Abstract
Phytochromes are photoreceptor proteins with a bilin chromophore that undergo photoconversion between two spectrally different forms, Pr and Pfr. In plants, phytochromes play a central role in growth and differentiation during the entire life cycle. Phytochromes of plants and other groups of archaeplastida have a common evolutionary origin in prokaryotes, but the exact prokaryotic origin is as yet uncertain. Two possibilities are presently discussed: either, archaeplastidal phytochromes arose from the last eukaryotic common ancestor (LECA) or they arose from the cyanobacterial endosymbiont that gave rise to plastids. We first constructed standard phylogenetic trees based on N-terminal protein sequences of the chromophore module. As usual, variation of algorithms and parameters led to different trees. A relationship between cyanobacteria and archaeplastida was observed in 7 out of 36 trees. The lack of consistency between results obtained from variation of parameters of tree constructions reflects the uncertainty of archaeplastidal origin. To gain more information about a possible cyanobacterial and archaeplastidal relationship, we performed phylogenetic studies based on the amino acids that line the chromophore pockets. These amino acids are highly conserved and could provide more accurate information about long evolutionary time scales, but the reduction of traits could also lead to insignificant results. From 30 selected chromophore-binding amino acids, 6 were invariant. The subsequent studies were thus based on the information dependent on 24 or fewer amino acid positions. Again, multiple trees were constructed to get information about the robustness of relationships. The very low number of information-containing traits resulted in low bootstrap values and many indistinguishable leaves. However, the major groups fungi, bacteria, cyanobacteria, and plants remained united. Without exception, cyanobacteria and archaeplastida were always closely linked. In this respect, the results were more robust than those of the classic approach, based on long contiguous sequences. We therefore consider cyanobacteria as the most likely origin of archaeplastidal phytochromes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Phytochromes are photoreceptors with a bilin chromophore, in which activation is initiated by a light-induced isomerization of the chromophore and the light-triggered switching between the red and far-red absorbing forms, Pr and Pfr, respectively. In plants, phytochromes control adaptation to specific light conditions through the entire life cycle. Central steps in development such as seed germination, flower induction, or shade avoidance are under phytochrome control. Plant phytochromes are therefore of major agricultural importance. Phytochromes are also present in green algae, prasinophytes, glaucophytes, and cryptophytes. These groups together belong to the supergroup archaeplastida [1,2,3], which unites organisms that originate from primary endosymbiosis. Other eukaryotes that contain phytochromes are slime molds [4], fungi [5], brown algae, and diatoms [6]. The latter two belong to the supergroup stramenopiles. Finally, many bacteria, including cyanobacteria, contain phytochromes [7]. The functions of non-plant phytochromes are less well established than those of plant phytochromes, although the knowledge of biochemical and structural properties of bacterial phytochromes is further advanced [7, 8].
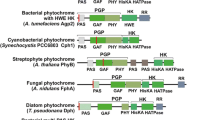
Typical phytochromes have three conserved domains in the N-terminal region that are involved in chromophore-binding and photoconversion, termed PAS, GAF, and PHY domains. All domains together are often denominated photosensory core module (PCM) [9]. The C-terminal part of bacterial, fungal, and stramenopile phytochromes is most often a histidine-kinase, whereas plant phytochromes have a histidine-kinase like region and two PAS domain insertions between this region and the PCM (Fig. 1A).
A Domain pattern of phytochromes. The present analysis was performed on the PCM region, which is common to all phytochromes. The covalent chromophore attachment site which is either in the GAF domain or close to the N-terminus is indicated by a red line. B Chemical structure of biliverdin, the chromophore of some cyanobacterial, non-cyanobacterial bacterial, fungal and stramenopile phytochromes, of phycocyanobilin, the chromophore of some cyanobacterial phytochromes, and of phytochromobilin, the chromophore of plant phytochromes. C biliverdin chromophore and chromophore-binding amino acid residues (within 4.5 Ǻ distance) of the bacterial phytochrome Agp1 crystal structure, drawn in ball and stick mode. Biliverdin carbon atoms are in green color, amino acids that were invariant in the present study are in blue, and the other amino acids in magenta color. PDB Code 5HSQ. The left and right panels represent different views, obtained by a rotation of 90 °C about the horizontal axis. D Degree of conservation of chromophore-binding amino acids presented by a Web Logo [40]. The positions in the structure of Agrobacterium phytochrome Agp1 are given below. Amino acids that interact with ring A or D are presented in green or orange, respectively
One of three different chromophores (Fig. 1B) is incorporated into the chromophore pocket of phytochromes [10]. This chromophore pocket is formed by the GAF domain of the protein [8, 11, 12], but the covalent attachment site can be outside the GAF domain. In non-cyanobacterial bacteria, a biliverdin chromophore is covalently attached to a Cys just N-terminal of the PAS domain [13]. Fungal and stramenopile phytochromes have the same N-terminal attachment site and probably incorporate biliverdin, as well [14]. In plants, a phytochromobilin chromophore is covalently attached to a conserved Cys in the GAF domain. Cyanobacterial phytochromes can either have the N-terminal Cys and a biliverdin chromophore or the Cys in the GAF domain and a phycocyanobilin chromophore [15]. In all cases reported so far, the Cys residues are exclusive: either the protein has the N-terminal Cys, which reacts with the ring A vinyl side chain of biliverdin, or there is the Cys in the GAF domain which reacts with the ethylidene side chain of phycocyanobilin or phytochromobilin [16]. In the present study, we also found sequences of cyanobacterial phytochromes with both Cys residues.
Here, we address the prokaryotic origin of archaeplastidal phytochromes. This question is still under debate. Due to the relevance of plant phytochromes for agriculture, it is important to clarify the evolutionary history of archaeplastidal phytochromes. Fungal, stramenopile, and slime mold phytochromes have most likely been inherited from the last eukaryotic common ancestor (LECA) [1, 17], an organism with archaeal and bacterial genes. According to present knowledge, archaeplastidal phytochromes can either be descendants of the cyanobacterium that gave rise to plastids, or can as well be traced back to the LECA.
In typical phylogenetic trees based on phytochrome sequences, each of the major eukaryotic groups forms a branch that is separated from other branches, in line with the monophyly of these species. Fungal and stramenopile phytochromes are often found in branches that divide from a common branch in such trees [3, 6], in line with their proposed common LECA origin. The archaeplastidal branch is placed next to branches of bacterial phytochromes, usually at different positions than the fungal or stramenopile branches. Therefore, these phylogenetic trees do not provide a clear answer to the origin of archaeplastidal phytochromes. We have performed phylogeny studies in which a set of phytochrome sequences was analyzed with various alignment programs, different phylogeny algorithms, and different parameters [18, 19]. These studies reflected the situations found in other published trees with respect to different insertion positions of the archaeplastidal branch, but in few trees, this branch was positioned next to the cyanobacterial branch [18, 19]. A comparable situation (one tree with the plant branch next to cyanobacteria and one with plants distant from cyanobacteria) was also described by Duanmu et al. [3].
Some cyanobacterial and almost all archaeplastidal phytochromes have common features by which they are distinguished from other lineages, namely the same chromophore attachment site in the GAF domain and a chromophore with an ethylidene side chain at ring A (Fig. 1). This is another argument for cyanobacterial ancestry of plant phytochromes, although the chromophore-binding cysteine can easily be switched by two amino acid replacements and could have evolved in both lineages independently in a convergent evolution. Here, we have tried to further clarify the evolutionary origin of plant phytochrome by generating phylogenetic trees based on the conserved amino acids of the chromophore pocket. The degree of conservation could allow evolutionary studies over long time ranges, but the low number of traits could cause low resolution and low stringency. With respect to the origin of archaeplastidal phytochromes, however, this approach gave clear results.
2 Methods
Phytochrome sequences of cyanobacteria, other bacteria, fungi, archaeplastida (mostly plants), and stramenopiles were isolated from the NCBI database by BLAST and stored in FASTA format. In this way, 368 sequences were collected. In some cases, the number of sequences was reduced to 356 by taking out 12 sequences that were often not placed in the appropriate group.
For subsequent alignments and phylogenetic tree constructions, the header of each FASTA sequence was replaced by a string of 6 letters, 5 lower-case letters derived from the species name, and one capital letter for the major group (B for non-cyanobacterial bacteria, C for cyanobacteria, P for archaeplastida, F for fungi, and S for stramenopiles). This code, the name of the strain, and a Gene-bank identifier by which the sequence can be found in the database are given in Supplementary Table S1. Alignments were either performed with Clustal Ω [20] or Muscle [21] using standard parameters. In some cases, gaps were removed using the trimal [22] command “trimal -i input.fasta -o output.fasta -gt 0.4 -st 0.001 -cons 50", in one case the parameters were “-gt 0.8 -st 0.001 -cons 60”.
In each sequence, the chromophore attaching Cys either N-terminal of the PAS domain or in the GAF domain was identified depending on whether it aligned with the respective Cys in Agp1 from Agrobacterium [13] or in cyanobacterial Cph1 [23], respectively. (Either Cys is easily recognized in the alignment.) For tree construction, the Maximum Likelihood, Neighbor Joining, Minimum Evolution, Parsimony or UPGMA programs of the software package Mega X [24] were used and different replacement matrices were used as indicated in Table 1. In two cases (trees 19 and 70), the optimum parameters for tree construction were obtained by the MEGA-X “find best model” command. Otherwise, default parameters and frequencies were used. The aim was to generate a selection of different trees to get an impression about robustness of results.
For the chromophore pocket, the amino acids of Agrobacterium fabrum phytochrome [11] that have at least one atom within 4 Å (22 amino acids) or 4.5 Å (30 amino acids) distance of the chromophore biliverdin were identified (see also Fig. 1C). These and the homologous amino acids in the columns of a Clustal Ω alignment were kept for further studies. All remaining amino acids were cut out. The choice of phylogeny programs and replacement matrices were similar as in the previous alignments. Each run included 100 bootstrapping cycles [25]; the relative bootstrap values (0–1) are represented by numbers with 1–4 significant digits, depending on the way of presentation. Trees 67–69 were constructed without bootstrapping.
All tree graphics were made with ITOL [26]. The graphics in Figs. 2, 3, 4, 5 were edited with CorelDRAW. In those cases, abbreviated species names were used, as listed in Table S1. The color codes red, green, magenta, and orange are used for bacteria, archaeplastida, fungi, and stramenopiles, respectively. Cyanobacterial phytochromes with the N-terminal chromophore-binding site are shown in dark blue, those with the binding site in the GAF domain in cyan, and those with both binding sites in turquoise. In the supplementary Figure S1, all trees are presented in black and white mode. In these cases, full species names and the accession codes of the proteins are given, in addition to the choice of alignment, and tree construction parameters of all trees are listed in Table 1.
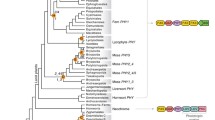
Unrooted maximum-likelihood tree of phytochrome PCM sequences, LG matrix. (Tree 1 of supplementary Fig. S1). Example for a tree with distant cyanobacterial and archaeplastidal branches. “Bacteria” (red) stands for bacteria outside cyanobacteria. Cyanobacterial phytochromes have two different chromophore binding Cys residues, those cyanobacterial sequences that contain the N-terminal Cys are presented in cyan, those with a Cys in the GAF domain in dark blue, and those with both Cys in turquoise. Sequences from Archaeplastida are drawn in green, those from fungi in orange, and those from stramenopiles in cyan. Bootstrap values are printed with one significant digit, and species names are printed with 5 letters (see Table S1 for full names)
Unrooted neighbor-joining tree of phytochrome PCM sequences, JTT matrix. (Tree 3 of supplementary Fig. S1). Example for a tree with split cyanobacterial and plant branches. Definition of “Bacteria”, color coding, representation of bootstrap values, and abbreviation of species names the same as in Fig. 2
3 Results and discussion
So far, phylogenetic trees presented in the literature have left the relationship between archaeplastidal and bacterial phytochromes an open issue. We performed a study in which we constructed several trees with different programs and parameters to get a selection of trees that can be compared in terms of the origin of archaeplastidal phytochromes. Such an approach employing multiple trees has been performed before by our group [18], and is repeated here as a control for the trees based on amino acid residues of the chromophore pockets (see below).
In this study, we have first selected phytochrome sequences from 368 species including 107 cyanobacterial sequences, 29 sequences from archaeplastida, 15 from fungi, and 4 from stramenopiles (Table S1) for tree construction. We focused our initial studies on the phytochrome chromophore module (PCM, Fig. 1A), which unifies the domains required for chromophore incorporation and photoconversion. We constructed 36 trees (Supplementary Fig. S1, tree 1–36) using different alignment programs, different phylogeny algorithms, and different replacement matrices with 368 or 356 sequences as listed in Table 1, which summarizes the results from all trees. For trees 19 and 70, we used an optimized replacement matrix as obtained with the “find best model” function of MEGA-X. Four characteristic trees are presented with abbreviated species labels in Figs. 2, 3, 4, 5.
In Table 1, the close relationship between archaeplastidal and cyanobacterial groups on the one hand and of fungal and stramenopile groups on the other hand are given in the 9th and 11th columns, respectively. In all PCM trees, fungi, archaeplastida, stramenopiles, and cyanobacteria formed their own branches and appeared separate from all other groups. In 21 out of 36 trees, fungi and stramenopiles appeared as branch partners, as in other studies [3, 6]. This suggested that phytochromes of both groups were inherited from the LECA [27].
The archaeplastida and cyanobacterial branches appeared in 7 trees as split branches. Note that the bootstrap values around these splits were usually between 0.5 and 1 (Table 1 column10 and Fig. 4), i.e., in more than half of the bootstrap datasets, the same split was obtained under the selected phylogeny parameters. This is a strong argument for the cyanobacterial/archaeplastidal relationship, but in the other 29 trees, no evidence for such a relationship was found. A split branch between archaeplastida and both other eukaryotic groups was found in one tree (Supplementary Fig. S1, tree 30). Cyanobacterial phytochromes with a chromophore-binding Cys N-terminal of the PAS domain or in the GAF domain were described in the literature [15, 16, 28] (see also Fig. 1). About half of the selected cyanobacterial sequences had a chromophore-binding Cys in the N-terminus; another half had a chromophore-binding Cys in the GAF domain (see also Supplementary Table S1). In addition, three phytochromes of different strains of Nostoc sp. Peltigeria membranacea had Cys residues at both positions. The two major groups of cyanobacterial phytochromes with respect to the position of the covalent chromophore-binding site were more or less separated in all PCM trees, but there were always insertions of members of one group in the other group (Figs. 2 and 3 and Supplementary Fig. S1, tree 1).
Unrooted neighbor joining tree based on amino acids of the phytochrome chromophore pockets, JTT matrix. (Tree 56 of supplementary Fig. S1). Definition of “Bacteria”, color coding, representation of bootstrap values, and abbreviation of species names the same as in Fig. 2
Unrooted maximum-likelihood tree based on amino acids of the phytochrome chromophore pockets, LG matrix. Two amino acids at the position of the two possible covalent chromophore-binding sites were removed from the alignment (Tree 60 of supplementary Fig. S1). Definition of “Bacteria”, color coding, representation of bootstrap values, and abbreviation of species names the same as in Fig. 2
The uncertainties that arise regarding the origin of archaeplastidal phytochromes are explained by weak similarities of amino acids in the alignments. Compared to, e.g., rRNA sequences, phytochrome sequences are highly variable. We therefore concentrated our subsequent studies on conserved amino acids of the proteins. The highest degree of conservation is found in the chromophore pockets of the PCM of phytochromes. In contrast to the general role of amino acids in stabilizing the secondary and tertiary structure of the protein, the amino acids of the chromophore pocket have additional functions, e.g., to determine the binding affinity between the chromophore and the polypeptide by specific interactions with the chromophore. For these reasons—higher similarities and additional functions—the selection of these amino acids could have a positive impact on the quality of the phylogenetic trees. We chose the amino acids of the chromophore pocket of the crystal structure of Agrobacterium fabrum Agp1, but kept the previous alignment for identification of homologous amino acids in other phytochromes. We found this to be a straightforward way to check out what result the selection could have. In the sequence of Agp1, we selected those 22 or 30 amino acid residues that are within 4.0 Å or 4.5 Å distance from the biliverdin chromophore [11], respectively. A Web Logo plot of the alignment based on the 30 amino acids that are within 4.5 Å distance from the chromophore is shown in Fig. 1D. The trees were constructed in a similar way as the PCM trees, see Table 1, Fig. 4 and Supplementary Fig. S1, trees 37–60.
Whereas the selection of highly conserved amino acids should have a positive effect on the quality of phylogenetic studies, we also considered that the reduction to few amino acids could affect the quality of a phylogenetic tree in a negative way, even more since six amino acids are indifferent and yield no information on the phylogeny. The amino acid selection resulted in groups of identical sequences and thus in a reduction the number of distinguishable leaves. (identical sequences were not eliminated; these appear with a branch length of 0 and no bootstrap values in the trees). In the chromophore-pocket trees, insertions of outliers from other groups were more often found than in the PCM trees. Bootstrap values were lower, often in the range of 0.1. The reduction of information in the chromophore pocket trees leads indeed to lower possibilities of differentiation.
With regard to the central question of this study, the bacterial origin of archaeplastidal phytochromes, the chromophore pocket trees yield, however, a clearer picture than the PCM trees, if the variations among different algorithms and parameters are considered. In the different chromophore pocket trees, members of cyanobacteria, archaeplastida, or fungi most often formed one branch each, in few cases two branches. The archaeplastidal branch was either inserted in the cyanobacterial branch or appeared as a neighboring branch of one cyanobacterial branch. Such results were obtained for all variants that were constructed with different parameters. In this respect, the results are more robust and clearer than the above results based on PCM sequences. In other words: archaeplastidal phytochrome chromophore pockets reveal a smaller distance to cyanobacterial chromophore pockets than to any other group, no matter which algorithm is used to calculate this distance.
The arrangements of chromophore-binding subtypes of cyanobacterial phytochromes were also principally different between chromophore pocket trees and PCM trees. Sequences with a chromophore-binding Cys close to the N-terminus were united in the chromophore-pocket trees, whereas sequences with different binding sites were more mixed in the PCM trees (Figs. 2, 3, 4, 5, Table 1 and Supplementary Fig. S1). Branches of archaeplastidal phytochromes were always next to branches of cyanobacterial phytochromes with a chromophore-binding site in the GAF domain.
The proposed common origin of stramenopile and fungal phytochromes was not reflected by the chromophore pocket trees (Table 1 and Supplementary Fig. S1, trees 37–66). The diversification of these groups could have occurred earlier than that between cyanobacteria and archaeplastida.
Results based on a low number of traits could be biased. Here, the chromophore chemistry could impose a certain bias. The plant chromophore phytochromobilin and the cyanobacterial chromophore phycocyanobilin are identical with the exception of one of the ring D side chains, which are vinyl and ethyl, respectively (Fig. 1B). It should also be considered that there are archaeplastidal phytochromes with a phycocyanobilin chromophore [29]. Phycocyanobilin differs from biliverdin in the ring A and D side chains, whereas phytochromobilin differs from biliverdin only in ring A.
Using our sequence alignments of 30 chromophore-binding amino acids, we tested what would happen to phylogenetic trees if the number of amino acids would be further reduced. For the trees 60–66, the alignments were reduced by removing the two positions of the chromophore-binding cysteins (20 and 249 in Fig. 1D). For trees 67–69, the alignments were reduced by removing those amino acids that are close to ring A (orange amino acids in Fig. 1D) (tree 67) or close to ring D (green amino acids in Fig. 1D) (tree 68) or both (tree 69). Although only few trees were constructed with further reduced numbers of amino acids, the outcome was clear: archaeplastidal phytochromes were always next to cyanobacterial phytochromes. Tree 69 was more disordered than the other trees, as sequences of phytochromes that were clearly unrelated to each other were often mixed. This showed that the continued reduction of traits came to a limit. Nonetheless, these results indicate that those chromophore pocket trees that support a relationship between cyanobacterial and archaeplastidal phytochromes are not biased by the chemical nature of the chromophore.
This present study based on the chromophore pocket of phytochromes shows that a selection of conserved amino acids can lead to additional helpful phylogenetic information. It became clear that the distance between chromophore pockets of archaeplastidal and cyanobacterial phytochromes is closer than the distance of archaeplastidal phytochromes to any other major group. This feature was robust against the variation of phylogeny programs and parameters. This supports the theory that archaeplastidal phytochromes were inherited from cyanobacteria via the endosymbiosis pathway.
In plants, phytochromes control many effects that are of agricultural interest. Because of this importance, phytochromes were discovered in plants much earlier [30, 31] than in cyanobacteria [28, 32, 33], other bacteria [34] or fungi [14]. A cyanobacterial origin of archaeplastidal phytochromes as proposed here would suggest common features of archaeplastidal and cyanobacterial phytochromes. One example is the common site of covalent chromophore attachment and the similar chromophore, but with respect to function, there are basic differences between both groups. Whereas plant phytochrome effects are conspicuous, cyanobacterial phytochromes control inconspicuous effects. So far, no clear phytochrome response has been observed in cyanobacteria [28, 35, 36]. The so-called cyanobacteriochromes, in contrast, control many conspicuous effects in cyanobacteria such as chromatic adaptation [32] or phototaxis [37]. Cyanoabacteriochromes are photoreceptors that evolved in cyanobacteria [36] and have one or several bilin-binding GAF domains which are combined with few other domains of various kind. Their GAF domains probably evolved from the GAF domain of phytochromes. This evolution must have started after the covalent chromophore attachment site in phytochromes has switched to the GAF domain. With this change, the covalent-binding site N-terminal of the PAS domain and the domain itself became dispensable. Typically, a cyanobacterium contains more than 10 cyanobacteriochromes and 1 or 2 phytochromes [38]. None of the cyanobacteriochromes are present in the plant lineage after plastidal endosymbiosis, whereas phytochrome evolved to the most important photoreceptor of plants. One possible explanation for this could be that cyanobacteriochromes were lost in favor of phytochromes, because there was a demand for regulation in the long-wavelength range in the host. Another possibility could be that cyanobacteriochromes were not yet present in the cyanobacterial endosymbiont. According to the latter hypothesis, the evolution of cyanobacteriochromes and endosymbiosis started soon after the transition of the covalent chromophore-binding site into the GAF domain.
In summary, we have shown that the chromophore pocket of archaeplastidal phytochromes is most similar to that of cyanobacteria, providing strong evidence for a cyanobacterial origin of archaeplastidal phytochromes via endosymbiosis. It is therefore expected that plant phytochromes have functionally more in common with cyanobacterial than with bacterial or fungal phytochromes. One common feature of bacterial and fungal phytochromes is that they act in darkness [7]. Plant phytochromes are inactive in darkness [39]. It will be interesting to see the activity pattern of cyanobacterial phytochromes in darkness. For a thorough comparison of cyanobacterial and archaeplastidal phytochromes, biological functions of cyanobacterial phytochromes must be uncovered and analyzed.
Data availability statement
All data are provided in the present manuscript or the supplement. The authors guarantee that all alignment files and tree files will be provided upon request.
References
Simpson, A. G. B., & Roger, A. J. (2004). The real “kingdoms” of eukaryotes. Current Biology., 14, R693–R696.
Mackievicz, P. G. P. (2014) Monophyly of Archaeplastida supergroup and relationships among its lineages in the light of phylogenetic and phylogenomic studies. Are we close to a consensus? Acta Societatis Botanicorum Poloniae, 263–280.
Duanmu, D. Q., Bachy, C., Sudek, S., Wong, C. H., Jimenez, V., Rockwell, N. C., Martin, S. S., Ngan, C. Y., Reistetter, E. N., van Baren, M. J., Price, D. C., Wei, C. L., Reyes-Prieto, A., Lagarias, J. C., & Worden, A. Z. (2014). Marine algae and land plants share conserved phytochrome signaling systems. Proceedings of the National Academy of Sciences of the United States of America., 111, 15827–15832.
Schaap, P., Barrantes, I., Minx, P., Sasaki, N., Anderson, R. W., Beinard, M., Biggar, K. K., Buehler, N. E., Bundschuh, R., Chen, X., Fronick, C., Fulton, L., Golderer, G., Jahn, N., Knoop, V., Landweber, L. F., Maric, C., Miller, D., Noegel, A. A., … Marwan, W. (2016). The physarum polycephalum genome reveals extensive use of prokaryotic two-component and metazoan-type tyrosine kinase signaling. Genome Biology and Evolution., 8, 109–125.
Purschwitz, J., Muller, S., Kastner, C., & Fischer, R. (2006). Seeing the rainbow: Light sensing in fungi. Current Opinion in Microbiology, 9, 566–571.
Fortunato, A. E., Jaubert, M., Enomoto, G., Bouly, J. P., Raniello, R., Thaler, M., Malviya, S., Bernardes, J. S., Rappaport, F., Gentili, B., Huysman, M. J. J., Carbone, A., Bowler, C., d’Alcala, M. R., Ikeuchi, M., & Falciatore, A. (2016). Diatom phytochromes reveal the existence of far-red-light-based sensing in the ocean. The Plant Cell, 28, 616–628.
Lamparter, T., Xue, P., Elkurdi, A., Kaeser, G., Sauthof, L., Scheerer, P., & Krauss, N. (2021). Phytochromes in Agrobacterium fabrum. Frontiers in Plant Science, 12, 642801.
Wagner, J. R., Brunzelle, J. S., Forest, K. T., & Vierstra, R. D. (2005). A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature, 438, 325–331.
Rockwell, N. C., & Lagarias, J. C. (2017). Phytochrome diversification in cyanobacteria and eukaryotic algae. Current Opinion in Plant Biology, 37, 87–93.
Lamparter, T., Michael, N., Caspani, O., Miyata, T., Shirai, K., & Inomata, K. (2003). Biliverdin binds covalently to Agrobacterium phytochrome Agp1 via its ring A vinyl side chain. Journal of Biological Chemistry, 278, 33786–33792.
Nagano, S., Scheerer, P., Zubow, K., Michael, N., Inomata, K., Lamparter, T., & Krauss, N. (2016). The crystal structures of the N-terminal photosensory core module of Agrobacterium phytochrome Agp1 as parallel and anti-parallel dimers. Journal of Biological Chemistry, 291, 20674–20691.
Schmidt, A., Sauthof, L., Szczepek, M., Lopez, M. F., Escobar, F. V., Qureshi, B. M., Michael, N., Buhrke, D., Stevens, T., Kwiatkowski, D., von Stetten, D., Mroginski, M. A., Krauss, N., Lamparter, T., Hildebrandt, P., & Scheerer, P. (2018). Structural snapshot of a bacterial phytochrome in its functional intermediate state. Nature Communications, 9, 4912.
Lamparter, T., Michael, N., Mittmann, F., & Esteban, B. (2002). Phytochrome from Agrobacterium tumefaciens has unusual spectral properties and reveals an N-terminal chromophore attachment site. Proceedings of the National academy of Sciences of the United States of America, 99, 11628–11633.
Blumenstein, A., Vienken, K., Tasler, R., Purschwitz, J., Veith, D., Frankenberg-Dinkel, N., & Fischer, R. (2005). The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Current Biology, 15, 1833–1838.
Quest, B., & Gärtner, W. (2004). Chromophore selectivity in bacterial phytochromes: Dissecting the process of chromophore attachment. European Journal of Biochemistry, 271, 1117–1126.
Lamparter, T., Carrascal, M., Michael, N., Martinez, E., Rottwinkel, G., & Abian, J. (2004). The biliverdin chromophore binds covalently to a conserved cysteine residue in the N-terminus of Agrobacterium phytochrome Agp1. Biochemistry, 43, 3659–3669.
Margulis, L., Dolan, M. F., & Guerrero, R. (2000). The chimeric eukaryote: Origin of the nucleus from the karyomastigont in amitochondriate protists. Proceedings of the National academy of Sciences of the United States of America, 97, 6954–6959.
Buchberger, T., & Lamparter, T. (2015). Streptophyte phytochromes exhibit an N-terminus of cyanobacterial origin and a C-terminus of proteobacterial origin. BMC Research Notes, 8, 144.
Kooss, S., & Lamparter, T. (2017). Cyanobacterial origin of plant phytochromes. Protoplasma, 254, 603–607.
Sievers, F., & Higgins, D. G. (2018). Clustal Omega for making accurate alignments of many protein sequences. Protein Science, 27, 135–145.
Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792–1797.
Capella-Gutiérrez, S., Silla-Martínez, J. M., & Gabaldón, T. (2009). trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics, 25, 1972–1973.
Lamparter, T., Mittmann, F., Gärtner, W., Borner, T., Hartmann, E., & Hughes, J. (1997). Characterization of recombinant phytochrome from the cyanobacterium Synechocystis. Proceedings of the National Academy of Sciences of the United States of America., 94, 11792–11797.
Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35, 1547–1549.
Felsenstein, J. (1985). Confidence-limits on phylogenies—An approach using the bootstrap. Evolution, 39, 783–791.
Letunic, I., & Bork, P. (2007). Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics, 23, 127–128.
Koonin, E. V. (2010). The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biology, 11, 209.
Lamparter, T., Mittmann, F., Gärtner, W., Borner, T., Hartmann, E., & Hughes, J. (1998). Characterization of recombinant phytochrome from the cyanobacterium Synechocystis (vol 94, pg 11792, 1997). Proceedings of the National Academy of Sciences of the United States of America., 95, 2714–2714.
Wu, S. H., McDowell, M. T., & Lagarias, J. C. (1997). Phycocyanobilin is the natural precursor of the phytochrome chromophore in the green alga Mesotaenium caldariorum. Journal of Biological Chemistry, 272, 25700–25705.
Butler, W. L., Norris, K. H., Siegelman, H. W., & Hendricks, S. B. (1959). Detection, assay, and preliminary purification of the pigment controlling photoresponsive development of plants. Proceedings of the National academy of Sciences of the United States of America, 45, 1703–1708.
Sage, L. C. (1992). Pigment of the imagination—a history of phytochrome research. Academic Press.
Kehoe, D. M., & Grossman, A. R. (1996). Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science, 273, 1409–1412.
Yeh, K. C., Wu, S. H., Murphy, J. T., & Lagarias, J. C. (1997). A cyanobacterial phytochrome two-component light sensory system. Science, 277, 1505–1508.
Davis, S. J., Vener, A. V., & Vierstra, R. D. (1999). Bacteriophytochromes: Phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science, 286, 2517–2520.
Hübschmann, T., Börner, T., Hartmann, E., & Lamparter, T. (2001). Characterisation of the Cph1 holo-phytochrome from Synechocystis sp. PCC 6803. European Journal of Biochemistry, 268, 2055–2063.
Ikeuchi, M., & Ishizuka, T. (2008). Cyanobacteriochromes: A new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochemical & Photobiological Sciences, 7, 1159–1167.
Yoshihara, S., Katayama, M., Geng, X., & Ikeuchi, M. (2004). Cyanobacterial phytochrome-like PixJ1 holoprotein shows novel reversible photoconversion between blue- and green-absorbing forms. Plant and Cell Physiology., 45, 1729–1737.
Nies, F., Worner, S., Wunsch, N., Armant, O., Sharma, V., Hesselschwerdt, A., Falk, F., Weber, N., Weiss, J., Trautmann, A., Posten, C., Prakash, T., & Lamparter, T. (2017). Characterization of Phormidium lacuna strains from the North Sea and the Mediterranean Sea for biotechnological applications. Process Biochemistry., 59, 194–206.
Casal, J. J., & Smith, H. (1989). The function, action and adaptive significance of phytochrome in light-grown plants. Plant, Cell and Environment, 12, 855–862.
Crooks, G. E., Hon, G., Chandonia, J. M., & Brenner, S. E. (2004). WebLogo: A sequence logo generator. Genome Research, 14, 1188–1190.
Funding
Open Access funding enabled and organized by Projekt DEAL. The funding has been received from Deutsche Forschungsgemeinschaft.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
This publication is dedicated to Prof. Silvia E. Braslavsky, a pioneer in photobiology and photobiophysics, on the occasion of her 80th birthday.
Supplementary Information
Below is the link to the electronic supplementary material.
43630_2022_271_MOESM1_ESM.pdf
Supplementary Table S1. List of species abbreviations and protein sequences with systematic origin and accession number for phytochrome sequence. The position of the chromophore attaching Cys residue (N-terminal or in GAF domain) is also given (PDF 353 kb)
43630_2022_271_MOESM2_ESM.pdf
Supplementary Fig. S1: All 70 phylogenetic trees based on phytochrome PCM and chromophore binding amino acid sequences. Trees were drawn with ITOL [26] using default parameters in black and white. The tree numbers (as given in Table 1) are identical with the page numbers and are also printed as large number in the upper left part of each graph. Each leaf is labeled with the major group to which the species belongs, the chromophore binding site, the species name and the accession number of the protein. Due to the large number of phytochrome sequences, each tree cannot be viewed as a hole on a page, it is necessary to enlarge relevant sections (PDF 3678 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gabriel, E., Krauß, N. & Lamparter, T. Evidence for evolutionary relationship between archaeplastidal and cyanobacterial phytochromes based on their chromophore pockets. Photochem Photobiol Sci 21, 1961–1974 (2022). https://doi.org/10.1007/s43630-022-00271-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43630-022-00271-7