Abstract
Corynebacterium glutamicum is a microbial production host established in the industry 60 years ago. It is mainly used for production of feed and food amino acids. As C. glutamicum strain development has been cutting edge since its discovery, it has been engineered for production of a plethora of valuable products. This review will focus on recent developments of C. glutamicum strain engineering for biotransformation and fermentation processes towards flavor and fragrance molecules as well as pigments and sweeteners.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fermentation of foods is known since ages. Besides preservation, sensory changes are introduced. Alternatively, sensory molecules such as sweeteners, fragrance and flavor compounds can be added to food to ameliorate their sensory perception. Fermentation processes with Corynebacterium glutamicum and Brevibacterium species that were reclassified as Corynebacterium species, e.g., B. flavum to C. glutamicum ssp. flavum [1] are established in biotechnology for more than 50 years. They primarily targeted the flavor enhancing sodium salt of the amino acid l-glutamate (MSG) and ribonucleoside monophosphates such as inosine monophosphate (IMP) or guanosine monophosphate (GMP) since these contribute to the flavor umami. The reader is referred to excellent reviews on the C. glutamicum-based production of amino acids [2] and many other products [3]. By contrast, this review focusses for the first time on the C. glutamicum-based production of flavor and fragrance molecules such as indole and patchoulol as well as pigments such as astaxanthin and sweeteners such as xylitol. Successful metabolic engineering strategies are highlighted for exemplary products and the key performance indicators of the established processes are listed in Table 1.
Flavors
MSG and ribonucleoside monophosphates
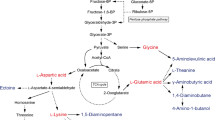
C. glutamicum is used at industrial scale to produce monosodium glutamate (MSG), the main component of the umami flavor. Fermentative production of l-glutamate started in late 1950s by Kyowa Hakko Kogyo Co Ltd. [26]. Nowadays, production of 3.2 million tons of l-glutamate/MSG by fermentation is estimated yearly, which accounts for over 40% of the amino acid market [2, 4]. Additionally, production of ribonucleoside monophosphates was demonstrated in C. glutamicum. Inosine monophosphate (IMP) and guanosine monophosphate (GMP) act synergistically with MSG to increase umami flavor. Microbial strain improvement for production of ribonucleoside monophosphates in different microorganisms such as Corynebacterium, Bacillus, or E. coli has recently been reviewed [27]. In C. glutamicum, deletion of conversion reactions from IMP to GMP and adenosine monophosphate (AMP) catalyzed by IMP-dehydrogenase (guaB2) and adenylosuccinate synthetase (purA) resulted in increased intracellular pool of IMP to 22 μmol g−1 CDW. Further engineering steps of deleting the glucose 6-phosphate isomerase decreased the IPM titer and accumulation of the corresponding nucleobase hypoxanthine was observed [6].
Indole
Recently fermentative production of indole was demonstrated by C. glutamicum. Indole is characterized by its specific odor which is described as pungent, floral, with a fecal and animalic musty character. Indole is used as a flavoring of tea and dairy products and as a fragrance in perfumery. Natural indole has limited availability and a high price; therefore, fermentative production offers an attractive alternative. In nature, indole is synthesized from l-tryptophan (l-Trp) in a hydrolytic β-elimination reaction catalyzed by tryptophanases (TNAs) or from indole-3-glycerol phosphate (IGP) in a retroaldol cleavage catalyzed by enzymes with IGP lyase (IGL) activity. Both biosynthetic routes were used in C. glutamicum to establish fermentative indole production.
A bioconversion process to produce indole from supplemented l-Trp has been established by the co-expression of the native aromatic amino acid permease gene aroP and a bacterial tryptophanase (tna) [8]. TNAs hydrolyze l-Trp in a β-elimination reaction yielding indole, pyruvate and ammonia. By mining of bacterial genome databases an array of TNAs were retrieved and the highest production of indole at 0.9 g L −1 in C. glutamicum chassis strain C1* was achieved based on tryptophanase from Providencia rettgeri. However, the product toxicity was observed as a major limiting factor for indole production by C. glutamicum. Indole toxicity was studied in C. glutamicum ATCC 13032 and the chassis strain C1* and the C1* strain showed higher tolerance to indole compared to its ancestor [28, 29]. Indole toxicity was further circumvented by in situ product recovery (ISPR) by which product is sequestered in the organic layer and maintained at sub-toxic concentrations in the growth medium. Sequestering indole in a food-grade organic phase, dibutyl sebacate, during the fermentation resulted in complete conversion of l-Trp and indole titer of 5.7 g L−1.
De novo production of indole in C. glutamicum from glucose was achieved in C. glutamicum by indole-3-glycerol phosphate lyases (IGLs) [7, 30] (Fig. 1). IGLs are considered to have evolved from the tryptophan synthase α-subunit (TSA). In tryptophan synthase (TS) two α- and two β-subunits are aligned. Indole is not released from TSA, but is efficiently channeled to tryptophan synthase β-subunit (TSB) for condensation of indole and l-serine to form l-Trp. In contrast, IGLs work as stand-alone enzymes and release free indole. Surprisingly, it was found that C. glutamicum TSA (trpA) functions as a bona fide IGL and can support fermentative indole production, while intrinsically low IGL activity of tryptophan synthase α-subunit (TSA) in the absence of the tryptophan synthase β-subunit was described. Bioprospecting of plant IGLs and bacterial TSAs resulted in identification of additional IGLs that could support fermentative indole production from IGP. Extensive shikimate pathway engineering was performed to increase IGP availability. Efficient conversion of anthranilate to IGP was achieved by deletion of chorismate mutase gene (csm) to avoid loss of the intermediate chorismate to the aromatic amino acids l-tyrosine and l-phenylalanine, by deletion of the l-glutamate export gene yygB and deletion of trpL which alleviates the endogenous trp operon from attenuation control. Using tributyrin as organic layer for in situ product recovery a final titer of 0.7 g L−1 indole was achieved by expressing either C. glutamicum TNA or IGL from wheat.
Indole (A) and vanillin (B) biosynthesis in metabolically engineered C. glutamicum. A De novo biosynthetic pathway of indole and B de novo biosynthetic pathway of vanillin. Arrows depicted in red indicate gene overexpression, while gene deletions are depicted with grey crosses. The abbreviations are as follows: E4P, erythrose-4-phosphate; PEP, phosphoenolpyruvate; DAHP, 3-deoxy-d-arabinoheptulosonate-7-phosphate; l-Phe, l-phenylalanine; l-Tyr,l- tyrosine; l-Gln, l-glutamine; l-Glu, l-glutamate; IGP, indole-3-glycerol phosphate; l-Trp, l-tryptophan; 4-HBA, 4-hydroxybenzoate; 4-HB aldehyde, 4-hydroxybenzaldehyde; 4-HB alcohol, 4-hydroxybenzyl alcohol; PCA, protocatechuate (3,4-dihydroxybenzoate); PC aldehyde, protocatechuic aldehyde; PC alcohol, protocatechuic alcohol; aroF, DAHP synthase; aroGEcD146N and aroGfbr, feedback-resistant DAHP synthase from E. coli; csm, chorismate mutase; trpE, anthranilate synthase; trpEEcS40F, feedback-resistant trpE from E. coli; trpECgS38R, feedback-resistant trpE from C. glutamicum; trpG, anthranilate synthase component I; yggB, MscS-type mechanosensitive channel; trpD, anthranilate phosphoribosyltransferase; trpDEc, trpD from E. coli; trpFC, N-(5′-phosphoribosyl)anthranilate isomerase; trpBA, tryptophan synthase; IGLTa IGP lyase from Triticum aestivum; trpA, tryptophan synthase α-subunit; ubiCPR, a product-resistant chorismate-pyruvate lyase from E. coli; pobA, 4-hydroxybenzoate hydroxylase; car, carboxylic acid reductase from Nocardia iowensis; creG, NAD+-dependent dehydrogenase; comtm, a mutant catechol O-methyltransferase from Rattus norvegicus; vanAB, vanillate demethylase; vdh, vanillin dehydrogenase; NCgl0324 aldehyde reductase. Adapted from [7, 16, 30]
l-theanine
l-theanine is the most abundant free amino acid in leaves of tea plants (Camellia sinensis) and contributes to the flavor of green tea and shows health benefits. l-theanine is a N-ethylated l-glutamate derivative produced in plants by the l-theanine synthetase from l-glutamate and ethylamine. Recently, C. glutamicum as well as E. coli and Pseudomonas putida have been engineered for l-theanine production [9, 31, 32]. In C. glutamicum l-theanine production with γ-glutamyl methylamide (GMA) synthetase (GMAS, EC 6.3.4.12) which catalyzes the ATP-dependent ligation of l-glutamate and ethylamine was established in an industrial l-glutamate overproducing strain GDK-9 [9]. Production of ethylamine endogenously by expressing plant-derived l-serine/l-alanine decarboxylases was unsuccessful, therefore a process with exogenous supplementation of ethylamine was established. The highest production at 19 g L−1 was established by overexpression of the codon optimized gmaS from the methylotrophic bacteria Methylovorus mays. The production was further increased by the deletion of the endogenous l-glutamate exporter gene yggB (NCg11221), resulting in production titer of about 42 g L−1 with ethylamine supplementation in 5L bioreactor. In comparison, production of up to 17 g L−1 of l-theanine from ethylamine was described in P. putida using different carbon sources [31]. In E. coli 16 g L−1 of l-theanine was produced de novo in strains where a biosynthetic pathway to provide endogenous ethylamine supply via transamination of acetaldehyde was established [32].
Fragrances
Terpenoid fragrances
Corynebacterium glutamicum was also proven as a suitable host organism for the production of volatile terpenoids (Fig. 2). This bacterium naturally produces carotenoids (Fig. 3). Thus, platform strain engineering for terpenoid-derived fragrance compounds relies on the deletion of carotenogenic genes and/or the deletion of prenyltransferases that compete with the heterologous pathways. The isoprenoid precursor molecules isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP) are synthesized in the methylerythritol phosphate (MEP) pathway that comprises nine enzymatic steps in C. glutamicum [10, 33,34,35]. Short-chain terpenoids such as α-farnesene, patchoulol, (+)-valencene and α-pinene, derive from farnesyl pyrophosphate (FPP) and geranyl pyrophosphate (GPP) for the latter one. Deletion of idsA, the major GGPP synthase gene as well as deletion of the carotenogenic operon(s) was shown to be efficient for establishment of short-chain terpenoid production. α-Pinene production could be shown by overexpression of GPP synthase gene from Abies grandies and pinene synthase gene from Pinus taeda [11]. Production of α-pinene could be improved to 27 µg g−1 CDW by an enhanced precursor biosynthesis and isoprenyl pyrophosphate balancing, namely by dxs and idi overexpression [11]. FPP biosynthesis relied on the FPP synthase gene ispA from E. coli. α-farnesene production was enabled by codon-optimized overexpression of the farnesene synthase gene from cultivated apple [10]. Interestingly, besides dxs and idi, the second enzyme Dxr (DXP reductoisomerase) was shown to improve α-farnesene by twofold with maximal titers of 75 mg L−1 [10]. The fragrance compound (+)-valencene was the first sesquiterpenoid being produced by C. glutamicum. It was shown that overexpression of ispA from E. coli and valencene synthase gene from Nootka cypress resulted in the best ( +)-valencene titer with 2.4 mg L−1 [15] that could be improved by induction of photo-caged IPTG instead of IPTG to maximal titers of 41 mg L−1 [14]. Production of patchoulol was shown by overexpression of ispA from E. coli with a patchoulol synthase gene from Pogostemon cablin. Although the patchoulol synthase gene was optimized for an alga l-production system, patchoulol titers of 0.46 mg L−1 were achieved in shake flask experiments from glucose and 0.25 mg L−1 from the alternative pentose sugar xylose [12, 13]. In this study maximal patchoulol titers were achieved in two-phase fed-batch fermentation with 50 mg L−1 [12, 13].
Vanillin
The aromatic aldehyde vanillin is an important flavoring compound. Several processes for vanillin production have been described starting from ferulic acid, eugenol, isoeugenol or glucose [36]. The first successful de novo production of vanillin in C. glutamicum was recently established [16] (Fig. 1). Production of aldehydes in microbial platforms poses a great challenge due to their rapid reduction to corresponding alcohols via endogenous aldehyde reductase activity. Bioprospecting revealed 27 putative aldehyde reductases in the C. glutamicum genome. In this study, a C. glutamicum strain producing 4-hydroxybenzyl alcohol was used as the model strain. The deletion of gene NCgl0324 strongly decreased reduction of 4-hydroxybenzaldehyde to 4-hydroxybenzyl alcohol in vivo. Next, a strain that produces protocatechuate (MA303) was engineered from 4-hydroxybenzoate-synthesizing C. glutamicum APS963. Vanillin production was engineered in strain MA303 by overexpressing of the vanillin/vanillyl alcohol biosynthetic pathway consisting of the carboxylic acid reductase from Nocardia iowensis (car) and a mutant catechol O-methyltransferase from Rattus norvegicus (comtm) genes. Deletion of the aldehyde reductase NCgl0324 in this strain resulted in the production of 0.31 g L−1 vanillin, while deletion of NCgl0324 in related strains enabled production of 1.18 g L−1 protocatechuic aldehyde and 1.36 g L−1 4-hydroxybenzaldehyde. Overcoming the observed product toxicity as e.g., vanillin accumulation lowered the specific growth rate, the final biomass and glucose consumption, holds the potential for further improvements.
Methyl anthranilate
Methyl anthranilate (MANT) is widely used to provide grape scent and flavor in cosmetics and food applications. Fermentative production of MANT was recently described in E. coli and C. glutamicum [17]. The key to the successful production in both hosts was the optimization of the expression of the anthranilic acid methyltransferase 1 aamt1 gene from Zea mays. Additionally, the anthranilate supply was successfully increased by the expression of the feedback-inhibition resistant mutant of 3-deoxy-d-arabinoheptulosonate 7-phosphate (DAHP) synthase from E. coli (aroGS80F). The availability of the cofactor S-adenosyl-l-methionine required by the AAMT1 was optimized by recycling of the SAM reaction product S-ribosyl-l-homocysteine (SAH) by activity of SAH hydrolase sahH. Fed-batch production in a bioreactor using two-phase cultivation in which tributyrin was used as organic layer led to the production of 4.47 and 5.74 g L−1 MANT in E. coli and C. glutamicum, respectively. In comparison, in Saccharomyces cerevisiae production of 414 mg L−1 methyl anthranilate was achieved [37]. Production of the related compound N-methyl anthranilate, a precursor of anticancer acridone alkaloids, in C. glutamicum was demonstrated at a final titer of 0.5 g L−1 [28, 29].
Pigments
C. glutamicum naturally produces the yellow carotenoid decaprenoxanthin. Decaprenoxanthin is a C50 carotenoid that derives from the isoprenoid precursor molecules isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP). In C. glutamicum, the methylerythritol phosphate (MEP) pathway synthesizes IPP and DMAPP from the central metabolites pyruvate and glyceraldehyde 3-phosphate (GAP) [33,34,35]. The genome comprises a major GGPP synthase gene idsA (cg2384) [33,34,35], one major carotenogenic operon (crtE-cg0722-crtB-crtI-crtYe-crtYf-crtEb) (cg0723-cg0717) [38, 39] as well as a second functional phytoene synthase gene cg2672 and a glycosyltransferase gene crtX (cg0730) [33,34,35]. Carotenoid biosynthesis is regulated by the MarR-like regulator CrtR [21]. CrtR binds the promoter of the crt operon to repress its transcription. GGPP was identified as the major inducer of CrtR (shorter diphosphates interfered less with CrtR binding promoter DNA), thus regulating carotenoid biosynthesis in a metabolite dependent manner [21, 40]. This regulation indicates a feedforward regulatory mechanism of carotenogenesis: first isoprenoid pyrophosphates are synthesized that act as effector molecules on the repressor CrtR and therefore induce transcription of the terminal carotenoid biosynthetic genes.
Decaprenoxanthin represents a rare C50 carotenoid that is not used in industry so far. However, a wide range of C40 carotenoids such as astaxanthin, β-carotene, canthaxanthin, lutein, lycopene and zeaxanthin, are used in the food and feed industries for decades. β-carotene and astaxanthin are dominating the market but the latter one is expected to be the leading product in the future due to diverse applications ranging from coloration of fish feeds to health protection in nutraceutical and cosmetic products due to its excellent antioxidant properties [41]. C. glutamicum was engineered for natural production of astaxanthin (Fig. 3). First, the genes crtEb, crtYe and crtYf were deleted in a prophage-cured MB001 strain to prevent biosynthesis of decaprenoxanthin and thus yields accumulation of the central C40 carotenoid lycopene. Moreover, the precursor supply was improved by the chromosomal overexpression of dxs under the strong constitutive Ptuf promoter [33,34,35]. 1-deoxy-5-xylulose phosphate synthase as the first enzyme in the MEP pathway plays a major role in the regulation of the MEP pathway [42,43,44]. It is long known that Dxs is feedback regulated by the products IPP and DMPP [43]. Moreover, the coordinated chromosomal overexpression of crtE-crtB-crtI as an artificial operon from the Ptuf promoter increased lycopene production [18]. Deregulation of the native carotenoid gene expression by deletion of crtR resulted in a threefold higher lycopene content (2 mg g−1 CDW) [21]. Based on endogenous lycopene production the production of cyclic C40 carotenoids can be established by overexpression of the lycopene β-cyclase CrtY. The lycopene cyclase CrtY from Pantoea ananatis is a non-redox flavoprotein that requires NAD(P)H for cyclization of ε-ends of lycopene to γ-carotene (one β-end) and β-carotene (two β-ends) [45]. β-carotene biosynthesis was proven to be very efficient in C. glutamicum with contents of up to 12 mg g−1 CDW [21]. The heterologous expression was established by a chromosomal integration under the Ptuf promoter that performed as good as the plasmid-driven overexpression of crtY [18]. It is hypothesized that β-carotene in contrast to lycopene could be integrated in a coordinated manner in the plasma membrane and thus can be produced with higher contents.
Terpenoids biosynthesis in metabolically engineered C. glutamicum. MEP-pathway (A). Enzyme abbreviations are given right to the reactions. Metabolites are shown in bold, cofactors are shown on the left side of the respective reaction. (GAP: Glyceraldehyde 3-phosphate; DXP: 1-deoxy-d-xylulose 5-phosphate; MEP: 2-methylerythritol 4-phosphate; CDP-ME: 4-diphosphocytidyl-2-methylerythritol; CDP-MEP: 4-diphosphocytidyl-2-methylerythritol 2-phosphate; ME-cPP: 2-methylerythritol 2,4-cyclopyrophosphate; HMBPP: 4-hydroxy-3-methyl-2-(E)-butenyl-4-pyrophosphate; IPP: Isopentenyl pyrophosphate; DMPP: Dimethylallyl pyrophosphate). Enzymes: Dxs (1-deoxy-d-xylulose 5-phosphate synthase), Dxr (1-deoxy-d-xylulose 5-phosphate reductoisomerase), IspD (2-C-methylerythritol 4-phosphate cytidylyltransferase), IspE (2-C-methylerythritol 4-diphosphocytidyl kinase), IspF (2-C-methylerythritol 2,4-cyclopyrophosphate synthase), IspG (4-hydroxyl-3-methyl-2-(E)-butenyl pyrophosphate synthase), IspH (4-hydroxyl-3-methyl-2-(E)-butenyl pyrophosphate reductase). Terpenoid biosynthesis (B). Biosynthesis of terpenoid intermediates GPP, FPP and GGPP on the basis of IPP and DMPP by GPP-, FPP- and GGPP synthases is illustrated. Different fragrance and pigment compounds are given as examples. Carotenoid biosynthesis (C). The biosynthesis of astaxanthin/decaprenoxanthin starting from the precursors IPP and DMPP are depicted. Red script indicates gene overexpressions, while gene deletions are depicted with grey crosses. Enzymes: IdsA/CrtE (GGPP synthase), CrtB (phytoene synthase), CrtI (phytoene desaturase), CrtEb (lycopene elongase), CrtYeYf (heterodimeric ε-cyclase), CrtYPa (lycopene β-cyclase from Pantoea ananatis), CrtWFp (β-carotene 4-ketolase from Fulvimarina pelagi), CrtZFp (β-carotene 3-hydroxylase from F. pelagi). The membrane fusion protein CrtZ ~ W (N-terminal CrtZ with 3 transmembrane helices and C-terminal CrtW with 5 transmembrane helices) is depicted
Based on the β-carotene producing platform strain astaxanthin biosynthesis was shown by three different strategies in C. glutamicum. First, it was shown in a combinatorial approach that coexpression of bacterial crtZ and crtW results in higher astaxanthin contents when both genes were expressed with high translation initiation rates. Second, it could be shown that separate expression of crtZ and crtW on a two-plasmid system indeed resulted in a higher astaxanthin [18]. Third, the engineering of a membrane-bound fusion protein harboring CrtZ and CrtW yielded the highest astaxanthin content under high glucose concentration (4%) [19]. A robust astaxanthin production under high glucose concentration is necessary for as the process design towards economic relevance is in favor of high cell density fermentations. The fusion protein CrtZ~W comprises the N-terminal CrtZ (3 TMH) and C-terminal CrtW (5 TMH) that are linked by an artificial ten amino acid linker sequence [19]. Although the exact mechanism of the robustness under high glucose concentration is unknown, it was shown very clear that the translational fusion CrtZ~W performed more than twofold better than the transcriptional fusion of CrtZ and CrtW resulting in 22 mg L−1 astaxanthin in the mL scale [19]. It was hypothesized that membrane integrity, protein stability or intermediate channeling could explain the superior production properties of the membrane-fusion protein. The astaxanthin derived from C. glutamicum represents a natural alternative to algal-based astaxanthin with different product specificities that rely on its free astaxanthin form. Thus, astaxanthin but also other C40 carotenoid production with C. glutamicum gain interest from industrial partners to meet the demand for natural pigments.
Sweeteners
Xylitol
Polyols find application as artificial sweeteners with a rising demand due to diabetes and the global epidemic obesity. The polyols contain no calories and they are used e.g., in sugar-free chewing gum as they give rise to a cooling sensation in the mouth [46]. Polyols such as xylitol are poorly absorbed, thus, hardly affecting blood sugar levels, and they do not promote caries as the oral microbiota is unable to digest them. C. glutamicum has been engineered for conversion of xylose to xylitol by implementation of xylose reductase from Rhodotorula mucilaginosa [23]. Xylitol was also produced from arabinose based on heterologous expression of the genes for l-arabinose isomerase, d-psicose 3-epimerase and l-xylulose reductase from Escherichia coli, Agrobacterium tumefaciens and Mycobacterium smegmatis, respectively. By combining both approaches with expression of a pentose transport gene from B. licheniformis, 27 ± 0.3 g L−1 xylitol was produced from pentose rich, acid pre-treated liquor of sorghum stover [23].
d-allulose
The zero-calory sugar d-allulose (or d-psicose) exhibits 70% relative sweetness compared to d-sucrose. A whole-cell biotransformation using C. glutamicum expressing d-allulose 3-epimerase (DAE) from Clostridium hylemonae allowed to produce 120 g L−1 of d-allulose from 400 g L−1 of d-fructose at 55 °C in 1.5 h [47]. The enzyme source appeared critical since DAE from C. hylemonae supported higher production as compared to DAE from Flavonifractor plautii [48] or a mixture of DAEs from Paenibacillus senegalensis, Clostridium cellulolyticum, and Ruminococcus sp. [22].
Further additives
Squalene
The triterpene squalene is valued as lipophilic antioxidant. Natural sources (including shark liver) are limited and its use in emulsion-based vaccine adjuvants recently boosted its demand. Upon expression of genes for squalene synthase from S. cerevisiae, FPP synthase IspA from E. coli, and endogenous Dxs and Idi, squalene production from glucose was enabled. When CRISPRi [49] was used to reduce transcription of the native GGPP synthase gene idsA, the highest product titer (0.1053 g L−1) was observed in Biolector cultures with CGXII medium containing 15 g L−1 tryptone and 40 g L−1 glucose [50]. Additional expression of endogenous ispDF increased the titer to about 0.14 g L−1 [10].
CoQ10
Coenzyme Q10 (CoQ10) is a lipid-soluble compound with important physiological functions and is sought after in the food and cosmetic industries owing to its antioxidant properties [51]. The menaquinone (MKn) containing C. glutamicum, which does not naturally synthesize any CoQ, was engineered for CoQ10 synthesis (Fig. 4). Prior knowledge on overproduction of aromatic compounds and prenyls provided a base strain overproducing both precursors, aromatic 4-hydroxybenzoate via the shikimate pathway and decaprenyl diphosphate via the MEP pathway [52]. Two aspects were important. First, expression of the genes encoding the Ubi complex accessory factors UbiJ and UbiK in addition to the ubi genes for the CoQ10 biosynthesis enzymes from E. coli improved production of CoQ isoprenologs from CoQ8 to CoQ11. Second, the replacement of the native polyprenyl diphosphate synthase IspB with a decaprenyl diphosphate synthase (DdsA) increased the CoQ10 titer, while the abundance of the unwanted isoprenologs CoQ8, CoQ9, and CoQ11 was reduced significantly [24].

Reproduced from [24]
Metabolic pathway of CoQn and MKn(H2) biosynthesis in engineered C. glutamicum. Enzymes are in bold, heterologous enzymes are underlined. As IspB mainly synthesizes NPP and OPP and DdsA mainly synthesizes DPP and UPP, the enzymes and corresponding direct products were marked with matching colors. The question mark indicates that the reaction attributed to Cgl0472 is not experimentally proven. MEP, methylerythritol phosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; FPP, farnesyl diphosphate; OPP, octaprenyl diphosphate; NPP, nonaprenyl diphosphate; DPP, decaprenyl diphosphate; UPP, undecaprenyl diphosphate; 4-HBA, 4-hydroxybenzoic acid; nP-HB, 3-n-prenyl-4-hydroxybenzoic acid; nP-Ph, 2-n-prenylphenol; CoQn, coenzyme Qn/ubiquinone-n; DHNA, 1,4-dihydroxy-2-naphthoic acid; DMKn, demethylmenaquinone-n; MKn, menaquinone-n; MKn(H2), dihydromenaquinone-n; IspA, farnesyl diphosphate synthase; IspB, polyprenyl diphosphate synthase; DdsA, decaprenyl diphosphate synthase; UbiC, chorismate-pyruvate lyase; UbiA, 4-hydroxybenzoate octaprenyltransferase; UbiD-X, 3-octaprenyl-4-hydroxybenzoate decarboxylase and flavin prenyltransferase; UbiIGHEF, 2-octaprenylphenol hydroxylase, 2-octaprenyl-6-hydroxyphenol/2-octaprenyl-3-methyl-5-hydroxy-6-methoxy-1,4-benzoquinol methyltransferase, 2-octaprenyl-6-methoxyphenol hydroxylase, ubiquinone/menaquinone biosynthesis methyltransferase, 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase; MenF, isochorismate synthase; MenA, 1,4-dihydroxy-2-naphthoate octaprenyltransferase; MenG, demethylmenaquinone methyltransferase; Cgl0472, putative menaquinone oxidoreductase.
Nisin
The ribosomally synthesized peptide antibiocin nisin shows antimicrobial activity against many Gram-positive bacteria including human pathogens such as Listeria monocytogenes and Staphylococcus aureus strains. This generally regarded as safe compound, which is approved by FDA and EFSA, is widely used in food preservation [53]. While nisin is typically produced using natural Lactococcus lactis strains, C. glutamicum has been engineered for synthesis, secretion, and maturation of nisin [25]. To this end, first, a synthetic operon comprising the genes nisZ, nisB, nisT and nisC of the nisin Z biosynthesis operon of L. lactis B1629 was expressed. The resulting pre-nisin required maturation, which was achieved by using a soluble form of the membrane-anchored L. lactis B1629 NisZ protease. LC–MS analysis revealed that about 0.5 g L−1 nisin was produced [25]. C. glutamicum was also engineered for production of other antimicrobial peptides, such as pediocin [54].
Concluding remarks and further directions
Corynebacterium glutamicum processes were successfully scaled to bioreactors with working volumes of several thousand cubic meters for the million-tons per year amino acid production. However, the titers, rates and yields reported here for most of the functional food additives/ingredients have to be improved to achieve and maintain viable economics. This may be achieved on the enzyme, cell or process level. Regarding the enzymes required for optimal pathway operation, new enzymes or variants may be identified by bioprospecting as shown for the IGL and TNA enzymes used for indole production [7, 8, 30]. Enzyme engineering has, for example, been applied to enzymes of aromatic biosynthesis in order to change the substrate scope [55] or alleviating product inhibition as recently shown for 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase [56] and chorismate-pyruvate lyase [57]. Enzyme fusions have been shown to improve, for example, astaxanthin production [19]. These enzyme-based approaches will certainly benefit from combining computational modelling with mechanistic experiments [58] and the rapidly increasing number of protein structures predicted by alphafold [59]. Machine learning has already been applied to enzyme evolution [60] and will be important to come to new-to-nature reactions [61], which eventually may be important to improve production of functional food additives/ingredients. In strain engineering, we see potential in the increasing number and versatility of CRISPR methods [62], including base editors for introduction of SNPs [63]. In this respect, it has to be noted that CRISPR methods can be multiplexed [64]. Genetically encoded biosensors can be used to identify mutants with improved production [65] or enzyme variants [66], or to dynamically control gene expression [7, 30, 67]. The design and application of microbial consortia is an option for division of labor, e.g., access to substrate by one bacterium and conversion to the target product by another [68]. Process intensification makes sure that processes are stable when scaled up and often involve medium optimization. For volatile products, two-layer approaches for in situ product removal (ISPR) offer to reach higher titers by avoiding possible inhibitory effects of the product on the catalyst cell. This concept has been employed, for example, in indole production [8]. The vigorous stirring in stirred tank bioreactors helps partitioning of the product into the organic layer as shown for patchoulol [12, 13]. For cell-bound products, process optimization aims at increasing biomass titers to high-cell density while maintaining high product content within these cells. Sometimes, it is of benefit to co-produce a cell-bond product such as astaxanthin and a secreted product such as lysine [12, 13] when targeting the same application, in this case animal feed additives. For secreted products, partitioning into a second phase is often not possible. One way to improve titers of a growth-inhibitory product is the selection of more tolerant strains as was applied to alleviate indole toxicity [28, 29]. Downstream processing has to comply with two aims: optimal product recovery and compatibility with regulatory issues of uses as food additives or ingredients. C. glutamicum, established for decades as excellent producer of food additives such as MSG, has recently been engineered for production of a wide variety of food additives and ingredients. We anticipate that this development takes up more momentum. First, production scales with C. glutamicum belong to the largest known in biotechnology. Second, its products are classified as GRAS and regulatory issues have been taken care of given the established amino acid production processes. Thus, with respect to the production of food additives and ingredients, C. glutamicum is competitive with E. coli and Baker’s yeast. The market and the consumers, thus, have more choices and may rely on better supply reliability.
References
Liebl W, Ehrmann M, Ludwig W, Schleifer KH. Transfer of Brevibacterium divaricatum DSM 20297T, “Brevibacterium flavum” DSM 20411, “Brevibacterium lactofermentum” DSM 20412 and DSM 1412, and Corynebacterium glutamicum and their distinction by rRNA gene restriction patterns. Int J Syst Bacteriol. 1991;41(2):255–60.
Wendisch VF. Metabolic engineering advances and prospects for amino acid production. Metab Eng. 2020;58:17–34.
Wolf S, Becker J, Tsuge Y, Kawaguchi H, Kondo A, Marienhagen J, Bott M, Wendisch VF, Wittmann C. Advances in metabolic engineering of Corynebacterium glutamicum to produce high-value active ingredients for food, feed, human health, and well-being. Essays Biochem. 2021;65(2):197–212.
Sanchez S, Rodríguez-Sanoja R, Ramos A, Demain AL. Our microbes not only produce antibiotics, they also overproduce amino acids. J Antibiot. 2018;71(1):26–36.
Wendisch VF, Bott M, Eikmanns BJ. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr Opin Microbiol. 2006;9(3):268–74.
Peifer S, Barduhn T, Zimmet S, Volmer DA, Heinzle E, Schneider K. Metabolic engineering of the purine biosynthetic pathway in Corynebacterium glutamicum results in increased intracellular pool sizes of IMP and hypoxanthine. Microb Cell Fact. 2012;11(1):138.
Ferrer L, Elsaraf M, Mindt M, Wendisch VF. l-serine biosensor-controlled fermentative production of l-tryptophan derivatives by Corynebacterium glutamicum. Biology. 2022;11(5):744.
Mindt M, Beyraghdar Kashkooli A, Suarez-Diez M, Ferrer L, Jilg T, Bosch D, Martins dos Santos V, Wendisch VF, Cankar K. Production of indole by Corynebacterium glutamicum microbial cell factories for flavor and fragrance applications. Microb Cell Fact. 2022;21(1):45.
Ma H, Fan X, Cai N, Zhang D, Zhao G, Wang T, Su R, Yuan M, Ma Q, Zhang C, Xu Q, Xie X, Chen N, Li Y. Efficient fermentative production of l-theanine by Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2020;104(1):119–30.
Lim H, Park J, Woo HM. Overexpression of the key enzymes in the methylerythritol 4-phosphate pathway in Corynebacterium glutamicum for improving farnesyl diphosphate-derived terpene production. J Agric Food Chem. 2020;68(39):10780–6.
Kang MK, Eom JH, Kim Y, Um Y, Woo HM. Biosynthesis of pinene from glucose using metabolically-engineered Corynebacterium glutamicum. Biotech Lett. 2014;36(10):2069–77.
Henke NA, Wichmann J, Baier T, Frohwitter J, Lauersen KJ, Risse JM, Peters-Wendisch P, Kruse O, Wendisch VF. Patchoulol production with metabolically engineered Corynebacterium glutamicum. Genes (Basel). 2018;9(4):219.
Henke NA, Wiebe D, Pérez-García F, Peters-Wendisch P, Wendisch VF. Coproduction of cell-bound and secreted value-added compounds: Simultaneous production of carotenoids and amino acids by Corynebacterium glutamicum. Bioresour Technol. 2018;247:722–52.
Binder D, Frohwitter J, Mahr R, Bier C, Grunberger A, Loeschcke A, Peters-Wendisch P, Kohlheyer D, Pietruszka J, Frunzke J, Jaeger KE, Wendisch VF, Drepper T. Light-controlled cell factories: employing photocaged isopropyl-beta-d-thiogalactopyranoside for light-mediated optimization of lac promoter-based gene expression and (+)-valencene biosynthesis in Corynebacterium glutamicum. Appl Environ Microbiol. 2016;82(20):6141–9.
Frohwitter J, Heider SA, Peters-Wendisch P, Beekwilder J, Wendisch VF. Production of the sesquiterpene (+)-valencene by metabolically engineered Corynebacterium glutamicum. J Biotechnol. 2014;191:205–13.
Kim H-S, Choi J-A, Kim B-Y, Ferrer L, Choi J-M, Wendisch VF, Lee J-H. Engineered Corynebacterium glutamicum as the platform for the production of aromatic aldehydes. Front Bioeng Biotechnol. 2022;10:880277.
Luo ZW, Cho JS, Lee SY. Microbial production of methyl anthranilate, a grape flavor compound. Proc Natl Acad Sci USA. 2019;116(22):10749–56.
Henke NA, Heider SA, Peters-Wendisch P, Wendisch VF. Production of the marine carotenoid astaxanthin by metabolically engineered Corynebacterium glutamicum. Mar Drugs. 2016;14(7):124.
Henke NA, Wendisch VF. Improved astaxanthin production with Corynebacterium glutamicum by application of a membrane fusion protein. Mar Drugs. 2019;17(11):621.
Li C, Swofford CA, Rückert C, Chatzivasileiou AO, Ou RW, Opdensteinen P, Luttermann T, Zhou K, Stephanopoulos G, Jones Prather KL, Zhong-Johnson EZL, Liang S, Zheng S, Lin Y, Sinskey AJ. Heterologous production of α-Carotene in Corynebacterium glutamicum using a multi-copy chromosomal integration method. Bioresour Technol. 2021;341: 125782.
Henke NA, Heider SAE, Hannibal S, Wendisch VF, Peters-Wendisch P. Isoprenoid pyrophosphate-dependent transcriptional regulation of carotenogenesis in Corynebacterium glutamicum. Front Microbiol. 2017;8:633.
Yang J, Tian C, Zhang T, Ren C, Zhu Y, Zeng Y, Men Y, Sun Y, Ma Y. Development of food-grade expression system for d-allulose 3-epimerase preparation with tandem isoenzyme genes in Corynebacterium glutamicum and its application in conversion of cane molasses to D-allulose. Biotechnol Bioeng. 2019;116(4):745–56.
Dhar KS, Wendisch VF, Nampoothiri KM. Engineering of Corynebacterium glutamicum for xylitol production from lignocellulosic pentose sugars. J Biotechnol. 2016;230:63–71.
Burgardt A, Pelosi L, Chehade MH, Wendisch VF, Pierrel F. Rational engineering of non-ubiquinone containing Corynebacterium glutamicum for enhanced coenzyme Q(10) production. Metabolites. 2022;12(5):428.
Weixler D, Berghoff M, Ovchinnikov KV, Reich S, Goldbeck O, Seibold GM, Wittmann C, Bar NS, Eikmanns BJ, Diep DB, Riedel CU. Recombinant production of the lantibiotic nisin using Corynebacterium glutamicum in a two-step process. Microb Cell Fact. 2022;21(1):11.
Sano C. History of glutamate production. Am J Clin Nutr. 2009;90(3):728S-732S.
Ledesma-Amaro R, Jiménez A, Santos MA, Revuelta JL. Biotechnological production of feed nucleotides by microbial strain improvement. Process Biochem. 2013;48(9):1263–70.
Walter T, Al Medani N, Burgardt A, Cankar K, Ferrer L, Kerbs A, Lee J-H, Mindt M, Risse JM, Wendisch VF. Fermentative N-methylanthranilate production by engineered Corynebacterium glutamicum. Microorganisms. 2020;8(6):866.
Walter T, Veldmann KH, Götker S, Busche T, Rückert C, Kashkooli AB, Paulus J, Cankar K, Wendisch VF. Physiological response of Corynebacterium glutamicum to indole. Microorganisms. 2020;8(12):1945.
Ferrer L, Mindt M, Suarez-Diez M, Jilg T, Zagorščak M, Lee JH, Gruden K, Wendisch VF, Cankar K. Fermentative indole production via bacterial tryptophan synthase alpha subunit and plant indole-3-glycerol phosphate lyase enzymes. J Agric Food Chem. 2022;70(18):5634–45.
Benninghaus L, Walter T, Mindt M, Risse JM, Wendisch VF. Metabolic engineering of Pseudomonas putida for fermentative production of l-theanine. J Agric Food Chem. 2021;69(34):9849–58.
Hagihara R, Ohno S, Hayashi M, Tabata K, Endo H, Kelly RM. Production of L-theanine by Escherichia coli in the absence of supplemental ethylamine. Appl Environ Microbiol. 2021;87(11):e00031-e121.
Heider SA, Peters-Wendisch P, Beekwilder J, Wendisch VF. IdsA is the major geranylgeranyl pyrophosphate synthase involved in carotenogenesis in Corynebacterium glutamicum. FEBS J. 2014;281(21):4906–20.
Heider SA, Peters-Wendisch P, Netzer R, Stafnes M, Brautaset T, Wendisch VF. Production and glucosylation of C50 and C40 carotenoids by metabolically engineered Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2014;98(3):1223–35.
Heider SA, Wolf N, Hofemeier A, Peters-Wendisch P, Wendisch VF. Optimization of the IPP precursor supply for the production of lycopene, decaprenoxanthin and astaxanthin by Corynebacterium glutamicum. Front Bioeng Biotechnol. 2014;2:28.
Martău GA, Călinoiu L-F, Vodnar DC. Bio-vanillin: towards a sustainable industrial production. Trends Food Sci Technol. 2021;109:579–92.
Kuivanen J, Kannisto M, Mojzita D, Rischer H, Toivari M, Jäntti J. Engineering of Saccharomyces cerevisiae for anthranilate and methyl anthranilate production. Microb Cell Fact. 2021;20(1):34.
Heider SA, Peters-Wendisch P, Wendisch VF. Carotenoid biosynthesis and overproduction in Corynebacterium glutamicum. BMC Microbiol. 2012;12:198.
Krubasik P, Kobayashi M, Sandmann G. Expression and functional analysis of a gene cluster involved in the synthesis of decaprenoxanthin reveals the mechanisms for C50 carotenoid formation. Eur J Biochem. 2001;268(13):3702–8.
Henke NA, Austermeier S, Grothaus IL, Götker S, Persicke M, Peters-Wendisch P, Wendisch VF. Corynebacterium glutamicum CrtR and its orthologs in actinobacteria: conserved function and application as genetically encoded biosensor for detection of geranylgeranyl pyrophosphate. Int J Mol Sci. 2020;21(15):5482.
Marketsandmarkets (2020). Carotenoids Market by Type (Astaxanthin, Beta-Carotene, Lutein, Lycopene, Canthaxanthin, and Zeaxanthin), Application (Feed, Food & Beverages, Dietary Supplements, Cosmetics, and Pharmaceuticals), Source, Formulation, and Region - Global Forecast to 2026.
Banerjee A, Sharkey TD. Methylerythritol 4-phosphate (MEP) pathway metabolic regulation. Nat Prod Rep. 2014;31(8):1043–55.
Banerjee A, Wu Y, Banerjee R, Li Y, Yan H, Sharkey TD. Feedback inhibition of deoxy-D-xylulose-5-phosphate synthase regulates the methylerythritol 4-phosphate pathway. J Biol Chem. 2013;288(23):16926–36.
Frank A, Groll M. The methylerythritol phosphate pathway to isoprenoids. Chem Rev. 2017;117(8):5675–703.
Hornero-Mendez D, Britton G. Involvement of NADPH in the cyclization reaction of carotenoid biosynthesis. FEBS Lett. 2002;515(1–3):133–6.
Schiweck H, Bär A, Vogel R, Schwarz E, Kunz M. Sugar Alcohols. In: Ullmann’s encyclopedia of industrial chemistry. Weinheim: Verlag Chemie; 1994.
Jeong S-H. Advanced whole-cell conversion for D-allulose production using an engineered Corynebacterium glutamicum. Biotechnol Bioprocess Eng. 2022;27:276–328.
Park C-S, Kim T, Hong S-H, Shin K-C, Kim K-R, Oh D-K. D-allulose production from D-fructose by permeabilized recombinant cells of Corynebacterium glutamicum cells expressing D-Allulose 3-Epimerase Flavonifractor plautii. PLoS ONE. 2016;11(7): e0160044.
Cleto S, Jensen JV, Wendisch VF, Lu TK. Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi). ACS Synth Biol. 2016;5(5):375–85.
Park J, Yu BJ, Choi J-I, Woo HM. Heterologous production of squalene from glucose in engineered Corynebacterium glutamicum using multiplex CRISPR interference and high-throughput fermentation. J Agric Food Chem. 2019;67(1):308–19.
Pierrel F, Burgardt A, Lee J-H, Pelosi L, Wendisch VF. Recent advances in the metabolic pathways and microbial production of coenzyme Q. World J Microbiol Biotechnol. 2022;38(4):58.
Burgardt A, Moustafa A, Persicke M, Sproß J, Patschkowski T, Risse JM, Peters-Wendisch P, Lee JH, Wendisch VF. Coenzyme Q(10) biosynthesis established in the non-ubiquinone containing Corynebacterium glutamicum by metabolic engineering. Front Bioeng Biotechnol. 2021;9: 650961.
Allison GE, Klaenhammer TR. Functional analysis of the gene encoding immunity to lactacin F, lafI, and its use as a Lactobacillus-specific, food-grade genetic marker. Appl Environ Microbiol. 1996;62(12):4450–60.
Goldbeck O, Desef DN, Ovchinnikov KV, Perez-Garcia F, Christmann J, Sinner P, Crauwels P, Weixler D, Cao P, Becker J, Kohlstedt M, Kager J, Eikmanns BJ, Seibold GM, Herwig C, Wittmann C, Bar NS, Diep DB, Riedel CU. Establishing recombinant production of pediocin PA-1 in Corynebacterium glutamicum. Metab Eng. 2021;68:34–45.
Mindt M, Heuser M, Wendisch VF. Xylose as preferred substrate for sarcosine production by recombinant Corynebacterium glutamicum. Bioresour Technol. 2019;281:135–42.
Jayaraman K, Trachtmann N, Sprenger GA, Gohlke H. Protein engineering for feedback resistance in 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase. Appl Microbiol Biotechnol. 2022;106:6505–17.
Han S-S, Kyeong H-H, Choi JM, Sohn Y-K, Lee J-H, Kim H-S. Engineering of the conformational dynamics of an enzyme for relieving the product inhibition. ACS Catal. 2016;6(12):8440–5.
Soler J, Gergel S, Klaus C, Hammer SC, Garcia-Borràs M. Enzymatic control over reactive intermediates enables direct oxidation of alkenes to carbonyls by a P450 iron-oxo species. J Am Chem Soc. 2022;144(35):15954–68.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–9.
Wittmann BJ, Johnston KE, Wu Z, Arnold FH. Advances in machine learning for directed evolution. Curr Opin Struct Biol. 2021;69:11–8.
Miller DC, Athavale SV, Arnold FH. Combining chemistry and protein engineering for new-to-nature biocatalysis. Nat Synth. 2022;1(1):18–23.
Schultenkämper K, Brito LF, Wendisch VF. Impact of CRISPR interference on strain development in biotechnology. Biotechnol Appl Biochem. 2020;67(1):7–21.
Wang Y, Zhao D, Sun L, Wang J, Fan L, Cheng G, Zhang Z, Ni X, Feng J, Wang M, Zheng P, Bi C, Zhang X, Sun J. Engineering of the translesion DNA synthesis pathway enables controllable C-to-G and C-to-A base editing in Corynebacterium glutamicum. ACS Synth Biol. 2022. https://doi.org/10.1021/acssynbio.2c00265.
Wang Y, Cheng H, Liu Y, Liu Y, Wen X, Zhang K, Ni X, Gao N, Fan L, Zhang Z, Liu J, Chen J, Wang L, Guo Y, Zheng P, Wang M, Sun J, Ma Y. In-situ generation of large numbers of genetic combinations for metabolic reprogramming via CRISPR-guided base editing. Nat Commun. 2021;12(1):678.
Binder S, Schendzielorz G, Stabler N, Krumbach K, Hoffmann K, Bott M, Eggeling L. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol. 2012;13(5):R40.
Kortmann M, Mack C, Baumgart M, Bott M. Pyruvate carboxylase variants enabling improved lysine production from glucose identified by biosensor-based high-throughput fluorescence-activated cell sorting screening. ACS Synth Biol. 2019;8(2):274–81.
Dixon AS, Schwinn MK, Hall MP, Zimmerman K, Otto P, Lubben TH, Butler BL, Binkowski BF, Machleidt T, Kirkland TA, Wood MG, Eggers CT, Encell LP, Wood KV. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem Biol. 2016;11(2):400–8.
Sgobba E, Wendisch VF. Synthetic microbial consortia for small molecule production. Curr Opin Biotechnol. 2020;62:72–9.
Acknowledgements
The authors would like to thank members of their groups for discussions.
Funding
Open Access funding enabled and organized by Projekt DEAL. Support in the framework of the ERA CoBioTech project INDIE (European Union’s Horizon 2020 research and innovation programme under grant agreement No. 722361) with national funding is acknowledged by KC (Dutch research council (NWO) grant number 053.80.732) and VFW (Renewable Resources Scheme (FNR) of the Federal Ministry of Food and Agriculture, Germany, grant number 22023517). NAH acknowledges funding by BMBF project KaroTec (grant number:03VP09460).
Author information
Authors and Affiliations
Contributions
KC, NAH, VFW: conceptualization and methodology; KC, NAH, VFW: literature research and manuscript writing; KC, NAH, VFW: manuscript revision; all the authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
All authors agree with their participation in this paper.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cankar, K., Henke, N.A. & Wendisch, V.F. Functional food additives/ingredients production by engineered Corynebacterium glutamicum. Syst Microbiol and Biomanuf 3, 110–121 (2023). https://doi.org/10.1007/s43393-022-00141-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-022-00141-4