing the most active compound with the standard drug, it can be seen that the most active compound has higher docking score than the standard drug which may be attributed to the kinds of interaction that occur between the active compound and the binding pocket of β-glucuronidase. Figure 5 shows the 2D structure of the visualized ligand-receptor 10.
Result of the design
Four new β-glucuronidase inhibitors were designed and found to bind tight in the binding pose of β-glucuronidase enzyme (Table 7). Addition of halo-substituted phenyl ring increased the binding energy of the designed compounds. The designed compounds were found to have higher binding energy than the template (− 9.5 kcal/mole) and the standard drug (5.7 kcal/mole). A design compound with serial number ID 4 was identified to have the highest binding energy among the designed compounds. It bound to the binding site of the β-glucuronidase enzyme via halogen bond with TYR505; electrostatic bond with GLU540, ASP207, GLU451, GLU451, and GLU451; hydrophobic bonds with ASP207; and hydrogen bond with GLU540 (1.86751 Å) and TYR505 (2.71161 Å). Figure 6 a and b show the 2D structure of the visualized ID 4 and the standard drug with their target enzyme.
Conclusion
A very high predictive QSAR model with good internal and external validation parameters: R2trng = 0.954942, Qcv2 = 0.925462, R2test = 0.855393, and LOF = 0.042924 was built and reported in this study. The model was further subjected to applicability domain to confirm the quality of the model in terms of its predictions and found to make good prediction. Docking investigation reveals that the most potent compound among all the data set bound to the binding pocket of β-glucuronidase enzyme via hydrophobic, electrostatic, and hydrogen bond. The design compound with serial number ID 4 was identified to have the highest binding energy of − 9.9 kcal/mole among the designed compounds. It bound in the binding site of the β-glucuronidase enzyme via halogen, hydrophobic, electrostatic, and hydrogen bond. The design compound was discovered to be better than the template used in the designing new compound and the standard drug. The aim of this research as stated was achieved.
Availability of data and materials
Not applicable to this research
Abbreviations
- QSAR:
-
Quantitative-structure activity relationship
- MLR:
-
Multi-linear regression
- GFA:
-
Genetic function approximation
- PDB:
-
Protein data bank
- PES:
-
Potential energy surface
- DTC:
-
Drug theoretic and cheminformatics
- VIF:
-
Variation inflation factor
- PaDEL:
-
Pharmaceutical Data Exploration Laboratory
References
Abdulfatai U, Uzairu A, Uba S (2017) Quantitative structure-activity relationship and molecular docking studies of a series of quinazolinonyl analogues as inhibitors of gamma amino butyric acid aminotransferase. J Adv Res 8:33–43
Abdullahi M, Uzairu A, Shallangwa GA, Mamza P, Arthur DE, Ibrahim MT (2020) In-silico modelling studies on some C14-urea-tetrandrine derivatives as potent anti-cancer agents against prostate (PC3) cell line. J King Saud Univ Sci 32:770–779
Ali F, Khan KM, Salar U, Iqbal S, Taha M, Ismail NH, Perveen S, Wadood A, Ghufran M, Ali B (2016) Dihydropyrimidones: as novel class of b-glucuronidase inhibitors. Bioorg Med Chem 24:3624–3635
Ambure P, Aher RB, Gajewicz A, Puzyn T, Roy K (2015) “NanoBRIDGES” software: open access tools to perform QSAR and nano-QSAR modeling. Chemom Intell Lab Syst 147:1–13
Amin SA, Gayen S (2016) Modelling the cytotoxic activity of pyrazolo-triazole hybrids using descriptors calculated from the open source tool “PaDEL-descriptor”. J Taibah Univ Sci 10:896–905
Baharudin MS, Taha M, Imran S, Ismail NH, Rahim F, Javid MT, Khan KM, Ali M (2017) Synthesis of indole analogs as potent β-glucuronidase inhibitors. Bioorg Chem 72:323–332
Beheshti A, Pourbasheer E, Nekoei M, Vahdani S (2016) QSAR modeling of antimalarial activity of urea derivatives using genetic algorithm–multiple linear regressions. J Saudi Chem Soc 20:282–290
Bouchikhi F, Anizon F, Moreau P (2008) Synthesis and antiproliferative activities of isoindigo and azaisoindigo derivatives. Eur J Med Chem 43:755–762
Chadha N, Silakari O (2017) Indoles as therapeutics of interest in medicinal chemistry: bird’s eye view. Eur J Med Chem 134:159–184
De Moreno de LeBlanc A, Perdigón G (2005) Reduction of b-glucuronidase and nitroreductase activity by yoghurt in a murine colon cancer model. Biocell 29:15–24
Gloux K, Berteau O, Béguet F, Leclerc M, Doré J (2011) A metagenomic β-glucuronidase uncovers a core adaptive function of the human intestinal microbiome. Proc Natl Acad Sci 108:4539–4546
Ibrahim MT, Uzairu A, Shallangwa GA, Uba S (2019) QSAR modelling and docking analysis of some thiazole analogues as⍺-glucosidase inhibitors. J Eng Exact Sci 5:0257–0270
Ibrahim MT, Uzairu A, Shallangwa GA, Uba S (2020c) In-silico activity prediction and docking studies of some 2, 9-disubstituted 8-phenylthio/phenylsulfinyl-9 h-purine derivatives as anti-proliferative agents. Heliyon 6:e03158
Ibrahim MT, Uzairu A, Uba S, Shallangwa GA (2020b) Computational modeling of novel quinazoline derivatives as potent epidermal growth factor receptor inhibitors. Heliyon 6:e03289
Ibrahim MT, Uzairu A, Umar AB, Bello AS, Isyaku Y (2020a) Molecular modelling, docking and pharmacokinetic studies of N-arylidenequinoline-3-carbohydrazides analogs as novel β-glucuronidase inhibitors. J Mex Chem Soc 64
Jalali-Heravi M, Kyani A (2004) Use of computer-assisted methods for the modeling of the retention time of a variety of volatile organic compounds: a PCA-MLR-ANN approach. J Chem Inf Comput Sci 44:1328–1335
Jorgensen WL (2004) The many roles of computation in drug discovery. Science 303:1813–1818
Kennard RW, Stone LA (1969) Computer aided design of experiments. Technometrics 11:137–148
Kitchen DB, Decornez H, Furr JR, Bajorath J (2004) Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov 3:935
Lee J-H, Lee J (2010) Indole as an intercellular signal in msicrobial communities. FEMS Microbiol Rev 34:426–444
Ojha Lokendra K, Rachana S, Rani BM (2013) Modern drug design with advancement in QSAR: a review. Int J Res Biosci 2:1–12
Patil SS, Dandagvhal KR (2016) Indole - an interesting scaffold in drug discovery. Int J Res Pharm Chem 6:301–311
Salar U, Miana GA, Khan KM, Naz F, Siddiqui NI, Taha M, Tauseef S, Khan S, Perveen S (2015) Biology-oriented syntheses (BIOS) of novel santonic-1, 3, 4-oxadiazole derivatives under microwave-irradiation and their antimicrobial activity. J Chem Soc Pak 37
Sharma R, Patil S, Maurya P (2014) Drug discovery studies on quinoline-based derivatives as potential antimalarial agents. SAR QSAR Environ Res 25:189–203
Taha M, Ismail NH, Imran S, Selvaraj M, Rashwan H, Farhanah FU, Rahim F, Kesavanarayanan KS, Ali M (2015) Synthesis of benzimidazole derivatives as potent β-glucuronidase inhibitors. Bioorg Chem 61:36–44
Tropsha, A. and Bajorath, J.r., Computational methods for drug discovery and design. 2015, ACS Publications.
Tropsha A, Gramatica P, Gombar VK (2003) The importance of being earnest: validation is the absolute essential for successful application and interpretation of QSPR models. Mol Inform 22:69–77
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461
Veerasamy R, Rajak H, Jain A, Sivadasan S, Varghese CP, Agrawal RK (2011) Validation of QSAR models-strategies and importance. Int J Drug Design Disc 3:511–519
Yap CW (2011) PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J Comput Chem 32:1466–1474
Acknowledgements
The authors acknowledge the technical effort of Ahmadu Bello University, Zaria—Nigeria.
Funding
This research did not receive any funding from anybody.
Author information
Authors and Affiliations
Contributions
MTI: contributed throughout the research work. SMT: gives directives and technical advices. ABU: partakes in technical activities. UA: also partakes in technical activities. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable to this research
Consent for publication
Not applicable to this research
Competing interests
All authors have declared that there is no conflict of interest regarding this submission.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ibrahim, M.T., Tahir, S.M., Umar, A.B. et al. Theoretical investigation and design of some indole derivatives as potent β-glucuronidase inhibitors. Bull Natl Res Cent 44, 114 (2020). https://doi.org/10.1186/s42269-020-00372-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42269-020-00372-2