Abstract
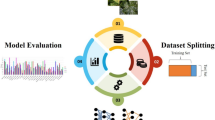
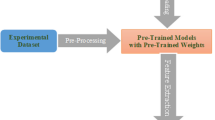
Supervised learning will be a bottleneck for developing plant disease identification since it relies on learning from massive amounts of carefully labeled images, which is costly and time-consuming. On the contrary, self-supervised learning has succeeded in various image classification tasks; however, it has not been applied broadly in the plant disease analysis process. This work, therefore, studies the effectiveness of self-supervised learning using contrastive pre-training with SimCLR for plant disease image classification. We investigated unsupervised pre-training scenarios on unlabeled plant images across multiple architectures, including supervised fine-tuning on labeled samples. In addition, we explored the label efficiency of the self-supervised approach, acquired by fine-tuning the models on various fractions of labeled images. Our results demonstrated that the performance of self-supervised learning on plant disease became comparable to that of the supervised training approach.





Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are courtesy of Bhattarai (2023) and available in the kaggle repository of https://www.kaggle.com/datasets/vipoooool/new-plant-diseases-dataset.
References
Alirezazadeh P, Schirrmann M, Stolzenburg F (2023) Improving deep learning-based plant disease classification with attention mechanism. Gesunde Pflanzen 75:49
Azizi S, Mustafa B, Ryan F, Beaver Z, Freyberg J, Deaton J, Loh A, Karthikesalingam A, Kornblith S, Chen T, Natarajan V, Norouzi M (2021) Big self-supervised models advance medical image classifications. In: Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV)
Bhattarai S (2023) New plant diseases dataset. Available at https://www.kaggle.com/datasets/vipoooool/new-plant-diseases-dataset.
Borhani Y, Khoramdel J, Najafi E (2022) A deep learning based approach for automated plant disease classification using vision transformer. Sci Rep. https://doi.org/10.1038/s41598-022-15163-0
Chen J, Chen J, Zhang D, Sun Y, Nanehkaran Y (2020a) Using deep transfer learning for image-based plant disease identification. Comput Electron Agric 173:105220
Chen T, Kornblith S, Swersky K, Norouzi M, Hinton G (2020b) Big self-supervised models are strong semi-supervised learners. Adv Neural Inform Process Syst 33
Chen T, Kornblith S, Norouzi, M, Hinton G (2020c) A simple framework for contrastive learning of visual representations. In: Proceedings of Machine Learning Research Vol. 119
Fang U, Li J, Lu X, Gao L, Ali M, Xiang Y (2021) Self-supervised cross-iterative clustering for unlabeled plant disease images. Neurocomputing 456:36
Güldenring R, Nalpantidis L (2021) Self-supervised contrastive learning on agricultural images. Comput Electron Agric 191:106510
Guo X, Qiu Y, Nettleton D, Schnable PS (2023) High-Throughput Field Plant Phenotyping: a self-supervised sequential CNN method to segment overlapping plants. Plant Phenom 5:2023
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: IEEE Conference on Computer Vision and Pattern Recognition (CVPR)
Howard A, Sandler M, Chu G, Chen LC, Chen B, Tan M, Wang W, Zhu Y, Pang R, Vasudevan V, Le QV, Adam H (2019) Searching for MobileNetV3. In: IEEE/CVF International Conference on Computer Vision (ICCV)
Lee S, Goëau H, Bonnet P, Joly A (2020) New perspectives on plant disease characterization based on deep learning. Comput Electron Agric 170:105220
Li L, Zhang S, Wang B (2021) Plant disease detection and classification by deep learning—a review. IEEE Access 9:105393
Li Y, Zhan X, Liu S, Lu H, Jiang R, Guo W, Chapman S, Ge Y, Solan B, Ding Y, Baret F (2023) Self-supervised plant phenotyping by combining domain adaptation with 3D plant model simulations: application to wheat leaf counting at seedling stage. Plant Phenom. https://doi.org/10.34133/plantphenomics.0041
Lin X, Li CT, Adams S, Kouzani AZ, Jiang R, He L, Hu Y, Vernon M, Doeven E, Webb L, Mcclellan T, Guskich A (2023) Self-supervised leaf segmentation under complex lighting conditions. Pattern Recogn 135:109021
Nagasubramanian K, Singh A, Singh A, Sarkar S, Ganapathysubramanian B (2022) Plant phenotyping with limited annotation: doing more with less. Plant Phenome J. https://doi.org/10.1002/ppj2.20051
Ogidi FC, Eramian MG, Stavness I (2023) Benchmarking self-supervised contrastive learning methods for image-based plant phenotyping. Plant Phenom. https://doi.org/10.34133/plantphenomics.0037
Reed C, Yue X, Nrusimha A, Ebrahimi S, Vijaykumar V, Mao R, Li B, Zhang S, Guillory D, Metzger S, Keutzer K, Darrell T (2022) Self-supervised pretraining improves self-supervised pretraining. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV)
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. In: International Conference on Learning Representations (ICLR)
Tan M, Le QV (2019) EfficientNet: rethinking model scaling for convolutional neural networks. In: International Conference on Machine Learning (ICML)
Verma A, Shekhar S, Garg H (2022) Plant disease classification using deep learning framework. In: International Conference on Computational Intelligence and Sustainable Engineering Solutions (CISES)
Yang G, Chen G, He Y, Yan Z, Guo Y, Ding J (2020) Self-Supervised collaborative multi-network for fine-grained visual categorization of tomato diseases. IEEE Access 8:211912
Zapata PAM, Roth S, Schmutzler D, Wolf T, Manesso E, Clevert DA (2021) Self-supervised feature extraction from image time series in plant phenotyping using triplet networks. Bioinformatics 37(6):861
Author information
Authors and Affiliations
Contributions
All authors contributed to this article equally.
Corresponding author
Ethics declarations
Conflicts of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bunyang, S., Thedwichienchai, N., Pintong, K. et al. Self-supervised learning advanced plant disease image classification with SimCLR. Adv. in Comp. Int. 3, 18 (2023). https://doi.org/10.1007/s43674-023-00065-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s43674-023-00065-z