Abstract
Early diagnosis of plant diseases is crucial for preventing plagues and mitigating their effects on crops. The most precise automatic methods for identifying plant diseases using images of plant fields are powered by deep learning. Big image datasets should always be gathered and annotated for these methods to work, which is often not technically or financially feasible. This paper offers one-shot learning (OSL) techniques for plant disease classification with limited datasets utilizing Siamese Neural Network (SNN). There are five different crop kinds in the dataset: grape, wheat, cotton, cucumber, and corn. Five sets of images showing both healthy and diseased crops are used to represent each of the new crops. The dataset's includes 25 classes with 875 leaf images. Data augmentation techniques are used to enhance the size and dimension of the plant leaf disease image dataset. To provide effective segmentation, this paper provides a unique method for region-based image segmentation that divides an image into its most prominent regions. It also addresses issues with earlier region-based segmentation methods. SVM-based classifiers have better generalization properties as their efficiency does not depend on the number of features. Such merit is beneficial in primary diagnostics decisions to check if the input image is included in the database or not to reduce the consumed time. OSL was applied and compared to standard fine-tuning transfer learning utilizing Siamese networks and triplet loss. Siamese provides superior classification accuracy and localization accuracy with minimal errors than other approaches. The proposed approach has a total processing time of 5 ms, which makes it appropriate for real-time applications. In terms of specificity, sensitivity, precision, accuracy, MCC, and F-measure, the proposed approach beats all current machine learning algorithms for small training sets.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
1 Introduction
It is crucial for agricultural, forestry, rural medical, and other commercial uses to identify and categorize plant leaf diseases. Plant leaf disease diagnostics are required for precision agriculture's automated weed detection. In the fields of the environment, forestry, rural medicinal plants, biodiversity, etc., there is a need for automated tree species identification solutions [1]. The resolution and level of expertise with which the leaf photos are obtained are problematic in all the categories.
Shape, texture, and color-based characteristics were the focus of most of the available literature on the categorization of plant leaf diseases. The learning across high-dimensional characteristics of smartphone leaf image data is rarely addressed [2] despite the existence of several large datasets for leaf classification studies. To combat this, there are five sets of photos showing both healthy and damaged crops. The dataset's final version contains 25 classes and 875 smartphone-captured photos of leaves in total [3].
Every season, a significant number of chemicals are employed as fungicides and pesticides to manage different crop diseases, which pose a serious threat to the environment and have a negative impact on human health owing to residual effects. To address this issue, different researchers have created essential technologies that combine image processing and artificial intelligence principles for illness diagnosis in the past [4].
Agriculture relies heavily on automatic plant disease categorization algorithms, for instance, to avoid or minimize plagues [5]. Due to a lack of the requisite infrastructure, it is still challenging in many regions of the world to identify plant diseases quickly. With current technology, high-definition smartphone cameras may capture photographs that are accessible worldwide. Consequently, the development of general-purpose, affordable, automated, and accurate plant disease Plant disease and plague management applications based on shots of field-captured plants will allow the adoption of plant disease and plague management applications everywhere [6]. Deep learning techniques based on convolutional neural networks (CNN), which have hundreds or even millions of programmable parameters, now produce the best results for categorizing images. These models face the problem of requiring sizeable annotated datasets to interpret the tiny visual elements of a disease in a plant image or the small differences between closely related plant species [7]. The lack of data (images) for various plant diseases is the main obstacle to developing autonomous plant disease classification systems based on CNNs [8].
Although certain illnesses are exceedingly rare in crops, they can have catastrophic effects, generating plagues with significant economic repercussions, especially in areas where such crops have just recently been introduced [6]. Additionally, adequately annotating an entire plant disease dataset takes a lot of time, requires professional human work, and is hence expensive [9]. Images of leaves are one of the most crucial sources of information for identifying and categorizing plant species and their diseases. Therefore, research has concentrated on creating automated techniques for classifying images of leaves [10].
Classic computer vision techniques were incorporated in several studies [11,12,13,14], while other researchers created tools and apps like MedLeaf [15], a medicinal plant identification app for smartphones. Meadleaf reports a classification accuracy of 56.33 percent for 30 species of Indonesian medicinal plants. 48 example images from each of the 30 species were used to build the model. In [16], researchers suggested a method for identifying woody species in Central Europe using a dataset of 151 species, each with at least 50 leaves. Using a kNN classifier, they were able to attain a success rate of 99.63% for 10 species. Other researchers [17] investigated common methods for detecting wheat diseases in the wild and incorporated the algorithm into a smartphone application. By applying CNNs, they recently enhanced their algorithms to accurately detect fungal infections in wheat at very early stages [18].
Deep learning-based algorithms are increasingly being used to replace machine learning-based techniques for classifying leaves in images [19,20,21,22,23]. Unfortunately, the creation of such algorithms has been hampered by the lack of sizable datasets with trustworthy annotations, particularly for extremely specific plant species and/or diseases. The effectiveness of various architectures for learning new classes with limited datasets, or few-shot learning (FSL), has been demonstrated by recent breakthroughs in deep learning technology [24, 25].
Metric learning, model initialization, and data hallucination approaches are a few examples of FSL techniques for image classification. In order to train classifiers for new classes from a small sample size, initialization strategies aim to discover efficient model initializations for the network's programmable parameters. Comparative learning methods are used in metrics. Once a network has mastered class comparison, it should be able to learn new classes given a small set of labeled samples. Numerous metric learning strategies, such as cosine similarity, Euclidean distance, and class-mean representations, have been researched [26], CNN relation modules [27], regression models [28], and graph artificial neural networks [29]. Finally, hallucination techniques make up for the lack of data by figuring out how to add more. To create data for new classes, these techniques use a generator that they have learned from the data in the base classes.
In FSL systems for categorizing plant leaves, adversarial models (hallucination models), Siamese networks with kNN classifiers, and contrastive loss have all recently made advancements. However, a metric learning-based FSL method for classifying plant leaves could be improved by training features with more sophisticated loss functions (such as triplet loss), which would allow for more efficient learning of the decision boundaries from sparse data [30]. Therefore, the goal of this study was to present an FSL model for classifying plant leaves based on Siamese networks and Triplet loss with an effective class boundary learning method based on multiclass support vector machines (SVM). This was accomplished by using a typical image classification system to classify plant leaves using contrastive and triplet loss to learn from a significant dataset. The models were then altered to learn from a small subset of images representing new plant leaf/disease classes, and their performance was evaluated and compared in relation to the number of images used to learn the new classes.
1.1 Motivation and contributions
Recent developments in deep learning have been particularly remarkable in neural network-based applications like facial recognition, signal processing, and fault and illness detection. The classification of plant diseases using field images is one of the most significant applications of deep learning. To shorten the time required for the classification process and output of the findings, SVM was used at the beginning of the suggested technique to do a preliminary check to see if the given images are present in the database or not. Furthermore, SVM-based classifiers have stronger generalization qualities than ANN-based classifiers since their effectiveness is independent of feature quantity. Because of the limitless number of features in this instance, this feature is important in first selection since it allows us to compute directly utilizing the original data without pre-processing it to extract its features. In context of this, the suggested OSL research using SNN in addition to SVM is a great alternative for classifying plant diseases in field images. This paper offers one-shot learning (OSL) techniques for plant diseaseclassification with limited datasets utilizing Siamese Neural Network (SNN). We opted for One-Shot Learning (OSL) due to its capacity to efficiently learn from a limited amount of labelled data, a crucial advantage in scenarios where amassing extensive datasets for our specific plant disease classification task is resource-intensive. OSL's rapid adaptability to new disease classes, inherent ability to handle diverse disease characteristics, and reduction in data annotation efforts further solidified its appeal. Additionally, it empowers our model to generalize to similar diseases and seamlessly incorporates transfer learning techniques, harnessing pre-trained models to enhance performance. In summary, OSL addresses the challenges of limited labelled data, the swift adaptation to new diseases, and the accurate classification of diverse diseases, making it a compelling choice for our plant disease classification system.
The following list summarizes this paper's main contributions:
-
This work investigates existing automatic approaches for detecting and diagnosing disease in field images, with the goal of introducing plant disease detection systems, particularly at the pre-harvest stage.
-
This paper offers one-shot learning (OSL) techniques for plant leaf disease classification with limited datasets utilizing deep learning.
-
The plant leaf disease image dataset's size and dimension are improved using data augmentation techniques.
-
To provide efficient segmentation and address issues with earlier region-based segmentation algorithms, this study provides a unique approach for region-based image segmentation that divides an image into its most prominent regions.
-
SVM-based classifiers have better generalization properties as their efficiency do not depend on the number of features. Hence, such merit is beneficial in primary diagnostics decision to check if the input image is included in the database or not to less the consumed time.
-
The proposed technique has a total processing time of 5 ms, which is acceptable for real-time applications.
Organization of paper is as follows: Section 2 presents SVM and Siamese schemes are used in the proposed diagnosis algorithm. The materials are highlighted in Section 3. Section 4 discusses the suggested diagnosis algorithm for the system under investigation. Section 5 presents the results and discussions. The paper is finally summarized in Section 6.
2 System architectures overview
SVM and Siamese schemes are used in the proposed diagnosis algorithm, so this section will examine them briefly.
2.1 Binary Support Vector Machine (BSVM)
SVMs are trained utilizing a learning bias-based optimization theory-based learning technique on a hyperplane space in a high-dimensional feature space. Data is represented by kernels, which express the degree of similarity or difference between data items [31]. By widening the gap between two or more training data sets' classes, the ideal separation hyperplane is found. This hyperplane is located in the middle of the margin and must
The following equation is then optimized to determine the solution to this problem:
where y the class label output result (y = 1, 1 for a binary classifier), C is the input feature vectors, and W is the collection of weights, one for each input feature. The Vs (support vectors) are the input training feature data points from C that have the largest margins above and below the hyperplane. The parameters b and in the output define a maximum margin solution. The categorization of an ambiguous vector Q is predicted by the decision function d (Q), which is positive for class 1 and negative for class 2. It is defined for the kernel (K) as follows:
2.2 Siamese learning architectures
The classification of objects is a popular issue in pattern learning and artificial intelligence research. The most important challenge for obtaining high accuracy, however, continues to be the extraction of useful information from the target objects for the identification tasks. A deep neural network approach is suggested in the present research to carry out the object categorization job. There are two main phases to completing this:
-
1.
Creating a solid learning dataset that includes several photos of various items seen in the actual workplace.
-
2.
Selecting the best machine learning approaches based on a task's difficulty and the volume of the data being utilized. In the great majority of domains, using a deep neural network trained on a large dataset and subsequently fine-tuning the selected deep classifier on the dataset is the most common approach to solving image classification challenges. One-shot learning, specifically Siamese neural networks (SNN), is the sole method for training a deep neural network on a tiny dataset [32]. Siamese networks were created for the first time by Bromley and LeCun in the early 1990s to solve the problem of object matching in signature authentication [33].
Numerous studies have shown the usefulness of using SNN to classify objects [34,35,36]. A fresh strategy for texture similarity measurement based on contemporary deep learning techniques was proposed by Hudec and Bencsova in [37]. They assessed how closely two regions of homogeneous and non-homogeneous textures in real-world photographs resembled one another. Based on the Siamese network concept, Zhang W et al. [38] suggested a reliable adaptive learning visual tracking system. To enhance the representation of the target object, they made full use of the three retrieved characteristics and adaptively combined them. In [39], Cuan et al. integrated deep convolutional neural networks with cosine similarity metric learning to produce a powerful deep Siamese network for appearance pairing matching. This model was able to recognize the same item again after a prolonged absence and could handle both partial and total occlusion. In [40], Wang et al. presented their study on a deep Siamese network with a hybrid convolutional feature extraction module for change detection based on multi-sensor remote sensing images.
The energy function lies at the top of the Siamese network, which is made up of two twin networks connected by a similarity layer. The outcome is invariant and ensures that very similar images cannot be at very different positions in the feature space since the weights of twins are linked (the same). The similarity layer determines the distance metric between embeddings or high-level feature representations of the input pair of images. Training on pairs is more advantageous since it generates quadratically more potential pairings of images for the model to be trained on, making overfitting more difficult. The encoder network, which is a "shoulder" of the trained one-shot model, is taken out and used as a feature extractor. The Multi-Layer Perceptron (MLP) method, which acts on the output feature vectors of the trained twin, serves as the classifier [32].
A feed-forward artificial neural network known as a multi-layer perceptron (MLP) consists of an input layer, a hidden layer, and an output layer (see Fig. 1). Every node, except the input nodes, is a neuron that employs a non-linear activation mechanism. MLP uses the supervised backpropagation learning technique for training. Due to its multiple layers and nonlinear activation, it differs from a linear perceptron since it can separate data that cannot be separated linearly. The most popular non-linear activation functions in MLP are Sigmoid, ReLU, and SoftMax. A typical Sigmoid activation function is defined as:
where \({y}_{i}\) is the weighted sum of the input connections (the input to a neuron) [41, 42]. The rectified linear unit (ReLU) is one of the potential solutions to the computing issues posed by the sigmoid distribution in recent deep learning advancements (for example, saturation and limited sensitivity). One definition of the rectified linear unit is [43],
The ReLU can only be used on the hidden layers of the neural network model, which is one of its limitations. The SoftMax method, which computes the probability distribution of events occurring over 'n', is occasionally compared to a generalized exponential function. Using the formula to define the common SoftMax function,
This method would estimate the likelihood of a target class over all potential classes expanding. The estimated probability is then used to determine the inputs' target class [44, 45].
3 Materials
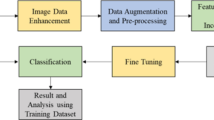
In the great majority of domains, using a deep neural network trained on a sizable dataset and fine-tuning the selected deep classifier on the studied dataset is the most common approach to solving image classification problems, as shown in Fig. 2. These images were collected realistically from the fields by the distinguished staff of JINR (Joint Institute for Nuclear Research/Russia). There are five different crop kinds in the dataset: grape, wheat, cotton, cucumber, and corn. Five sets of images are used to represent each of the five new crops: grape leaves (healthy, esca, chlorosis, powdery mildew, and black rot), wheat leaves (diseases: black chaff, brown rust, powdery mildew, yellow rust), cotton (Verticillium wilt, Powdery mildew, Nutrient deficiency, Alternaria leaf blight, and healthy); and cucumber: (Powdery milde (diseases: downy mildew, eyespot, northern leaf blight, southern rust, and healthy). The final collection consists of 25 classes and 875 leaf images in total. Without performing any extra image processing, all images for the studies were downsized to 256 × 256 pixels. A training set (80%) and a test set (20%) were created from both datasets. The proportions of the train and test runs for each class were maintained throughout all stratified data divisions, which were done at random. Multiple images from the same leaf were pushed into the same partition to prevent data leakage.
4 The proposed methods
In the proposed methodology for plant disease classification using field images and deep learning, a comprehensive approach is outlined. It encompasses various critical stages, starting with data augmentation to enhance the dataset, followed by image preprocessing to improve image quality and segmentation accuracy. A novel region-based segmentation method is introduced to address over-segmentation issues. Feature extraction combines YCbCr histograms, Local Binary Pattern (LBP), Local Zernike Moments (LZM), and texture features, all equally weighted for a balanced representation. A Siamese neural network architecture is employed for feature extraction due to its effectiveness in handling diverse datasets. Binary Support Vector Machine (BSVM) is used for quick availability checking, followed by a deep learning model, potentially a Siamese Neural Network, for precise plant disease classification based on extracted features. This holistic approach offers a promising solution for automated and real-time plant disease identification, catering to the challenges of limited data and diverse disease characteristics in agriculture (Fig. 3).
4.1 Image augmentation
Data augmentation is used to extract more data from the current dataset. In this situation, it makes perturbed duplicates of the existing photos. The principal objective is to strengthen the neural network with different diversities, which results in a network that can differentiate between significant and irrelevant properties in the dataset. Several ways may be used to enhance images. When necessary, augmentation methods are effectively used to follow the quantity and quality of available data. Our approach combines a variety of strategies to support a sizable dataset for various situations. This approach has several strategies that are Gaussian blurring, zoom, rotation and shear. The initial strategy is the Gaussian blurring. A Gaussian filter may be used to remove high-frequency elements, resulting in a blurred images version. In zoom stage, the enlarging or decreasing the image's zoom would scale the image's size. Thirdly, a rotation of between 10° and 180° is applied to the image. Finally, image shearing may be done using rotation and the imitation factor for the third dimension. The datasets were expanded and utilized in the training phase using these techniques. The testing set won't be expanded, though, throughout the testing period. This would demonstrate the architecture's resilience and prevent over-fitting.
4.2 Image preprocessing
The image preparation phase is crucial for the input image's contrast and contour to be improved. Poor background quality is one of numerous subtleties in the image captured by sensors and cameras that could influence segmentation accuracy. Images must be preprocessed to raise their quality and increase the dependability of the feature extraction stage. There are numerous operations in the preparation stage, including image filtering, image enhancement, and image scaling. The noise removal technique is crucial for detecting plant diseases by enhancing image quality and clarity. Plant disease images are known to frequently have many artifacts and noise because of the complicated imaging environment and imaging principles such as low contrast. To enable effective testing, the chosen images will be resized. Image enrichment techniques are broadly used in many applications of image processing where the content quality of the images is important for human understanding. Contour and contrast, among other entities, are important factors in evaluating any content of image quality. To reduce this noise, several filters are employed. Background noise is reduced using an anisotropic filter, while salt and pepper noise is reduced using a weighted median filter. Wavelet-based de-noising methods skew wavelet and scaling coefficients.
4.3 Proposed region-based segmentation
The research presents a unique region-based segmentation method to overcome the over-segmentation problem. In order to remove the disparities in color hues that occurred across comparable areas, this approach first applies morphological procedures to each image in the dataset. First, area labeling is done, then the regions are sorted according to size, and lastly, a certain resolution is chosen to integrate tiny regions with bigger ones according to the color criteria.We chose to employ the YCbCr color model in our segmentation technique for several reasons. Firstly, the YCbCr model's separation of luminance (Y) and chrominance (Cb and Cr) components enables independent processing of brightness and color information, which is beneficial for accurate segmentation tasks. Secondly, it offers robustness to lighting variations, with the luminance component being less affected by changes in illumination compared to chrominance components, making it well-suited for real-world scenarios where lighting conditions can vary. Additionally, the YCbCr model provides discriminative color information, particularly valuable when segmenting objects with distinct color characteristics. Moreover, its reduced correlation between color channels enhances segmentation precision, and its compatibility with established techniques and tools in image processing simplifies implementation and integration into existing workflows, ultimately leading to more effective and versatile segmentation results.
4.3.1 Morphological operations
Using a structuring element, morphological processes create an output image that is the same size as the input image. Each pixel value in the output image is obtained by comparing the relevant pixel's neighbors in the input image. By changing the neighborhood's size and shape, a morphological operation can be developed to be sensitive to certain features in the input image. The two most fundamental morphological processes are dilation and erosion. By applying a rule to the appropriate pixel and its neighbors in the input image, one may deduce a pixel in the output image. The opening and closing are two more crucial morphological actions. Erosion and dilation, employing the same structural element for both processes, are the two steps in the morphological opening of an image. Opening often removes thin protrusions, splits narrow isthmuses, and smooths out contour objects. Dilation and erosion of the same structural element follow each other to form the morphological close of an image. In contrast to opening, closing often mixes short breaks and long, thin gulfs, closes tiny holes, and fills in contour gaps. Closing also tends to smooth out certain contour parts. According to Fig. 4, an opening and a closing are applied to each image. This eliminates stem marks and black spots while attenuating bright and dark artifacts.
4.3.2 YCbCr Color Quantization
Each element of the YCbCr color space is quantized into a predetermined number of areas before being combined to form the histogram. The YCbCr histogram divides the ycbcr color space, with each division standing in for a histogram bin and containing shades of closely related hues. The ycbcr color quantization is used in this framework to eliminate the disparities in color shades that occurred between comparable areas, merging the adjacent parts with the same color characteristics. The Y is split into sixteen areas in an implementation of the YCbCr histogram, whereas the cb and cr are each divided into eight. The three-color components are connected thereafter, resulting in a (16 × 8x8) histogram with 1024 bins, as shown in Fig. 5.
4.3.3 Over-segmentation handling
Due to many local minima in the input image, the prior region-based segmentation approach results in a large number of tiny basins. A minimum of the gradient corresponds to each catchment basin. These minima are created by minute fluctuations brought on by noise in the values of the gray level. The effects of over-segmentation are somewhat mitigated by morphological processes and ycbcr color quantization. Controlling the number of segmented sections is necessary to fully resolve this issue. After performing area labeling and sorting the regions by size, a certain resolution is chosen to mix tiny regions with bigger ones based on the color criterion.
Region labeling
An image is segmented into a number of homogenous sections. Giving these divided parts a unique identity (ID) is known as region labeling.
Region sorting
Sorting is the process of ranking a group of things according to predetermined standards. The collection to be sorted is made up of a list of segmented areas, and the size (number of pixels) of each segment serves as the sorting criterion. The segmented areas are shown in decreasing order in Fig. 6.
Region combination
The technique of joining nearby areas according to certain criteria is known as region combination. In this method, the region combination is made using a color criterion. An area is joined with its neighbor with the minimum color difference. By selecting a group of regions with a large size and joining additional smaller regions to them, the number of segmented regions may be managed. The regions that have been discovered are selected to be the first items in the segmented regions sorted list.
4.4 Feature extraction
This section employs a multifaceted approach to capture essential characteristics from images for plant disease classification. Feature extraction plays a pivotal role in distilling pertinent information from images, enabling effective disease classification. The following subsections outline the distinct techniques utilized for feature extraction in this methodology:
4.4.1 The proposed YCbCr histograms
The histogram is based on an underlying YCbCr color space and uses its components to assess the likelihood that a specific color occurs in an image. Each component of the YCbCr histogram is quantized into a specific number of regions when creating a YCbCr histogram. An important aspect of building a color histogram is the quantization of the color space into certain levels that represent the histogram bins. The number of quantized levels affects both the similarity measure between colors and the size of the feature vector. Too many bins mean that similar colors with tiny shads are treated as dissimilar and generate a large size feature vector that needs large storage space. Too few bins mean that dissimilar colors are treated as similar and generate a small-size feature vector that needs small storage space. To generate a YCbCrhistogram, the luminance (Y) is quantized into eight regions, while the chromic components Cb and Cr are quantized into four sections each. The three quantized color components are then linked, yielding a 64-bin (8 × 4x4) histogram.
4.4.2 Texture feature
The effective term for a texture operator is Local Binary Pattern (LBP). From a gray image, it may gather details about the texture of the immediate neighborhood. The main features of LBP are their computational simplicity and tolerance to light variations. LBP begins by figuring out the binary relationship between each image pixel and its nearby grayscale neighbors. An LBP code was created by weighing binary associations according to specific guidelines. The LBP histogram series is described as an image feature that is extracted from an image sub-region. The LBP operator extracts numerous different texture primitives, such as spots, the line ends, edges, and corners, often gathered into a histogram over a region, to record local texture information. After identifying the objects, the image is separated into regions from which the LBP feature distributions are extracted and combined to create an improved feature vector that may be used as a descriptor. The LBP is utilized as a strong local descriptor that can withstand scaling, rotation, changes in lighting, and background clutter.
4.4.3 Shape feature
The global Zernike Moments generate a single moment value for the whole image, but the local Zernike Moments (LZM) are based on the assessment of the moments for each area. In terms of image recognition, LZM is shown to be robust and to have great success. A section of an image's local properties are represented by Zernike moments. In order to calculate the complex moment coefficients of Zernike moments, Zernike moments are defined over a collection of complex polynomials. The closest match between a test image's vector and all other vectors in the database is determined by calculating the test image's vector of moments as well. However, these holistic moments were deemed insufficient for the images, and as a result, a unique representation technique known as Local Zernike Moments (LZM) was shown to be effective in recognition. For each pixel, the moments surrounding it are calculated locally using the LZM algorithm. In the end, each moment component's complex moment image is estimated. By splitting each moment image into non-overlapping sub areas, the recovered phase-magnitude histograms from each sub-region are combined to create the final feature vector. Low-resolution images' local form differences are crucial for identification. Due to this, LZMs are used as local phase-magnitude histograms. In this framework, LZM is used. It is a shape descriptor that offers a local invariant description and is resistant to occlusion and geometrical distortion.
4.4.4 Feature fusion
The proposed system combines the YCbCr histogram, LBP, and LZM descriptors. To extract the color information from each pixel in the image, the YCbCr histogram descriptor is applied globally to the entire image. The LBP extracts the texture information, while the LZM shape descriptor extracts the shape information. The weights assigned to feature fusion determine how much each individual feature contributes to the final fused representation. All feature weights are equally set and equal to 1, it signifies that each feature has an equal influence on the fused representation. This approach offers advantages such as simplicity, fairness, avoidance of feature bias, robustness to feature variations, preservation of information from all features, and flexibility for future adjustments if needed. Essentially, equal weights of 1 indicate an unbiased and balanced fusion of all available features, ensuring no particular feature is favored or underrepresented in the final representation.
4.4.5 Siamese extractor feature
The Siamese neural network with L layers, h1,l hidden vector for the first twin, and h2,l hidden vector for the second twin is the proposed model in the current study. Early layers utilize the rectified linear unit (ReLU), while later layers use the (Softmax) unit. The number of convolutional filters is specified as a multiple of 16 in order to enhance the performance of the SNN. The generated feature maps are then max-pooled with a filter size and stride of 2, after which the ReLU activation function is used to activate the maps. Thus, each layer's kth filter map has the following shape.
where the three-dimensional tensor \({W}_{l-1,l}\) represents the feature mappings for layer l. Here, "*" is assumed to represent a convolutional operation that is legal. As a result, only those output units are returned that are the outcome of a complete overlap between each convolutional filter and the input feature maps.
Loss function and optimization
Let M be the batch size, and (i) represent the batch's index. Additionally, \(y\left({x}_{1}^{\left(i\right)},{x}_{2}^{\left(i\right)}\right)\) is an M-dimensional vector that contains the labels for the batch, with \(y\left({x}_{1}^{\left(i\right)},{x}_{2}^{\left(i\right)}\right)\) = 1, if x1 and x2 are from the same character class, otherwise it equals zero. On the binary classifier, the following regularized cross-entropy goal was used [32]:
where \({\lambda }^{T}{\left|w\right|}^{2}\) are the regularization terms (Tikhonov regularization) and p is the class's probability. These factors speed up computation and training, guard against overfitting, and guarantee that the network will generalize well to new input [46]. This objective is combined with a standard back-propagation technique, in which the connected weights cause the gradient to be additive across the twin networks. A batch size of 32 was set, and we layer-wise specified the learning rate \({\eta }_{j}\), momentum \({\mu }_{j}\), and \({L}_{1}\) regularisation weights \({\lambda }_{j}\). As a result, the update rule at epoch T is as follows:
where \(\Delta {w}_{kj}^{\left(T\right)}\) is the partial derivative about the weight between the j-th neuron in one layer and the k-th neuron in the next layer. The convolutional layer network weights were all initialized from a normal distribution with a mean of zero and a standard deviation of 10–2. A normal distribution was used to initialize the biases as well, but with a mean of 0.5 and a standard deviation of 10–2. Although each layer was permitted to have a different learning rate, learning rates evenly decreased by 1% every epoch throughout the network, culminating in \({\eta }_{j}^{\left(T\right)}=0.99{\eta }_{j}^{\left(T-1\right)}\). The network was demonstrated to be more easily able to converge to local minima without becoming trapped in the error surface by annealing the learning rate. Every layer's momentum was programmed to begin at 0.5 and increase linearly every epoch until it reached the value \({\mu }_{j}\), which represents the distinct momentum term for the jth layer. Each network was trained for a maximum of 100 epochs while the reduction in a one-shot validation error was tracked to prevent overfitting. The technique was stopped and the parameters of the model at the best epoch, based on the one-shot validation error, were used when the validation error did not reduce after 20 epochs. If the validation error continues to fall throughout the whole learning schedule, the final state of the model created using this technique is saved, and the number of epochs in the subsequent run is increased.
4.5 Check availability using Binary Support Vector Machine (BSVM)
In the proposed approach, the utilization of a Binary Support Vector Machine (BSVM) involves a binary classification process aimed at ascertaining whether the input image belongs to specific predefined categories, typically denoting the health condition of plants, such as "healthy" or "diseased." During the training phase, the BSVM undergoes training on a dataset that includes labeled images, each associated with a specific class label. The BSVM's objective during this training process is to determine an optimal decision boundary, visually represented as a hyperplane, which effectively segregates the two classes while maximizing the margin between them. Subsequently, when the system encounters an unlabeled image for classification, typically during the testing or inference phase, the BSVM employs its acquired knowledge to evaluate which side of the decision boundary the image aligns with. If the algorithm's decision function places the image on one side of the boundary, it categorizes the image as belonging to the corresponding class ("healthy" or "diseased"). This binary classification step plays a critical role in assessing the presence of the input image within the predefined dataset classes, thus aiding in accurate plant disease classification, as elaborated in the paper. Consequently, this capability proves valuable in the initial diagnostic decision-making process, determining whether the input image is part of the database and thereby reducing processing time. Notably, the proposed technique boasts an overall processing time of 5 ms, rendering it well-suited for real-time applications.
4.6 Classification using the proposed deep learning model
In the proposed methodology, the subsequent phase involves the classification of plant diseases using a developed deep learning model. Following the initial stages of data preprocessing, feature extraction, and the determination of image availability using Binary Support Vector Machine (BSVM), the input image is prepared for more detailed categorization into specific plant disease classes. This classification task is carried out using a deep learning model, likely built upon the Siamese Neural Network (SNN) architecture mentioned earlier. Siamese networks are renowned for their effectiveness in handling complex and diverse datasets, particularly those encountered in the challenging field of plant disease classification. The deep learning model has been trained to identify and classify plant diseases based on the extracted features and patterns from the input images. This training process typically leverages a labeled dataset encompassing various plant diseases and their corresponding images. During training, the model learns to differentiate between different types of diseases and assign appropriate class labels. Once the deep learning model has undergone training, it becomes capable of real-time inference. When presented with an unlabeled image of a plant leaf, the model utilizes its acquired knowledge to predict the specific disease category or class to which the leaf belongs. This classification step furnishes invaluable information to farmers and agricultural experts, empowering them to swiftly implement targeted measures to control disease spread and safeguard crop yields. To underline its suitability for real-time applications, it's noteworthy that the proposed approach boasts a total processing time of 5 ms. In summary, the classification step, enabled by the developed deep learning model and complemented by one-shot learning (OSL) techniques for plant leaf disease classification with limited datasets, plays a pivotal role in automating the precise identification of plant diseases based on extracted features and patterns from input images. This automation significantly contributes to the effective management of plant diseases in agriculture.
4.7 Performance measures
In evaluating the effectiveness of classifying plant diseases, various performance measures are employed. These measures include sensitivity, specificity, precision, accuracy, F-measure, MCC (Matthews Correlation Coefficient), and the use of a confusion matrix. These metrics collectively provide insights into the classification accuracy and reliability of plant disease detection systems.
where the number of successfully identified negative cases is indicated by the output "TN," which stands for True Negative. Similar to that, "TP" stands for True Positive and indicates how many accurately identified positive occurrences there were. The acronym "FP" stands for "actual negative cases classified as positive," while "FN" stands for "actual positive cases classed as negative." One of the most frequently used criteria when categorizing is accuracy.
5 Results and discussions
In this section, we present the outcomes of our evaluation for the proposed methodology, conducted on a robust computational platform. Leveraging Python as the primary software environment, our study was carried out on hardware equipped with a Xeon-X5650 processor, 24 GB of RAM, and an NVIDIA GeForce GTX 760 graphics card, ensuring the efficient execution of system algorithms.
Figure 7 represents the training results displayed using t-SNE and confounding matrix. A high-dimensional data visualization tool (t-SNE) [47], seeks to reduce the Kullback–Leibler divergence between the joint probability of the low-dimensional embedding and the high-dimensional data by converting data point similarities into joint probabilities. Since the t-SNE cost function is not convex, different initializations can produce different results. Figures 8 and 9 show Examples of correct and incorrect predictions using test images.
There are five different crop kinds in the dataset: grape, wheat, cotton, cucumber, and corn. Five sets of photos showing both healthy and diseased crops are used to represent each of the new crops. The final collection consists of 25 classes and 875 leaf photos in total. Tables 1, 2, 3, 4 and 5 show the analyzed 10 machine learning methods against the five datasets.
The proposed approach outperforms all the machine learning algorithms shown in Table 1 for the wheat dataset: sensitivity, specificity, accuracy, precision, F-measure, and MCC all have values of 0.999, 0.001, 0.998, 0.989, 0.999, and 0.999, respectively.
For the grape dataset, the proposed approach beats all the machine learning algorithms given in Table 2: sensitivity, specificity, accuracy, precision, F-measure, and MCC all have values of 0.981, 0.0032, 0.988, 0.988, and 0.987, respectively.
The proposed approach outperforms all machine learning algorithms listed in Table 3 for the cucumber dataset, with values for sensitivity, specificity, accuracy, precision, F-measure, and MCC of 0.981, 0.0012, 0.988, 0.978, 0.985, and 0.977, respectively.
The proposed approach outperforms every machine learning algorithm listed in Table 4 for the cotton dataset, with scores of 0.998, 0.001, 0.997, 0.979, 0.995, and 0.989 for sensitivity, specificity, accuracy, precision, F-measure, and MCC, respectively.
The proposed approach outperforms all machine learning algorithms shown in Table 5 for the corn dataset, with values for sensitivity, specificity, accuracy, precision, F-measure, and MCC of 0.990, 0.0018, 0.981, 0.988, 0.999, and 0.998, respectively.
6 Conclusions
This paper introduces innovative one-shot learning (OSL) techniques for the classification of plant leaves using deep learning models, despite working with limited datasets. The dataset encompasses five distinct crop types, including grapes, wheat, cotton, cucumbers, and corn. Each crop category is represented by five sets of images, depicting both healthy and diseased crops. Consequently, the dataset comprises a total of 25 classes and 875 leaf images. The research leverages OSL and compares it to conventional fine-tuning transfer learning methods that involve Siamese networks and Triplet loss functions. Furthermore, the paper presents a novel region-based image segmentation approach, effectively isolating the most significant sections within an image for precise segmentation. This method addresses shortcomings in previous region-based segmentation techniques. The adoption of Support Vector Machine (SVM)-based classifiers is highlighted for their superior generalization capabilities, irrespective of the number of features involved. This quality is particularly advantageous in initial diagnostic decisions, aiding in the verification of whether the input image exists within the database, thereby reducing processing time. The experimental results demonstrate the superior performance of the proposed model compared to the most recent methods employed in the classification of plant diseases. However, it's worth noting that the suggested model may not be suitable for mobile platforms. Consequently, there is an ongoing effort to develop lightweight models for leaf disease identification. Additionally, exploring more efficient feature extraction methods may further streamline model complexity. Future directions include evaluating the model's recognition performance with more complex datasets and considering other cutting-edge deep learning approaches. The potential applications of the proposed models extend to the identification of quality attributes in various vegetables and fruits in real-time settings, leveraging the Internet of Things. Moreover, there is the possibility of transforming this concept into an Android smartphone application, enabling farmers to capture real-time images of their plants and receive prompt disease detection reports.
Data availability
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
References
Sinha BB, Dhanalakshmi R (2022) Recent advancements and challenges of Internet of Things in smart agriculture: A survey. Futur Gener Comput Syst 126:169–184
Camacho A, Arguello H (2018) Smartphone-based application for agricultural remote technical assistance and estimation of visible vegetation index to farmer in Colombia: AgroTIC. In: Remote Sensing for Agriculture, Ecosystems, and Hydrology XX, vol. 10783. SPIE, pp 137–148
Perez H, Tah JH (2021) Deep learning smartphone application for real-time detection of defects in buildings. Structural Control Health Monitoring 28(7):e2751
Kavindi Gunasinghe ULD, Malaviarachchi HW, Konthasinghe VT, Diwantha KS, Sriyarathna D, Kasthurirathna D (2022) GreenHubLK: A machine learning driven solution for crop disease detection and post-harvest crisis. In: Future of Information and Communication Conference. Cham: Springer International Publishing, pp 273–293
Sladojevic S, Arsenovic M, Anderla A, Culibrk D, Stefanovic D (2016) Deep neural networks based recognition of plant diseases by leaf image classification. Comput Intell Neurosci 2016
Argüeso D et al (2020) Few-Shot Learning approach for plant disease classification using images taken in the field. Comput Electron Agric 175:105542
Ghassemi N, Shoeibi A, Rouhani M (2020) Deep neural network with generative adversarial networks pre-training for brain tumor classification based on MR images. Biomed Sig Process Control 57:101678
Lu J, Tan L, Jiang H (2021) Review on convolutional neural network (CNN) applied to plant leaf disease classification. Agriculture 11(8):707
Picon A et al (2019) Deep convolutional neural networks for mobile capture device-based crop disease classification in the wild. Comput Electron Agric 161:280–290
Mohanty SP, Hughes DP, Salathé M (2016) Using deep learning for image-based plant disease detection. Front Plant Sci 7:1419
Rumpf T et al (2010) Early detection and classification of plant diseases with support vector machines based on hyperspectral reflectance. Comput Electron Agric 74(1):91–99
Sannakki SS et al (2011) Leaf disease grading by machine vision and fuzzy logic. Int J Comp Tech Appl 2(5):1709–1716
Martinelli F et al (2015) Advanced methods of plant disease detection. A review. Agron Sustain Dev 35(1):1–25
Al-Hiary H et al (2011) Fast and accurate detection and classification of plant diseases. Int J Comput Appl 17(1):31–38
Prasvita DS, Herdiyeni Y (2013) MedLeaf: mobile application for medicinal plant identification based on leaf image. Int J Adv Sci Eng Inf Technol 3(2):5–8
Novotný P, Suk T (2013) Leaf recognition of woody species in Central Europe. Biosys Eng 115(4):444–452
Johannes A et al (2017) Automatic plant disease diagnosis using mobile capture devices, applied on a wheat use case. Comput Electron Agric 138:200–209
Abbass MY, Kwon KC, Alam MS et al (2021) Image super resolution based on residual dense CNN and guided filters. Multimed Tools Appl 80:5403–5421
Chug A et al (2022) A novel framework for image-based plant disease detection using hybrid deep learning approach. Soft Comput 27(18):13613–13638
Wang G, Sun Y, Wang J (2017) Automatic image-based plant disease severity estimation using deep learning. Comput Intell Neurosci 2017
Ferentinos KP (2018) Deep learning models for plant disease detection and diagnosis. Computers electronics in agriculture 145:311–318
Geetharamani G, Pandian A (2019) Identification of plant leaf diseases using a nine-layer deep convolutional neural network. Comput Electr Eng 76:323–338
Chen J et al (2020) Using deep transfer learning for image-based plant disease identification. Computers Electronics in Agriculture 173:105393
Chen WY, Liu YC, Kira Z, Wang YCF, Huang JB (2019) A closer look at few-shot classification. arXiv preprint arXiv:1904.04232
Larochelle H (2020) Few-shot learning. Computer vision: a reference guide
Snell J, Swersky K, Zemel R (2017) Prototypical networks for few-shot learning. Adv Neural Inf Process 30
Sung F, Yang Y, Zhang L, Xiang T, Torr PH, Hospedales TM (2018) Learning to compare: Relation network for few-shot learning. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1199–1208
Bertinetto L, Henriques JF, Torr PH, Vedaldi A (2018) Meta-learning with differentiable closed-form solvers. arXiv preprint arXiv:1805.08136
Kim J, Kim T, Kim S, Yoo CD (2019) Edge-labeling graph neural network for few-shot learning. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 11–20
Hu G et al (2019) A low shot learning method for tea leaf’s disease identification. Comput Electron Agric 163:104852
Le VNT, Apopei B, Alameh K (2019) Effective plant discrimination based on the combination of local binary pattern operators and multiclass support vector machine methods. Inf Process Agric 6(1):116–131
Koch G, Zemel R, Salakhutdinov R (2015) Siamese neural networks for one-shot image recognition. In ICML deep learning workshop 2:1
Bromley J, Guyon I, LeCun Y, Säckinger E, Shah R (1993) Signature verification using a" siamese" time delay neural network. Adv Neural Inf Process 6
Goncharov P, Ososkov G, Nechaevskiy A, Uzhinskiy A, Nestsiarenia I (2019) Disease detection on the plant leaves by deep learning. In: Advances in Neural Computation, Machine Learning, and Cognitive Research II: Selected Papers from the XX International Conference on Neuroinformatics, October 8-12, 2018, Moscow, Russia. Springer International Publishing, pp 151–159
Abbass MY, Kwon KC, Kim N et al (2021) A survey on online learning for visual tracking. Vis Comput 37:993–1014. https://doi.org/10.1007/s00371-020-01848-y
Uzhinskiy A, Ososkov G, Goncharov P, Nechaevskiy A (2019) Multifunctional platform and mobile application for plant disease detection. In CEUR Workshop Proc 2507:110–114
Hudec L, Bencsova W (2018) Texture similarity evaluation via siamese convolutional neural network. In: 2018 25th International Conference on Systems, Signals and Image Processing (IWSSIP). IEEE, pp 1–5
Zhang W et al (2021) Robust adaptive learning with Siamese network architecture for visual tracking. Vis Comput 37(5):881–894
Cuan B, Idrissi K, Garcia C (2018) Deep siamese network for multiple object tracking. In 2018 IEEE 20th International Workshop on Multimedia Signal Processing (MMSP). IEEE, pp 1–6
Wang M et al (2020) A deep siamese network with hybrid convolutional feature extraction module for change detection based on multi-sensor remote sensing images. Remote Sens 12(2):205
Han J, Moraga C (1995) The influence of the sigmoid function parameters on the speed of backpropagation learning. In International workshop on artificial neural networks. Berlin, Heidelberg: Springer Berlin Heidelberg, pp 195–201
Dombi J, Jónás T (2022) The generalized sigmoid function and its connection with logical operators. Int J Approx Reason 143:121–138. https://doi.org/10.1016/j.ijar.2022.01.006
Jahan I et al (2022) Self-gated rectified linear unit for performance improvement of deep neural networks. ICT Express 9(3):320–325. https://doi.org/10.1016/j.icte.2021.12.012
Abbass MY, Kwon KC, Kim N et al (2021) Visual tracking using convolutional features with sparse coding. Artif Intell Rev 54:3349–3360. https://doi.org/10.1007/s10462-020-09905-7
Abbass MY, Sadic N, Ashiba HI et al (2022) An Efficient Technique for Non-Uniformity Correction of Infrared Video Sequences with Histogram Matching. J Electr Eng Technol 17:2971–2983. https://doi.org/10.1007/s42835-022-01010-9
Palaiahnakote S, di Baja GS, Wang L, Yan WQ (Eds.) (2020) Pattern recognition: 5th asian conference, ACPR 2019, Auckland, New Zealand, November 26–29, 2019, Revised Selected Papers, Part I, vol. 12046. Springer Nature.
Van der Maaten L, Hinton G (2008) Visualizing data using t-SNE. Journal of machine learning research 9(11)
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Saad, M.H., Salman, A.E. A plant disease classification using one-shot learning technique with field images. Multimed Tools Appl 83, 58935–58960 (2024). https://doi.org/10.1007/s11042-023-17830-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-17830-4