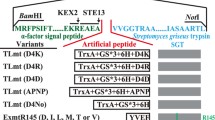
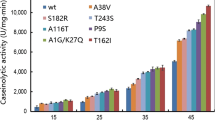
Abstract
Streptomyces griseus trypsin (SGT) is a bacteria-sourced trypsin that could be potentially applied to industrial insulin productions. However, SGT produced by microbial hosts displayed low catalytic efficiency and undesired preference to lysine residue. In this study, by engineering the α signal peptide in Pichia pastoris, we increased the SGT amidase activity to 67.91 U mL−1 in shake flask cultures. Afterwards, we engineered SGT by evolution-guided mutagenesis and obtained three variants A45S, V177I and E180M with increased catalytic efficiencies. On this basis, we performed iterative combinatorial mutagenesis and constructed a mutant A45S/V177I/E180M which the amidase activity reached 98 U mL−1 in shake flasks and 2506 U mL−1 in 3-L fed-batch cultures. Moreover, single mutation T190 to S190 increased the substrate catalytic preference of R to K and the R/K value was improved to 7.5, which was 2 times better than the animal-sourced trypsin.







Similar content being viewed by others
Availability of data and materials
All data generated or analyzed during this study are included in this article and e-supplementary data for this work can be found in e-version of this paper online.
References
Carter P, Wells JA. Dissecting the catalytic triad of a serine protease. Nature. 1988;332(6164):564–8. https://doi.org/10.1038/332564a0.
Kraut J. Serine proteases: structure and mechanism of catalysis. Annu Rev Biochem. 1977;46:331–58. https://doi.org/10.1146/annurev.bi.46.070177.001555.
Williams JA. Trypsin. In: Johnson LR, editor. Encyclopedia of gastroenterology. New York: Elsevier; 2004. p. 533–4.
Mao Y, Krischke M, Hengst C, Kulozik U. Comparison of the influence of pH on the selectivity of free and immobilized trypsin for β-lactoglobulin hydrolysis. Food Chem. 2018;253:194–202. https://doi.org/10.1016/j.foodchem.2018.01.151.
Ketnawa S, Benjakul S, Martínez-Alvarez O, Rawdkuen S. Fish skin gelatin hydrolysates produced by visceral peptidase and bovine trypsin: bioactivity and stability. Food Chem. 2017;215:383–90. https://doi.org/10.1016/j.foodchem.2016.07.145.
Gudmundsdóttir Á, Hilmarsson H, Stefansson B. Potential use of atlantic cod trypsin in biomedicine. Biomed Res Int. 2013;2013: 749078. https://doi.org/10.1155/2013/749078.
Liu H, Zhou X, Tian S, Hao X, You J, Zhang Y. Two-step transpeptidation of the insulin precursor expressed in Pichia pastoris to insulin ester via trypsin-catalyzed cleavage and coupling. Biotechnol Appl Biochem. 2014;61(4):408–17. https://doi.org/10.1002/bab.1186.
Lombardi J, Woitovich Valetti N, Picó G, Boeris V. Obtainment of a highly concentrated pancreatic serine proteases extract from bovine pancreas by precipitation with polyacrylate. Sep Purif Technol. 2013;116:170–4. https://doi.org/10.1016/j.seppur.2013.05.047.
Grishina Z, Ostrowska E, Halangk W, Sahin-Tóth M, Reiser G. Activity of recombinant trypsin isoforms on human proteinase-activated receptors (PAR): mesotrypsin cannot activate epithelial PAR-1, -2, but weakly activates brain PAR-1. Br J Pharmacol. 2005;146(7):990–9. https://doi.org/10.1038/sj.bjp.0706410.
EFSA Panel on Food Contact Materials E, Aids P, Lambré C, Barat Baviera JM, Bolognesi C, Cocconcelli PS, et al. Safety evaluation of the food enzyme trypsin from porcine pancreas. EFSA J. 2022;20(1):e07008. https://doi.org/10.2903/j.efsa.2022.7008.
Zhang Y, Liang Q, Zhang C, Zhang J, Du G, Kang Z. Improving production of Streptomyces griseus trypsin for enzymatic processing of insulin precursor. Microb Cell Fact. 2020;19(1):88. https://doi.org/10.1186/s12934-020-01338-9.
Walmsley SJ, Rudnick PA, Liang Y, Dong Q, Stein SE, Nesvizhskii AI. Comprehensive analysis of protein digestion using six trypsins reveals the origin of trypsin as a significant source of variability in proteomics. J Proteome Res. 2013;12(12):5666–80. https://doi.org/10.1021/pr400611h.
Page MJ, Wong S-L, Hewitt J, Strynadka NCJ, MacGillivray RTA. Engineering the primary substrate specificity of Streptomyces griseus trypsin. Biochemistry. 2003;42(30):9060–6. https://doi.org/10.1021/bi0344230.
Oh EA, Kim M-S, Chi W-J, Kim J-H, Hong S-K. Characterization of the sgtR1 and sgtR2 genes and their role in regulating expression of the sprT gene encoding Streptomyces griseus trypsin. FEMS Microbiol Lett. 2007;276(1):75–82. https://doi.org/10.1111/j.1574-6968.2007.00907.x.
Ling Z, Ma T, Li J, Du G, Kang Z, Chen J. Functional expression of trypsin from Streptomyces griseus by Pichia pastoris. J Ind Microbiol Biotechnol. 2012;39(11):1651–62. https://doi.org/10.1007/s10295-012-1172-3.
Ling Z, Ma T, Li J, Du G, Kang Z, Chen J. Functional expression of trypsin from Streptomyces griseus by Pichia pastoris. J Ind Microbiol Biotechnol. 2012;39:1651–62. https://doi.org/10.1007/s10295-012-1172-3.
Ling Z, Liu Y, Teng S, Kang Z, Zhang J, Chen J, et al. Rational design of a novel propeptide for improving active production of Streptomyces griseus Trypsin in Pichia pastoris. Appl Environ Microbiol. 2013;79(12):3851–5. https://doi.org/10.1128/AEM.00376-13.
Zhang Y, Ling Z, Du G, Chen J, Kang Z. Improved production of active Streptomyces griseus trypsin with a novel auto-catalyzed strategy. Sci Rep. 2016;6(1):23158. https://doi.org/10.1038/srep23158.
Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38(7):3022–7. https://doi.org/10.1093/molbev/msab120.
Wang Z, Wang Y, Zhang D, Li J, Hua Z, Du G, et al. Enhancement of cell viability and alkaline polygalacturonate lyase production by sorbitol co-feeding with methanol in Pichia pastoris fermentation. Biores Technol. 2010;101(4):1318–23. https://doi.org/10.1016/j.biortech.2009.09.025.
Zhang Y, Huang H, Yao X, Du G, Chen J, Kang Z. High-yield secretory production of stable, active trypsin through engineering of the N-terminal peptide and self-degradation sites in Pichia pastoris. Bioresour Technol. 2018;247:81–7. https://doi.org/10.1016/j.biortech.2017.08.006.
Ben Azoun S, Belhaj AE, Gongrich R, Gasser B, Kallel H. Molecular optimization of rabies virus glycoprotein expression in Pichia pastoris. Microb Biotechnol. 2016;9(3):355–68. https://doi.org/10.1111/1751-7915.12350.
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–303. https://doi.org/10.1093/nar/gky427.
Kleffner R, Flatten J, Leaver-Fay A, Baker D, Siegel JB, Khatib F, et al. Foldit standalone: a video game-derived protein structure manipulation interface using Rosetta. Bioinformatics. 2017;33(17):2765–7. https://doi.org/10.1093/bioinformatics/btx283.
Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–61. https://doi.org/10.1002/jcc.21334.
Lin-Cereghino GP, Stark CM, Kim D, Chang J, Shaheen N, Poerwanto H, et al. The effect of α-mating factor secretion signal mutations on recombinant protein expression in Pichia pastoris. Gene. 2013;519(2):311–7. https://doi.org/10.1016/j.gene.2013.01.062.
Chahal S, Wei P, Moua P, Park SPJ, Kwon J, Patel A, et al. Structural characterization of the α-mating factor prepro-peptide for secretion of recombinant proteins in Pichia pastoris. Gene. 2017;598:50–62. https://doi.org/10.1016/j.gene.2016.10.040.
Huang C Jr, Damasceno LM, Anderson KA, Zhang S, Old LJ, Batt CA. A proteomic analysis of the Pichia pastoris secretome in methanol-induced cultures. Appl Microbiol Biotechnol. 2011;90(1):235–47. https://doi.org/10.1007/s00253-011-3118-5.
Liang Q, Shi J, Jin X, Du G, Kang Z. Optimization of enterokinase secretion in Pichia pastoris. Sheng Wu Gong Cheng Xue Bao. 2020;36(8):1689–98. https://doi.org/10.13345/j.cjb.190577.
Bae J-H, Sung BH, Seo J-W, Kim CH, Sohn J-H. A novel fusion partner for enhanced secretion of recombinant proteins in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2016;100(24):10453–61. https://doi.org/10.1007/s00253-016-7722-2.
Jiang N, Xu C, Zhang L, Chen J. “Resurrected” human-source urate oxidase with high uricolytic activity and stability. Enzyme Microb Technol. 2021;149: 109852. https://doi.org/10.1016/j.enzmictec.2021.109852.
Stolterfoht H, Steinkellner G, Schwendenwein D, Pavkov-Keller T, Gruber K, Winkler M. Identification of key residues for enzymatic carboxylate reduction. Front Microbiol. 2018;9:250. https://doi.org/10.3389/fmicb.2018.00250.
Spilliaert R, Gudmundsdóttir Á. Atlantic cod trypsin Y—member of a novel trypsin group. Mar Biotechnol. 1999;1(6):598–607. https://doi.org/10.1007/PL00011815.
Evnin L, Vásquez J, Craik C. Substrate specificity of trypsin investigated by using a gentic selection. Proc Natl Acad Sci USA. 1990;87:6659–63. https://doi.org/10.1073/pnas.87.17.6659.
Weiner SJ, Seibel GL, Kollman PA. The nature of enzyme catalysis in trypsin. Proc Natl Acad Sci. 1986;83(3):649. https://doi.org/10.1073/pnas.83.3.649.
Reetz MT, Carballeira JD. Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nat Protoc. 2007;2(4):891–903. https://doi.org/10.1038/nprot.2007.72.
Liao X, Zhao J, Liang S, Jin J, Li C, Xiao R, et al. Enhancing co-translational folding of heterologous protein by deleting non-essential ribosomal proteins in Pichia pastoris. Biotechnol Biofuels. 2019;12:38. https://doi.org/10.1186/s13068-019-1377-z.
Burgard J, Grünwald-Gruber C, Altmann F, Zanghellini J, Valli M, Mattanovich D, et al. The secretome of Pichia pastoris in fed-batch cultivations is largely independent of the carbon source but changes quantitatively over cultivation time. Microb Biotechnol. 2020;13(2):479–94. https://doi.org/10.1111/1751-7915.13499.
Maiolo M, Zhang X, Gil M, Anisimova M. Progressive multiple sequence alignment with indel evolution. BMC Bioinform. 2018;19(1):331. https://doi.org/10.1186/s12859-018-2357-1.
Labas YA, Gurskaya NG, Yanushevich YG, Fradkov AF, Lukyanov KA, Lukyanov SA, et al. Diversity and evolution of the green fluorescent protein family. Proc Natl Acad Sci. 2002;99(7):4256. https://doi.org/10.1073/pnas.062552299.
Acknowledgements
This work was financially supported by the Jiangsu Province Natural Science Fund for Distinguished Young Scholars (BK20200025), a grant from the Key Technologies R & D Program of Jiangsu Province (BE2019630), the China Postdoctoral Science Foundation (2021M691286).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shi, J., Duan, C., Pang, B. et al. Enhanced catalytic efficiency and substrate specificity of Streptomyces griseus trypsin by evolution-guided mutagenesis. Syst Microbiol and Biomanuf 3, 287–297 (2023). https://doi.org/10.1007/s43393-022-00107-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-022-00107-6