Abstract
In mammals, mitofusin 2 (MFN2) is involved in mitochondrial fusion, and suppresses the virus-induced RIG-I-like receptor (RLR) signaling pathway. However, little is known about the function of MFN2 in non-mammalian species. In the present study, we cloned an MFN2 ortholog (LcMFN2) in large yellow croaker (Larimichthys crocea). Phylogenetic analysis showed that MFN2 emerged after the divergence of amphioxus and vertebrates. The protein sequences of MFN2 were well conserved from fish to mammals. LcMFN2 was expressed in all the tissues/organs examined at different levels, and its expression was upregulated in response to poly(I:C) stimulation. Overexpression of LcMFN2 inhibited MAVS-induced type I interferon (IFN) promoter activation and antiviral gene expression. In contrast, knockdown of endogenous LcMFN2 enhanced poly(I:C) induced production of type I IFNs. Additionally, LcMFN2 enhanced K48-linked polyubiquitination of MAVS, promoting its degradation. Also, overexpression of LcMFN2 impaired the cellular antiviral response, as evidenced by the increased expression of viral genes and more severe cytopathic effects (CPE) in cells infected with spring viremia of carp virus (SVCV). These results indicated that LcMFN2 inhibited type I IFN response by degrading MAVS, suggesting its negative regulatory role in cellular antiviral response. Therefore, our study sheds a new light on the regulatory mechanisms of the cellular antiviral response in teleosts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The constant threat of invading pathogens into host cells has driven the evolution of the innate immune system to quickly detect and respond to conserved pathogen-associated molecular patterns (PAMPs) (Buchmann 2014; Kumar et al. 2011; Medzhitov and Janeway 2002; Netea et al. 2004). Cytoplasmic retinoic acid-inducible gene I-like receptors (RLRs) are a type of pattern recognition receptors (PRRs) that sense virus-derived dsRNA and trigger antiviral responses in immune and other cells, such as epithelial and gonad cells (Chen and Hur 2022; Kawai and Akira 2008; Takeuchi and Akira 2009). The RLR family, which belongs to the DExD/H box RNA helicases superfamily, includes three members: RIG-I, melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2). Members of this family sense viral nucleic acids using a C-terminal helicase domain (Bruns and Horvath 2012; Takahasi et al. 2009). Upon viral recognition, RIG-I and MDA5 activate the downstream mitochondrial antiviral-signaling protein (MAVS), a mitochondrial outer membrane protein (Baril et al. 2009; Seth et al. 2005), and promote its oligomerization (Buskiewicz et al. 2016; Onomoto et al. 2021; Wu and Hur 2015). Conversely, LGP2 interacts with RIG-I or MDA5 to control signal transduction (Gong et al. 2022; Satoh et al. 2010; Zhang et al. 2018). Then, MAVS recruits TNF receptor-associated factor 3 (TRAF3) and TANK-binding kinase 1 (TBK1), which phosphorylate and activate interferon regulatory factor 3 (IRF3) and IRF7 (Liu et al. 2015). After being phosphorylated, IRF3 and IRF7 homodimerize or heterodimerize and translocate into the nucleus, where they bind to the interferon-stimulated response element (ISRE) within the IFN promoter, leading to the early production of type I IFN (Dalskov et al. 2020; Ding et al. 2016; Feng et al. 2016; Honda and Taniguchi 2006). The secretion of type I IFNs then activates the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway, resulting in the transcription of hundreds of IFN-stimulated genes (ISGs), including many antiviral effectors, such as myxovirus resistance protein (Mx), virus inhibitor protein, endoplasmic reticulum-related factor, interferon-inducible factor (Viperin), protein kinase R (PKR), and ISG15 (Au-Yeung et al. 2013; Raftery and Stevenson 2017).
Mitochondria, which are the double-membrane organelles commonly referred to as “powerhouses” of eukaryotic cells, maintain a constantly changing morphology through fission and fusion events that require the coordination of a complex group of proteins (Berman et al. 2008; Davey and Clark 1996; Mills et al. 2017; Sabouny and Shutt 2020). Mitochondrial fusion is mediated by outer and inner mitochondrial membrane proteins, including the GTPases MFN1 and MFN2, which are localized on the outer mitochondrial membrane (OMM), and the Opa1, in the inner mitochondrial membrane (IMM) (Chen et al. 2003a; Lee et al. 2004). Conversely, mitochondrial elongation and fission are regulated by the interaction of dynamin-related protein 1 (Drp1) and other OMM proteins, such as MiD51 and Fis1 (Lee et al. 2004; Palmer et al. 2011; Yu et al. 2019).
Growing evidence suggests that mitochondrial fusion is linked to the antiviral response and modulation of RLR signaling (Castanier et al. 2010; Kim et al. 2018). For example, mitochondrial fragmentation reduces virus-induced IRF3 and NF-κB-dependent signal transmissions and impairs host antiviral activity against RNA viruses (Onoguchi et al. 2010). Furthermore, in fusion-deficient mouse embryo fibroblasts lacking both MFN1 and MFN2, virus-induced RLR-dependent production of IFN-β and cytokine interleukin-6 (IL-6) was drastically reduced, resulting in the increased viral replication (Chen et al. 2003a). There are some discrepancies between MFN1 and MFN2. Although deletion of MFN2 in mouse embryonic fibroblast (MEF) cell lines reduces vesicular stomatitis virus (VSV)-induced IFN-β production, manipulating MFN1 expression levels did not yield similar phenotypes (Yasukawa et al. 2009). This suggests that MFN2 functions as a negative regulator of RLR signaling. Also, recent study has revealed that inhibiting MFN2 expression with epigoitrin leads to upregulation of influenza virus-induced MAVS and IFN-β activation (Luo et al. 2019). Additionally, MFN2 has been shown to inhibit the RIG-I/IRF7 signaling pathway, resulting in decreased Seneca Valley virus (SVV)-induced IFN-λ3 production (Deng et al. 2022).
While the importance of MFN proteins in modulating RLR signaling pathways has been extensively studied in mammals, their function in non-mammalian species remains largely unknown. In the current study, we cloned an MFN2 ortholog (LcMFN2) from a large yellow croaker (Larimichthys crocea), which is a commercially important mariculture species in East Asia. The fish suffer from infectious diseases caused by Pseudomonas plecoglossicida, Cryptocaryon irritans, and large yellow croaker iridovirus (Chen et al. 2003b; Li et al. 2020a, b; Nelapati et al. 2012; Yin et al. 2019). The expression patterns of LcMFN2 under normal and poly(I:C)-stimulated-conditions were examined. Moreover, we demonstrated that LcMFN2 plays a negative regulatory role in MAVS-mediated type I IFN signaling. Mechanically, LcMFN2 interacts with LcMAVS and promotes its degradation through the ubiquitin–proteasome pathway. These findings contribute significantly to our understanding of the regulatory mechanisms of the cellular antiviral response in teleosts.
Results
Sequence characterization and phylogenetic analysis
A search was performed in the genomic database (GCF_000972845.2) (Ao et al. 2015) for MFN2, named LcMFN2 in large yellow croaker, using TBLASTN, and its sequence was confirmed by Sanger sequencing. The open-reading frame (ORF) of LcMFN2 was 2277 base pairs (bp) in length, encoding 759 amino acids (aa) with a calculated molecular mass of 86 kDa. Analysis of the genome data indicates that LcMFN2 is located on primary assembly XV, and spans an approximately 11.81 kb chromosome contig. In addition, the coding sequence of LcMFN2 is arranged in 17 exons and 16 introns, with all intron–exon boundaries following the typical GT/AG pattern, similar to its mammalian counterparts (Table 3). Domain analysis revealed that the LcMFN2 protein contains highly conserved functional domains, including an N-terminal GTPase domain, a heptad repeat 1 (HR1) domain, a proline-rich (PR) domain, a transmembrane (TM) domain, and a C-terminal HR2 domain (Supplementary Fig. S1A and B). The protein structure of LcMFN2 is comparable to that of human MFN2, with average root-mean-square deviations (RMSD) of 4.701 (rigid alignment) and 2.41 (flexible alignment), respectively (Supplementary Fig. S1C). However, the amino acid sequence of LcMFN2 shows marked differences compared to that of vertebrate MFN1 and non-chordate metazoa MFN2/MFN2-like, particularly at the C-terminus, with a lowest similarity of < 60% (Fig. 1A).
Sequence comparison and phylogenetic relationships of MFNs from different species. A Sequence comparison of MFN coding regions among various species, encompassing non-chordate metazoa and chordates. The color green denotes 100% similarity, yellow–green denotes 80–100% similarity, yellow denotes 60–80% similarity, and gray denotes less than 60% similarity. B Phylogenetic relationships among MFN sequences. The phylogenetic tree was inferred using the neighbor-joining method in MEGA11 with the Jones–Taylor–Thornton (JTT) model, and 10,000 bootstrap replicates. The branches were labeled with the bootstrap values
Phylogenetic analysis indicates that MFN genes from non-chordate metazoans formed the basal branch, suggesting that these genes encode ancestral MFN proteins (Fig. 1B). Nevertheless, these genes were incorrectly annotated as MFN2/MFN2-like in the NCBI (National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/) database. MFN1 and MFN2 from urochordates (Styela clava), cephalochordates (amphioxus), and vertebrates (cartilaginous fish, bony fish, amphibians, reptiles, birds, and mammals) formed the sister group of the MFN genes from non-chordate metazoans. Two sequences from B. floridae and S. clave, annotated as MFN2-like, were found cluster outside the vertebrate MFN genes.
The time-calibrated phylogenetic tree shows that the chordate MFN genes diverged from the ancestral MFN genes ~ 522 million years ago (MYA), whereas vertebrate MFN genes emerged ~ 394 MYA (Fig. 2). Vertebrate MFN1 and MFN2 diverged at ~ 356 MYA and 310 MYA, respectively (Fig. 2). Taking together, our analysis suggested that MFN2 emerged after the divergence of amphioxus and vertebrates, and its amino acid sequence has remained highly conserved from fish to mammals, indicating that its functional role has been maintained across a wide range of species over a long period of evolutionary time.
Expression analysis of the LcMFN2 gene
Gene expression analysis showed that LcMFN2 was expressed in all examined tissues and organs with different expression levels. LcMFN2 expression level was highest in the liver, followed by the gill, skin, heart, muscle, brain, stomach, blood, and spleen, with the lowest level detected in the intestine and head kidney (Fig. 3A).
The expression pattern of the LcMFN2 gene. A The expression levels of LcMFN2 in tissues/organs were measured and normalized to the expression of the housekeeping gene β-actin, and the results were reported in arbitrary units. The lowest level of LcMFN2 expression was observed in the head kidney, which was defined as 1. The abbreviations are L, Liver; G, gill; SK, skin; HE, heart; MU, muscle; BR, brain; ST; stomach; BL, blood; SP, spleen; IN, intestine; HK, head kidney. B–D The relative mRNA level of LcMFN2 was measured at 3, 6, 12, 24, and 48 h post-stimulation/infection and compared to a control group treated with PBS. The expression of LcMFN2 in each tissue was normalized to that of β-actin and presented as fold changes relative to control fish injected with the same volume of PBS. The data are presented as mean ± SD (N = 3), with asterisks (*) denoting a significant difference (*P < 0.05, **P < 0.01, ***P < 0.001) above each bar
Challenge experiments were carried out to assess whether LcMFN2 could be induced by poly(I:C) stimulation. As shown in Fig. 3B–D, poly(I:C) stimulation led to significantly increased levels of LcMFN2 expression in the head kidney, spleen, and intestine after 6 h post-infection (hpi), with fold increases of 1.63, 1.99, and 1.70, respectively, compared to the control group. By 12 hpi, LcMFN2 expression was consistently higher in all of the tested tissues/organs, with fold increases of 5.66 (head kidney), 7.69 (spleen), and 2.49 (intestine) compared to the control group. At 24 hpi, LcMFN2 expression was persistently elevated in all the examined tissues/organs, being 15.02 (head kidney), 48.09 (spleen), and 9.47 (intestine)-fold higher than in the control group, respectively. At 48 hpi, significant increases were also observed in all the tested tissues/organs, with the head kidney being 20.73-fold, the spleen being 26.34-fold, and the intestine being 6.67-fold higher than the control, respectively.
LcMFN2 inhibits LcMAVS-induced type I IFN signaling
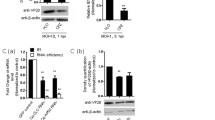
To investigate whether MFN2 plays a role in poly I:C-induced IFN production, we employed siRNA to silence LcMFN2. Our results showed that MFN2 expression was reduced by ~ 63% (siMFN2-2) in large yellow croaker head kidney (LYCK) cells (Wang et al. 2014) transfected with siRNA oligos compared to control cells (Fig. 4A). Subsequently, qPCR analysis demonstrated that MFN2 knockdown enhanced poly(I:C)-induced expression of IFNi (Fig. 4B) and IFNd (Fig. 4C). Previous studies have shown the regulatory role of mammalian MFN2 in the antiviral responses elicited by MAVS (Yasukawa et al. 2009). Therefore, we investigated how MFN2 regulates the activation of MAVS-induced IFN promoters in fish. Luciferase activity assay was performed, employing LcIFNi and LcIFNd as reporter genes, with LcIFNi being classified as a group II fish IFN and LcIFNd being classified as a group I fish IFN (Chen et al. 2021, 2022). As shown in Fig. 4D, E, both LcMAVS and LcTBK1 significantly activated LcIFN promoters. However, co-transfection with LcMFN2 decreased LcMAVS-induced activation of LcIFN promoters, whereas LcTBK1 had no effect. Previous studies have shown that overexpressing fish MAVS may boost the antiviral activity of epithelioma papulosum cyprini (EPC) cells (Chen et al. 2017; Zhou et al. 2015). Thus, we investigated the impact of LcMFN2 on MAVS-induced antiviral gene expression. Our results showed that overexpression of LcMFN2 significantly inhibited LcMAVS-induced EPC antiviral gene expression (Fig. 4F). Then, we tested the interaction between LcMFN2 and LcMAVS. Human embryonic kidney 293T (HEK293T) cells were co-transfected with FLAG-tagged LcMFN2 and His-tagged LcMAVS. The Co-IP experiment revealed that LcMFN2 could interact with LcMAVS, suggesting that this inhibitory impact may result from protein–protein interaction (Fig. 4G). Taken together, these findings suggest strongly that MFN2 serves as a negative regulator of RLRs signaling molecules, particularly MAVS-induced IFN production.
LcMFN2 interacts with LcMAVS and inhibits LcMAVS-mediated signaling. A Effects of RNA interference on the expression of endogenous MFN2. LYCK cells were transfected with three different sequences of LcMFN2 siRNA or control siRNA (siNC) for 24 h, and then collected for qPCR. B, C Effects of siRNA-treated LYCK cells in the poly(I:C)-induced IFNs transcription. Cells were transfected with 100 nM si-MFN 2-2 for 24 h, and then transfected with poly(I:C) (1 μg/mL) for 24 h. The transcription of IFNi (B) and IFNd (C) was detected by qPCR. D, E Overexpression of LcMFN2 selectively inhibits LcMAVS, but not LcTBK1, mediated IFN promoter activation. EPC cells were co-transfected with 400 ng LcIFN promoter reporters, LcIFNipro-luc (D) and LcIFNdpro-luc (E), and plasmids expressing His-tagged LcMFN2 (250 ng) or LcTBK1 (250 ng), in combination with Flag-tagged LcMAVS or empty vector pcDNA™3.1/myc-His at 250 ng each. Luciferase activities were measured 48 h after transfection. F Overexpression of LcMFN2 suppresses LcMAVS-induced expression of antiviral genes in EPC cells. Cells were transfected with LcMAVS-Flag (2 μg) along with His-LcMFN2 (2 μg) or empty vector (2 μg) for 24 h. The expression levels of Mx1, Viperin, IFNa, PKR, β2-microglobulin (β2M), and PKR were measured by qRT-PCR and normalized to β-actin. The results were standardized to 1 in cells transfected with the empty vector. Error bars indicate the standard deviation of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. G Analysis of the association between LcMAVS and LcMFN2 by immunoprecipitation. HEK293T cells were cultured in 10-cm dishes and co-transfected with 5 μg of LcMAVS-Flag and 5 μg of LcMFN2-His or an empty vector. Following 24 h of transfection, cells were immunoprecipitated with anti-FLAG antibody and subjected to western blotting using the indicated antibodies
LcMFN2 promotes the degradation of LcMAVS by enhancing K48-linked ubiquitination
Based on our previous finding that LcMFN2 interacts with LcMAVS and inhibits type I IFN production, we speculated that LcMFN2 could degrade LcMAVS. To investigate the role of LcMFN2 in regulating LcMAVS protein expression, we co-transfected EPC cells with LcMFN2 and LcMAVS expression plasmids and examined LcMAVS protein expression using immunoblot assays with anti-His-tag antibody. Our results showed that co-transfection with LcMFN2 promoted the degradation of LcMAVS in a dose-dependent manner (Fig. 5A). To further confirm the effect of LcMFN2 on the protein stability of LcMAVS, we conducted a cycloheximide (CHX) assay to measure the degradation rate of LcMAVS. As shown in Fig. 5B, the overexpression of LcMFN2 expedited the degradation of LcMAVS in the presence of CHX, resulting in a reduction of LcMAVS's half-life. Eukaryotic cells mainly undergo protein degradation through two pathways, the autophagolysosome pathway and the ubiquitin–proteasome pathway. Therefore, we determined the degradation pathway of LcMAVS by using specific inhibitors, MG132 for the ubiquitin–proteasome system and NH4CI for the autophagosome–lysosome system (Guo et al. 2016; D’Eletto et al. 2009). As illustrated in Fig. 5C, LcMFN2-mediated degradation of LcMAVS could be suppressed by MG132 but not by NH4CI, demonstrating that overexpression of LcMFN2 results in the degradation of LcMAVS via the ubiquitin–proteasome pathway. An additional discovery supporting this conclusion is that overexpression of LcMFN2 enhanced the polyubiquitination level of LcMAVS (Fig. 5D). To investigate which type of Ub chain structure was affecting LcMAVS, we utilized two mutants: Ub-K48 and Ub-K63, in which six of the seven lysine residues in the Ub molecule were replaced with arginine, leaving only one lysine residue intact. Our results showed a significant increase in K48-linked ubiquitination of LcMAVS under the regulation of LcMFN2, with K63-linked Ub chains remaining unchanged (Fig. 5E, F). These data suggest strongly that LcMFN2 specifically induces K48-ubiquitination of LcMAVS, ultimately leading to its proteasomal degradation, which is consistent with previous research that has underscored the importance of K48-ubiquitin chains in directing protein substrates toward proteasomal degradation.
LcMFN2 degrades LcMAVS through the ubiquitin–proteasome pathway. A LcMFN2 mediated the degradation of LcMAVS. EPC cells were transfected with 2 μg LcMAVS-Flag together with 2 μg empty vector or LcMAVS-His for 24 h. The protein level of LcMAVS was detected by western blotting. B CHX chase analysis of exogenous LcMAVS in LcMFN2 transfected cells or control cells. EPC cells were treated with 100 μg/mL CHX for the specified time period, and the expression levels of LcMAVS were quantified. C The protein degradation of LcMAVS was prevented by treatment with MG132. EPC cells were transfected with 2 μg of Flag-LcMAVS and 2 μg of an empty vector or His-LcMAVS for 24 h, and then treated with either MG132 (20 μmol/L) or NH4Cl (20 mmol/L) for 6 h before immunoblot analysis. The western blotting signals were quantified by analyzing the densitometric grayscale using ImageJ software. D–F Overexpression of LcMFN2 facilitates the K48-linked ubiquitination of LcMAVS. HEK293T cells cultured in 10-cm dishes were co-transfected with LcMAVS-Flag and LcMFN2-His in the presence of Ub-HA-WT (D), Ub-HA-K48 (E) or Ub-HA-K63 (F). After 24 h of transfection, the cells were immunoprecipitated with an anti-FLAG antibody, followed by western blotting using the appropriate antibodies
Overexpression of LcMFN2 impairs the cellular antiviral response
To investigate the impact of LcMFN2 on the cellular antiviral response, EPC cells were transfected with an FLAG-tagged LcMFN2 expression plasmid or a CMV-FLAG empty vector as a control and subsequently infected with SVCV. At 72 hpi, control cells showed a moderate CPE caused by SVCV, whereas cells transfected with LcMFN2 expression plasmid exhibited a severe CPE (Fig. 6A, B). The effect of LcMFN2 overexpression on the expression of viral genes was also examined (Fig. 6C), and the results showed that viral glycoprotein (G) was increased by 4.38-fold; RNA-dependent RNA polymerase (L) was increased by 7.01-fold; matrix protein (M) was increased by 5.53-fold; nucleo-protein (N) was increased by 4.55-fold; and phosphoprotein (P) was increased by 4.04-fold in cells expressing LcMFN2.
Overexpression of LcMFN2 impairs the cellular antiviral response. A EPC cells were transfected with LcMFN2 expression plasmid and infected with SVCV. After 72 h post-infection, the CPE was observed microscopically. B The CPE was detected by staining the cells with 1% crystal violet, and the cell viability was assessed by measuring the optical density at a wavelength of 562 nm. C The impact of LcMFN2 overexpression on the expression of SVCV genes. EPC cells were transfected with LcMFN2-His for 24 h, and then infected with SVCV (1000 TCID50) for another 24 h. The mRNA levels of SVCV genes were subsequently quantified using real-time PCR. The error bars represent the standard error of the mean ± SD of three independent experiments. *P < 0.05
Discussion
Recent evidence highlights the mitochondrial fusion protein MFN2 in regulating host immune responses, especially in the virus-induced RLR signaling pathway (Eisenächer and Krug 2012; Tan et al. 2022; Xu et al. 2020). In this study, we observed an increase in the expression level of LcMFN2 following poly(I:C) stimulation, suggesting its potential involvement in the regulation of antiviral immune responses.
Since MFN2 gene identification is scarce in non-mammalian species, we analyzed the sequence of MFN2 in the large yellow croaker. Our analysis showed that the protein structure of the large yellow croaker MFN2 is similar to that of human MFN2, suggesting that fish MFN2 may have similar physiological functions to mammalian MFN2. It may be inferred that the well-conserved protein sequences of MFN2 from fish to mammals are subject to purifying selection, which eliminates deleterious mutations over time and reduces the genetic diversity (Cvijović et al. 2018). However, the C-terminal region of MFN2 in vertebrates is highly divergent from MFN1 in vertebrates and MFN proteins in early metazoans. The accumulation of non-synonymous mutations in the C-terminus may have contributed to the functional diversification of MFNs (Hughes 1994). Interestingly, our investigation has revealed that the MFN genes may have undergone two rounds of duplication. In early chordates, such as tunicates (Styela clava) and cyclostomes (lampreys), only one copy of the MFN gene was found, whereas most basic metazoans had two copies, indicating that the first MFN gene duplication occurred before the emergence of chordates. Furthermore, according to the results of phylogenetic analysis, the ancestral MFN genes constituted the basal branch, with MFN1 and MFN2 clustering together in vertebrates. Thus, it is likely that the subsequent duplication event occurred after the amphioxus–vertebrate split, leading to the divergence of MFN1 and MFN2, which underwent subfunctionalization or neofunctionalization (Chen et al. 2020; Naon and Scorrano 2014).
The current study demonstrated that LcMFN2 participates in regulating the RLR-induced type I IFN signaling by negatively regulating the MAVS-mediated signaling transduction, suggesting that the function of LcMFN2 is similar to its mammalian counterparts (Deng et al. 2022; Yasukawa et al. 2009). Interestingly, LcMFN2 selectively inhibits the promoter activity induced by LcMAVS but has minimal effect on the promoter activity induced by LcTBK1, which may be attributed to the upstream molecular position of LcMFN2 concerning LcTBK1. This observation is consistent with a previous report demonstrating that the knockdown of MFN2 did not affect the expression of NF-κB induced by TRAF6 or the expression of IFN-β induced by TBK1 (Yasukawa et al. 2009). The current study also provides evidence that LcMFN2 could promote the degradation of LcMAVS via the ubiquitin–proteasome pathway in a dose-dependent manner, suggesting that LcMFN2 may function as a fine-tuner of MAVS activity, providing a mechanism for regulating the intensity and duration of antiviral responses. Generally, K63-linked ubiquitination enhances substrate protein stability, whereas K48-linked ubiquitination promotes substrate protein degradation (Mallette and Richard 2012; Newton et al. 2008; Tracz and Bialek 2021). The ubiquitination of MAVS is controlled by a variety of E3 ubiquitin ligases and deubiquitinases. For example, it has been demonstrated that ring finger protein 125 (RNF125) and tripartite motif-containing protein 25 (TRIM25), two E3 ubiquitin ligases, may target MAVS for K48-linked ubiquitination and degradation (Castanier et al. 2012; You et al. 2009). Recently, Hou et al. (2021) reported that deubiquitinase ubiquitin-specific peptidase 18 (USP18) could stabilize MAVS by promoting K63-linked polyubiquitin chains from MAVS, thereby preventing its proteasomal degradation. The limitation of our current study is that we did not identify the individual E3 ubiquitinates responsible for MAVS degradation. Future research should aim to determine the specific ubiquitinates involved in this process.
As degraded MAVS is typically associated with reduced cellular antiviral capability, we finally investigated the influence of LcMFN2 on virus replication. Consistent with the reduced IFN response, overexpression of LcMFN2 resulted in CPE caused by SVCV infection. However, the effect of MFN2 on other viruses and cell types remains unclear. Further research is necessary to determine the generalizability of these findings and gain a comprehensive understanding of the role of the mitochondrial network in regulating antiviral signaling.
In conclusion, the MFN2 gene was cloned and identified in L. crocea, and its expression was upregulated by poly(I:C) treatment. LcMFN2 negatively regulated the MAVS-mediated signaling pathway by degrading LcMAVS through the ubiquitin–proteasome pathway, inhibiting the expression of type I IFNs and ISGs (Fig. 7). These findings provide new insights into the mechanisms governing the cellular antiviral response in teleost.
MFN2 regulates the MAVS-mediated antiviral response in large yellow croaker. Upon recognizing viral PAMPs such as dsRNA, RLRs recruit MAVS to activate TBK1, which phosphorylates and activates IRF3. Then, homodimerized IRF3 translocates to the nucleus, binding to ISRE in the IFN promoter, leading to early production of type I IFN. By binding to MAVS on the mitochondrial outer membrane, MFN2 enhances the ubiquitination level of MAVS, resulting in MAVS degradation and inhibition of its downstream signaling. The new findings in the current study were marked with red lines
Materials and methods
Bioinformatics analysis
The conserved domain structures were annotated by querying against the Conserved Domain Database (CDD) of the NCBI, and manually checked by comparing with the protein domain of human MFN2. Protein structure prediction was performed using Alphafold2, and the model with the highest confidence (pLDDT) was selected for further analysis (Mirdita et al. 2022). Rigid and flexible comparisons of protein structure were carried out with PyMOL software (version 2.5) and FATCAT 2.0 web service (https://fatcat.godziklab.org/), respectively (Li et al. 2020a, b). Multiple sequence alignment was performed using the Geneious alignment plugin in Geneious Prime (version 2022.1.1) (Kearse et al. 2012). The best-fit substitution model for phylogenetic analysis was determined by MEGA11 software based on the aligned sequences. Neighbor-joining trees were constructed using MEGA11 with the Jones–Taylor–Thornton (JTT) model and 10,000 bootstrap replicates (Tamura et al. 2021). The time-based phylogenetic tree was generated using the RelTime-ML method in MEGA11, which is effective in calculating divergence times for duplicated genes. Three calibration constraints were applied, including the estimated divergence time of: (1) Danio rerio and Lepisosteus oculatus between 298.8 and 342.5 MYA, retrieved from the TimeTree5 database (http://timetree.org); (2) Hydra vulgaris and Acropora digitifera between 543.0 and 610.0 MYA; and (3) Podarcis muralis and Gallus gallus between 276.0 and 286.8 MYA. Two MFN genes from Amphimedon queenslandica were used as outgroups. The trees were edited for visualization in Adobe Illustrator CC2019.
Animal, cell lines, and virus
Large yellow croakers weighing 100 g were obtained from a mariculture farm in Ningde, Fujian, China, and acclimatized for one week in aquaria with water at 2% salinity and 22 °C as approved by the College of Life Science at Fujian Agriculture and Forestry University. For challenge studies, 45 fish were divided into three groups of 15 and injected intraperitoneally with either poly(I:C) (Sigma-Aldrich, 0.2 mL/100 g) in phosphate-buffered saline (PBS, pH 7.4) or PBS alone. Tricaine methanesulfonate (MS-222, 100 mg/L, Sigma-Aldrich) was used to anesthetize the fish during sample collection. Tissues/organs, including the head kidney, spleen, and intestine, were collected at 3, 6, 12, 24, and 48 h post-injection and promptly frozen in liquid nitrogen until use.
HEK293T (ATCC®CRL-3216 ™) were cultured in Dulbecco modified Eagle medium (DMEM, Gibco) with 10% fetal bovine serum (FBS) and 2% penicillin–streptomycin (Gibco) in a humidified incubator at 37 °C and 5% CO2. EPC cells (ATCC®CRL-2872 ™) were cultured in medium 199 (M199, Gibco) supplemented with 10% FBS and 2% penicillin–streptomycin (Gibco) in a humidified incubator at 27 °C and 5% CO2. LYCK cells were cultured in L-15 medium (Life Technologies, Carlsbad, CA, USA) with 10% FBS and 2% penicillin–streptomycin (Gibco) at 27 °C. SVCV was gifted from Professor Yong-An Zhang at Huazhong Agricultural University and propagated in EPC cells in M199 containing 2% FBS.
RNA extraction, cDNA synthesis, and qRT-PCR analysis
Total RNA was extracted using Trizol regent (Invitrogen), the manufacturer's protocol. The concentration of total RNA was determined by calculating the absorbance ratio at 260/280 using a Nanodrop Spectrophotometry (Thermo Scientific, USA), and the quality of the RNA was determined by electrophoresis on a 1% agarose gel. To generate the first-strand cDNA for gene amplification and expression analysis, 2 μg of total RNA was reverse-transcribed using the HiScript III first-strand cDNA Synthesis Kit (+ gDNA wiper) (Vazyme Biotech).
Real-time PCR was performed using QuantStudio™ 5 Real-time PCR system (Applied Biosystems). Amplification reactions were carried out in triplicate, and each reaction containing 10 μL of 2 × SYBR qPCR master mix (Vazyme Biotech), 1 μL of the diluted cDNA, 0.4 μL of each primer, and 8.2 μL ddH2O. The Cp value of each sample was computed automatically by software, and relative fold changes were calculated using the 2−ΔΔCt method. The PCR amplification efficiency was calculated using the standard curve method, and optimized to over 95%. Primer specificity was evaluated by examining the melt curves.
Gene clone and plasmid construction
Specific primers for the full-length ORF of LcMFN2 were designed based on the predicted 5′ upstream and 3′ downstream sequences of LcMFN2 (GenBank accession No. XM_019259949.2). The ORF of LcMFN2 was amplified from the spleen cDNA template according to the instructions of Ex-Taq DNA polymerase (Takara). After gel purification, the PCR products were cloned into the pMD19-T vector (Takara) and subsequently transformed into Escherichia coli DH5α. Putative positive clones were sequenced using the vector-specific primers M13F/M13R.
The ORF of LcMFN2 was inserted into the EcoR V/Kpn I site of pcDNA™3.1/myc-His (−) A (Invitrogen) to construct expression plasmid. The LcIFNipro-Luc and LcIFNdpro-Luc reporter constructs have been described previously (Chen et al. 2021; Ding et al. 2016). The expression plasmids of LcMAVS and LcTBK1 were cloned and constructed according to the previous studies (Shen et al. 2016; Zhang et al. 2015). The plasmids pRK5-HA-Ubiquitin (Ub-HA-WT), pRK5-HA-Ubiquitin-K48 (Ub-HA-K48), and pRK5-HA-Ubiquitin-K63 (Ub-HA-K63) were procured from Addgene (Watertown, MA, USA). All plasmids were verified by sequencing.
RNA interference experiments
Small interfering RNAs (siRNAs) targeting LcMFN2 were designed using Invitrogen BLOCK-iT™ RNAi Designer and synthesized by Tsingke Biological Technology Co., Ltd. LYCK cells were seeded in 6-well plates overnight and transfected with 100 nmol/L of either siRNA or negative control (siNC) using Lipofectamine 2000 transfection reagent as per the manufacturer’s recommended protocol. The effects on the expression of the target gene were evaluated using qPCR.
Infection assays
The infection assays were performed as previously described (Guan et al. 2020; Wei et al. 2021). Briefly, EPC cells seeded in 24-well plates (8 × 105 per well) were transfected with 2 μg recombinant plasmids expressing LcMFN2 or empty vector as control using Lipofectamine 2000 (Invitrogen). At 24 h post-transfection, the supernatant medium was changed with fresh serum-free L-15 culture medium, incubated with SVCV (TCID50 1000) for 2 h, and then replaced with fresh L-15 medium containing 2% FBS and 1% methylcellulose (Beyotime). On day 3 post-infection, the surviving cells were fixed with 10% paraformaldehyde, and quantified by measuring the optical density at a wavelength of 562 nm following crystal violet staining.
Luciferase activity assays
EPC cells seeded in 24-well plates (8 × 105 cells/well) overnight were co-transfected with various overexpression plasmids as indicated in figures, together with control vector (pRL-TK, 25 ng) and luciferase reporter plasmids (LcIFNi-pro or LcIFNd-pro, 400 ng). Empty vector DNA was employed to maintain the equivalent amount of transfected DNA. After 48 h of transfection, cells were washed twice with ice-cold PBS, and the lysate was prepared by treating 100 μL of diluted (1 ×) Passive Lysis Buffer for 10 min. The cell lysate was centrifuged, and the luciferase activity was measured in 20 μL of the supernatant using a GloMax Discover (Promega) according to the instructions of the dual-luciferase reporter assay kit (Promega). Each experiment was carried out in triplicate.
Co-immunoprecipitation and immunoblot analysis
HEK293T cells seeded in 10-cm plates (1 × 107 cells/well) were co-transfected with 5 μg FLAG-tagged LcMFN2 and 5 μg His-tagged LcMAVS or empty vector. After 24 h, cells were lysed with prechilled lysis buffer (50 mmol/L Tris HCL, pH 7.4, 150 mmol/L NaCl, 1 mmol/L EDTA, 1% Triton-X-100, and 1 mmol/L PMSF). Proteins were precipitated using BeyoMag™ Anti-Flag Magnetic Beads (Beyotime) at 4 °C for 6 h. Following three times washing with TBS/T, the immunocomplex was eluted with 3 X Flag Peptide (sigma). For immunoblotting, denatured proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and electro-imprinted on 0.2 μmol/L PVDF membrane (Millipore, Bedford, MA) using a micro-trans-Blot cell system (Bio-Rad), and then blocked with Quick block blocking buffer (Beyotime). The membrane was incubated overnight at 4 °C with primary antibodies against FLAG-Tag (MBL, M185-3L, 1:10,000 dilution in TBS/T containing 1% non-fat dry milk) or His-Tag (1:3000 dilution in TBS/T containing 1% non-fat dry milk), followed by washing three times with TBS/T. Then, the membrane was probed with the HRP-labeled goat anti-mouse secondary antibody (Proteintech, SA0001-1, 1:10,000). Signals were detected using an ECL chemiluminescent system captured by a gel imaging system (GE Healthcare).
Statistical analysis
Data were compared using a one-way analysis of variance (One-Way ANOVA) followed by Duncan's multiple post hoc comparison test (SPSS Statistics, Version 17.0). All experiments were repeated three times, and the results were presented as the mean ± standard deviation (SD).
Data availability
Data will be made available on request.
References
Ao JQ, Mu YN, Xiang LX, Fan DD, Feng MJ, Zhang SC, Shi Q, Zhu LY, Li T, Ding Y, Nie L, Li QH, Dong WR, Jiang L, Sun B, Zhang XH, Li MY, Zhang HQ, Xie SB, Zhu YB et al (2015) Genome sequencing of the perciform fish Larimichthys crocea provides insights into molecular and genetic mechanisms of stress adaptation. PLoS Genet 11:e1005118
Au-Yeung N, Mandhana R, Horvath CM (2013) Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT 2:e23931
Baril M, Racine ME, Penin F, Lamarre D (2009) MAVS dimer is a crucial signaling component of innate immunity and the target of hepatitis C virus NS3/4A protease. J Virol 83:1299–1311
Berman SB, Pineda FJ, Hardwick JM (2008) Mitochondrial fission and fusion dynamics: the long and short of it. Cell Death Differ 15:1147–1152
Bruns AM, Horvath CM (2012) Activation of RIG-I-like receptor signal transduction. Crit Rev Biochem Mol Biol 47:194–206
Buchmann K (2014) Evolution of innate immunity: clues from invertebrates via fish to mammals. Front Immunol 5:459
Buskiewicz IA, Montgomery T, Yasewicz EC, Huber SA, Murphy MP, Hartley RC, Kelly R, Crow MK, Perl A, Budd RC, Koenig A (2016) Reactive oxygen species induce virus-independent MAVS oligomerization in systemic lupus erythematosus. Sci Signal 9:ra115
Castanier C, Garcin D, Vazquez A, Arnoult D (2010) Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep 11:133–138
Castanier C, Zemirli N, Portier A, Garcin D, Bidère N, Vazquez A, Arnoult D (2012) MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol 10:1–13
Chen YG, Hur S (2022) Cellular origins of dsRNA, their recognition and consequences. Nat Rev Mol Cell Biol 23:286–301
Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC (2003a) Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol 160:189–200
Chen XH, Lin KB, Wang XW (2003b) Outbreaks of an iridovirus disease in maricultured large yellow croaker, Larimichthys crocea (Richardson), in China. J Fish Dis 26:615–619
Chen SN, Zou PF, Nie P (2017) Retinoic acid-inducible gene I (RIG-I)-like receptors (RLR s) in fish: current knowledge and future perspectives. Immunology 151:16–25
Chen W, Sun Y, Sun Q, Zhang JJ, Jiang MX, Chang CW, Huang XL, Wang CY, Wang PX, Zhang ZR, Chen XJ, Wang Y (2020) MFN2 plays a distinct role from MFN1 in regulating spermatogonial differentiation. Stem Cell Rep 14:803–817
Chen XJ, Guan YY, Li KX, Luo T, Mu YN, Chen XH (2021) IRF1 and IRF2 act as positive regulators in antiviral response of large yellow croaker (Larimichthys crocea) by induction of distinct subgroups of type I IFNs. Dev Comp Immunol 118:103996
Chen JJ, Guan YY, Guan HX, Mu YN, Ding Y, Zou J, Ouyang SY, Chen XH (2022) Molecular and structural basis of receptor binding and signaling of a fish type I IFN with three disulfide bonds. J Immunol 209:806–819
Cvijović I, Good BH, Desai MM (2018) The effect of strong purifying selection on genetic diversity. Genetics 209:1235–1278
Dalskov L, Narita R, Andersen LL, Jensen N, Assil S, Kristensen KH, Mikkelsen JG, Fujita T, Mogensen TH, Paludan SR, Hartmann R (2020) Characterization of distinct molecular interactions responsible for IRF3 and IRF7 phosphorylation and subsequent dimerization. Nucleic Acids Res 48:11421–11433
Davey GP, Clark JB (1996) Threshold effects and control of oxidative phosphorylation in nonsynaptic rat brain mitochondria. J Neurochem 66:1617–1624
D’Eletto M, Farrace MG, Falasca L, Reali V, Oliverio S, Melino G, Griffin M, Fimia GM, Piacentini M (2009) Transglutaminase 2 is involved in autophagosome maturation. Autophagy 5:1145–1154
Deng H, Zhu S, Zhu L, Sun J, Ding Y, Li F, Jian Z, Zhao J, Deng L, Deng J, Deng Y, Guo H, Sun X, Lai SY, Tang H, Cui H, Ge LP, Xu Z (2022) Mfn2 is responsible for inhibition of the RIG-I/IRF7 pathway and activation of NLRP3 inflammasome in Seneca Valley virus-infected PK-15 cells to promote viral replication. Front Immunol 13:955671
Ding Y, Ao JQ, Huang XH, Chen XH (2016) Identification of two subgroups of type I IFNs in perciforme fish large yellow croaker Larimichthys crocea provides novel insights into function and regulation of fish type I IFNs. Front Immunol 7:343
Eisenächer K, Krug A (2012) Regulation of RLR-mediated innate immune signaling—it is all about keeping the balance. Eur J Cell Biol 91:36–47
Feng H, Zhang QM, Zhang YB, Li Z, Zhang J, Xiong YW, Wu M, Gui JF (2016) Zebrafish IRF1, IRF3, and IRF7 differentially regulate IFNΦ1 and IFNΦ3 expression through assembly of homo-or heteroprotein complexes. J Immunol 197:1893–1904
Gong XY, Zhang QM, Zhao X, Li YL, Qu ZL, Li Z, Dan C, Gui JF, Zhang YB (2022) LGP2 is essential for zebrafish survival through dual regulation of IFN antiviral response. Iscience 25:104821
Guan YY, Xj C, Luo T, Ao JQ, Ai CX, Chen XH (2020) Molecular characterization of the interferon regulatory factor (IRF) family and functional analysis of IRF11 in the large yellow croaker (Larimichthys crocea). Fish Shellfish Immunol 107:218–229
Guo N, Peng ZL, Zhang JW (2016) Proteasome inhibitor MG132 enhances sensitivity to cisplatin on ovarian carcinoma cells in vitro and in vivo. Int J Gynecol Cancer 26:839–844
Honda K, Taniguchi T (2006) IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol 6:644–658
Hou JX, Han LL, Zhao Z, Liu HQ, Zhang L, Ma C, Yi F, Liu BY, Zheng Y, Gao CJ (2021) USP18 positively regulates innate antiviral immunity by promoting K63-linked polyubiquitination of MAVS. Nat Commun 12:2970
Hughes AL (1994) The evolution of functionally novel proteins after gene duplication. Proc Biol Sci 256:119–124
Kawai T, Akira S (2008) Toll-like receptor and RIG-1-like receptor signaling. Ann N Y Acad Sci 1143:1–20
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Kim SJ, Ahn DG, Syed GH, Siddiqui A (2018) The essential role of mitochondrial dynamics in antiviral immunity. Mitochondrion 41:21–27
Kumar H, Kawai T, Akira S (2011) Pathogen recognition by the innate immune system. Int Rev Immunol 30:16–34
Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ (2004) Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol Biol Cell 15:5001–5011
Li CW, Wang SL, Ren QL, He TL, Chen XH (2020a) An outbreak of visceral white nodules disease caused by Pseudomonas plecoglossicida at a water temperature of 12 C in cultured large yellow croaker (Larimichthys crocea) in China. J Fish Dis 43:1353–1361
Li Z, Jaroszewski L, Iyer M, Sedova M, Godzik A (2020b) FATCAT 2.0: towards a better understanding of the structural diversity of proteins. Nucleic Acids Res 48:W60–W64
Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, Chen ZJ (2015) Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347:aaa2630
Luo Z, Liu LF, Wang XH, Li W, Jie C, Chen H, Wei FQ, Lu DH, Yan CY, Liu B, Kurihara H, Li YF, He RR (2019) Epigoitrin, an alkaloid from Isatis indigotica, reduces H1N1 infection in stress-induced susceptible model in vivo and in vitro. Front Pharmacol 10:78
Mallette FA, Richard S (2012) K48-linked ubiquitination and protein degradation regulate 53BP1 recruitment at DNA damage sites. Cell Res 22:1221–1223
Medzhitov R, Janeway CA Jr (2002) Decoding the patterns of self and nonself by the innate immune system. Science 296:298–300
Mills EL, Kelly B, O’Neill LA (2017) Mitochondria are the powerhouses of immunity. Nat Immunol 18:488–498
Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M (2022) ColabFold: making protein folding accessible to all. Nat Methods 19:679–682
Naon D, Scorrano L (2014) At the right distance: ER-mitochondria juxtaposition in cell life and death. Biochim Biophys Acta 1843:2184–2194
Nelapati S, Nelapati K, Chinnam BK (2012) Vibrio parahaemolyticus—an emerging foodborne pathogen—a review. Vet World 5:48–62
Netea MG, Van der Meer JW, Kullberg BJ (2004) Toll-like receptors as an escape mechanism from the host defense. Trends Microbiol 12:484–488
Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, Komuves L, French DM (2008) Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134:668–678
Onoguchi K, Onomoto K, Takamatsu S, Jogi M, Takemura A, Morimoto S, Julkunen I, Namiki H, Yoneyama M, Fujita T (2010) Virus-infection or 5′ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLoS Pathog 6:e1001012
Onomoto K, Onoguchi K, Yoneyama M (2021) Regulation of RIG-I-like receptor-mediated signaling: interaction between host and viral factors. Cell Mol Immunol 18:539–555
Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT (2011) MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep 12:565–573
Raftery N, Stevenson NJ (2017) Advances in antiviral immune defence: revealing the importance of the IFN JAK/STAT pathway. Cell Mol Life Sci 74:2525–2535
Sabouny R, Shutt TE (2020) Reciprocal regulation of mitochondrial fission and fusion. Trends Biochem Sci 45:564–577
Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O (2010) LGP2 is a positive regulator of RIG-I–and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A 107:1512–1517
Seth RB, Sun L, Ea CK, Chen ZJ (2005) Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell 122:669–682
Shen B, Hu YW, Zhang SY, Zheng JL, Zeng L, Zhang JS, Zhu AY, Wu CW (2016) Molecular characterization and expression analyses of three RIG-I-like receptor signaling pathway genes (MDA5, LGP2 and MAVS) in Larimichthys crocea. Fish Shellfish Immunol 55:535–549
Takahasi K, Kumeta H, Tsuduki N, Narita R, Shigemoto T, Hirai R, Yoneyama M, Horiuchi M, Ogura K, Fujita T, Fuyuhiko Inagaki F (2009) Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J Biol Chem 284:17465–17474
Takeuchi O, Akira S (2009) Innate immunity to virus infection. Immunol Rev 227:75–86
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027
Tan J, Yi SQ, Xiao ZJ, Huang KL, Gao YJ, Yan HY, Chen SX, Wu H (2022) Mitochonic acid 5 promotes the migration of mouse microglial BV-2 cells in the presence of LPS-induced inflammation via Mfn2-associated mitophagy. Acta Neurobiol Exp 82:442–447
Tracz M, Bialek W (2021) Beyond K48 and K63: non-canonical protein ubiquitination. Cell Mol Biol Lett 26:1
Wang XH, Wang KR, Nie P, Chen XH, Ao JQ (2014) Establishment and characterization of a head kidney cell line from large yellow croaker Pseudosciaena crocea. J Fish Biol 84:1551–1561
Wei ZY, Li XF, Li WR, Fu QL, Mu YN, Chen XH (2021) Molecular characterization and role in virus infection of Beclin-1 in large yellow croaker (Larimichthys crocea). Fish Shellfish Immunol 116:30–41
Wu B, Hur S (2015) How RIG-I like receptors activate MAVS. Curr Opin Virol 12:91–98
Xu F, Qi H, Li JQ, Sun L, Gong JJ, Chen YY, Shen A, Li W (2020) Mycobacterium tuberculosis infection upregulates MFN2 expression to promote NLRP3 inflammasome formation. J Biol Chem 295:17684–17697
Yasukawa K, Oshiumi H, Takeda M, Ishihara N, Yanagi Y, Seya T, Kawabata S, Koshiba T (2009) Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal 2:ra47
Yin F, Bao PB, Liu X, Yu YB, Wang L, Wang LM (2019) Antiparasitic effect of copper alloy surface on Cryptocaryon irritans in aquaculture of Larimichthys crocea. Appl Environ Microbiol 85:e01982-e2018
You FP, Sun H, Zhou X, Sun WX, Liang SM, Zhai ZH, Jiang ZF (2009) PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol 10:1300–1308
Yu R, Jin SB, Lendahl U, Nistér M, Zhao J (2019) Human Fis1 regulates mitochondrial dynamics through inhibition of the fusion machinery. EMBO J 38:e99748
Zhang DL, Chen J, Fan S, Wang ZY (2015) Expression profiles and interaction suggest TBK1 can be regulated by Nrdp1 in response to immune stimulation in large yellow croaker Larimichthys crocea. Fish Shellfish Immunol 46:745–752
Zhang QM, Zhao X, Li Z, Wu M, Gui JF, Zhang YB (2018) Alternative splicing transcripts of zebrafish LGP2 gene differentially contribute to IFN antiviral response. J Immunol 200:688–703
Zhou W, Zhou JJ, Lv Y, Qu YX, Chi MD, Li J, Feng H (2015) Identification and characterization of MAVS from black carp Mylopharyngodon piceus. Fish Shellfish Immunol 43:460–468
Acknowledgements
This work was supported by National Key Research and Development Program of China under Grant No. 2022YFD2401001, National Natural Science Foundation of China under Grant No. U1905204, China Agriculture Research System of MOF and MARA under Grant No. CARS-47, and Fujian Science and Technology Department under Grant No. 2021N5008, Institute of Oceanology of Fuzhou (2021F02).
Author information
Authors and Affiliations
Contributions
WXL: writing—conceptualization, methodology, and writing—original draft. XHW: data curation and data analysis. YJL: data curation and data analysis. YYZ: data curation and data analysis. JL: data curation. XYZ: data curation and data analysis. XHC: writing—review and editing, supervision, and funding acquisition.
Corresponding author
Ethics declarations
Conflict of interest
The authors confirm that no conflicts of interest are known to be involved with this publication.
Animal and human rights statements
All the animal experiments were approved by the college of life science at Fujian Agriculture and Forestry University. This study does not include any research involving human participants.
Additional information
Edited by Xin Yu.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, WX., Wang, XH., Lin, YJ. et al. Large yellow croaker (Larimichthys crocea) mitofusin 2 inhibits type I IFN responses by degrading MAVS via enhanced K48-linked ubiquitination. Mar Life Sci Technol 5, 359–372 (2023). https://doi.org/10.1007/s42995-023-00189-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42995-023-00189-8