Abstract
Basic helix-loop-helix (bHLH) transcription factors are widely distributed in eukaryotes, and in plants, they regulate many biological processes, such as cell differentiation, development, metabolism, and stress responses. Few studies have focused on the roles of bHLH transcription factors in regulating growth, development, and stress responses in maize (Zea mays), even though such information would greatly benefit maize breeding programs. In this study, we cloned the maize transcription factor gene ZmbHLH36 (Gene ID: 100193615, GRMZM2G008691). ZmbHLH36 possesses conserved domains characteristic of the bHLH family. RT-qPCR analysis revealed that ZmbHLH36 was expressed at the highest level in maize roots and exhibited different expression patterns under various abiotic stress conditions. Transgenic Arabidopsis (Arabidopsis thaliana) plants heterologously expressing ZmbHLH36 had significantly longer roots than the corresponding non-transgenic plants under 0.1 and 0.15 mol L−1 NaCl treatment as well as 0.2 mol L−1 mannitol treatment. Phenotypic analysis of soil-grown plants under stress showed that transgenic Arabidopsis plants harboring ZmbHLH36 exhibited significantly enhanced drought tolerance and salt tolerance compared to the corresponding non-transgenic plants. Malondialdehyde contents were lower and peroxidase activity was higher in ZmbHLH36-expressing Arabidopsis plants than in the corresponding non-transgenic plants. ZmbHLH36 localized to the nucleus when expressed in maize protoplasts. This study provides a systematic analysis of the effects of ZmbHLH36 on root growth, development, and stress responses in transgenic Arabidopsis, laying a foundation for further analysis of its roles and molecular mechanisms in maize.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants initiate a series of stress responses when facing adverse conditions, such as drought, high temperature, salinity, and other stresses (Gong et al. 2020). Transcription factors play crucial roles in these adaptive responses by regulating the expression of stress-related genes, thereby helping plants acclimate to harsh environments (Baillo et al. 2019). Numerous stress-related transcription factors have been investigated in plants (Erpen et al. 2018). These transcription factors regulate stress-responsive genes by binding to specific elements in their promoters. For example, AP2/ERF transcription factors bind to dehydration response elements (DREs/CRTs) (Xie et al. 2019), WRKY transcription factors to W-box sequences (Van Aken et al. 2013), NAC transcription factors to NAC response elements (Tran et al. 2004), MYB transcription factors to MYB binding sites (Baldoni et al. 2015), and NF-Y transcription factors to CCAAT sequences (Zhang et al. 2023).
The basic helix-loop-helix (bHLH) transcription factor family is widely conserved in eukaryotes. bHLH transcription factors have a bHLH domain containing an N-terminal basic region and a C-terminal HLH region. The basic region, typically located at the N terminus, is composed of a stretch of basic amino acids and is crucial for DNA binding and transcriptional activation. The C-terminal HLH region consists of two α helices and a short loop. One of the α helices binds to DNA and recognizes specific DNA sequences, while the other α helix interacts with other proteins, forming dimers or multimeric complexes (Hao et al. 2021).
The bHLH transcription factors represent one of the largest transcription factor families in plants, where they play important roles in signal transduction and the regulation of growth, development, and responses to environmental stress (Guo et al. 2021). For example, in A. thaliana, bHLH57, together with the abscisic acid (ABA)–related protein REDUCED DORMANCY 1 (AtRDO1), inhibits the expression of NCED, which encodes a key enzyme involved in ABA biosynthesis, thereby breaking seed dormancy (Liu et al. 2020). The phytochrome-interacting factor (PIF) photoreceptors are bHLH proteins that integrate external and internal signals by interacting with various factors to regulate seed germination, phytohormonal crosstalk, and associated signaling pathways (Pham et al. 2018). Four bHLH proteins (FBH1, FBH2, FBH3, and FBH4) act as transcriptional activators of the CONSTANS gene, a key factor that converts the biological clock signal into a flowering signal (Ito et al. 2012). bHLH transcription factors also regulate the growth and development of plant roots. In rice (Oryza sativa), a bHLH transcription factor mutant, Osrhl1-1, had shorter root hairs than the corresponding non-transgenic plants under low-temperature conditions (Ding et al. 2009). Exogenous overexpression of the sorghum (Sorghum bicolor) gene SbbHLH85 in Arabidopsis led to a significant increase in root hair formation in the corresponding non-transgenic plants and restored the root hair defects in single and double mutants (Song et al. 2022).
The bHLH transcription factors also play important roles in plant resistance to abiotic stress. Arabidopsis plants overexpressing bHLH122 had significantly higher ABA levels and exhibited greater tolerance to drought, salt, and osmotic stress compared to non-transgenic plants (Liu et al. 2014). Heterologous overexpression of the wheat (Triticum aestivum) TabHLH1 gene enhanced the tolerance of transgenic tobacco (Nicotiana tabacum) to drought and salt stress by upregulating the ABA signaling genes PYR1/PYL/RCAR1-LIKE 12 (NtPYL12) and SUCROSE NON-FERMENTING1-RELATED PROTEIN KINASE 21 (NtSAPK21), as well as regulating intracellular substance accumulation and reactive oxygen species (ROS) homeostasis (Yang et al. 2016). Heterologous overexpression of CgbHLH001 from Chenopodium glaucum (Oxybasis glauca) enhanced drought tolerance in maize. Under drought stress, maize plants expressing CgbHLH001 showed increased soluble sugar and proline accumulation, increased antioxidant enzyme (superoxide dismutase [SOD], peroxidase [POD], and catalase [CAT]) activity, and upregulated expression of stress-related genes, demonstrating a drought-tolerant phenotype (Li et al. 2010). Overexpressing the bHLH family transcription factor gene ZmPTF1 in maize increased root length, root hair quantity, and root surface area, enhancing the plant’s water absorption capacity and drought tolerance (Li et al. 2019). A signaling pathway involving the transcription factor ZmbHLH32, the Aux/IAA family gene ZmIAA9, and AUXIN RESPONSE FACTOR 1 (ZmARF1) positively regulates salt tolerance and inhibits the expression of ROS scavenging genes in maize (Yan et al. 2023). In rice, the cold-induced bHLH transcription factor INDUCER OF CBF EXPRESSION 1 (OsICE1), positively regulates cold tolerance in rice seedlings (Liu et al. 2023). Heterologously overexpressing the rice bHLH transcription factor gene OrbHLH001, which has high sequence similarity to ICE1, enhanced the freezing and salt tolerance of the model plant Arabidopsis (Chen et al. 2013; Li et al. 2010). In maize, ZmICE1 in maize plays a crucial role in regulating the expression of amino acid metabolism genes, thereby affecting cold tolerance (Jiang et al. 2022).
Although previous studies have investigated the molecular mechanisms of various bHLH transcription factors, little research has been conducted on bHLH family transcription factors in the monocotyledonous model crop plant maize. We previously performed root phenotype analysis and transcriptome sequencing (RNA-seq) of maize at key stages of growth, including the 6-leaf stage (V6), 12-leaf stage (V12), tasseling stage (VT), and milk-mature stage (R3). ZmbHLH36 was highly expressed during periods of vigorous root growth, such as the V6 and V12 stages, pointing to its potential involvement in regulating root growth and development (Zhang et al. 2020). In this study, to elucidate the roles of ZmbHLH36 in regulating root growth and development as well as plant responses to stress, we cloned the ZmbHLH36 gene. Using heterologous expression in Arabidopsis, we initiated a functional analysis of the roles of this transcription factor in root development and stress tolerance. Our findings lay a foundation for further in-depth investigation of the biological functions of ZmbHLH36 in maize.
Results
Cloning and structural analysis of the maize transcription factor gene ZmbHLH36
Structural domain analysis of the predicted protein encoded by ZmbHLH36 revealed the presence of a conserved domain characteristic of the bHLH family (Fig. 1A). We amplified the coding sequence (CDS) of ZmbHLH36 by RT-PCR. The full-length CDS was found to be 1038 bp long, encoding a protein of 345 amino acids (Fig. 1B). Analysis of the physicochemical properties of the deduced ZmbHLH36 protein sequence revealed a molecular weight of 36.39 kDa, a molecular formula of C1567H2548N486O486S13, and a theoretical isoelectric point (pI) of 8.59.
In the predicted secondary structure of ZmbHLH36, α-helixes comprised the highest proportion of structures (48.99%), followed by irregular coils (37.68%). The predicted tertiary structure of ZmbHLH36 is shown in Fig. 1C, and the amino acid sequence contains no predicted transmembrane structures (Fig. 1D). Analysis of the 2-kb sequence upstream of the start codon (ATG) of ZmbHLH36 revealed the presence of potential cis-acting elements in the promoter region, including an ABRE (involved in the ABA response), a CAT-box (cis-regulatory element associated with organ-specific expression), a CGTCA-motif (jasmonic acid methyl ester response cis-regulatory element), a G-box (responsive to ABA, light, and UV radiation), a TGA-element (auxin response cis-regulatory element), and a low-temperature response element, among others.
The expression of ZmbHLH36 in different maize tissues
We examined the relative expression of ZmbHLH36 in various maize tissues, including young roots and leaves of plants at the three-leaf stage; roots and leaves of plants at the tassel stage; and silks, tassels, and young embryos 10 days after pollination, via reverse-transcription quantitative PCR (RT-qPCR). The expression level in mature leaves was set to 1. ZmbHLH36 was expressed in all eight tissues examined, with higher expression in roots than in the other tissues, particularly in young roots of plants at the three-leaf stage (Fig. 2A). The V6 and V12 stages represent periods of active root growth and development. The expression level of ZmbHLH36 was higher in the V6 and V12 stages compared to VT and R3 (Fig. 2B). These results suggest that ZmbHLH36 may be involved in regulating root growth and development.
Analysis of ZmbHLH36 expression in different tissues and during key stages of maize root growth and development. A Tissue-specific expression of ZmbHLH36 determined by reverse-transcription quantitative PCR (RT-qPCR). The analyzed tissues included young roots and leaves; roots, stems, leaves, male inflorescences, and stamens at the tassel stage; and young embryos 1 week after pollination. B Relative expression levels of ZmbHLH36 in maize roots at growth stages V6, V12, VT, and R3. Data are means ± SD (Student’s t tests, *P < 0.05, **P < 0.01, n = 3 biological replicates)
Expression of ZmbHLH36 under different abiotic stresses
Under osmotic stress, induced by mannitol treatment (Fig. 3A), ZmbHLH36 was upregulated in roots, reaching its highest level of expression, approximately 9.67-fold higher than the control, at 5 h. Subsequently, ZmbHLH36 expression sharply decreased, reaching approximately 0.1-fold that of the control at 24 h. In leaves, the expression level of ZmbHLH36 showed an initial increase, followed by a decrease, with the highest level observed at 2 h, approximately 3.43-fold higher than the control. Under dehydration treatment (Fig. 3B), the expression of ZmbHLH36 in leaves reached its highest peak at 24 h (approximately 20.5-fold higher than the control). The expression level in roots showed an initial increase, followed by a decrease, under this treatment.
Relative expression level of ZmbHLH36 under different abiotic stress conditions. A–D The expression pattern of ZmbHLH36 in maize roots and leaves under A 0.2 mol L−1 mannitol treatment, B dehydration treatment, C 4 °C treatment, and D 0.2 mol L−1 NaCl treatment. Three independent biological experiments were conducted using nine individual maize seedlings at the three-leaf stage. The seedlings were subjected to 0.2 mol L−1 NaCl, 0.2 mol L−1 mannitol, 4 °C, or natural dehydration treatment for the specified time periods. Leaves and roots were collected for RNA extraction and RT-qPCR analysis. Data are means ± SD (Student’s t tests, *P < 0.05, **P < 0.01, n = 3 biological replicates)
Under cold treatment (Fig. 3C), the expression of ZmbHLH36 in roots slightly decreased, followed by an increase at 2 h, reaching approximately 10.53-fold higher than the control. ZmbHLH36 expression subsequently decreased, reaching its lowest value at 24 h (approximately 0.07-fold of the control). In leaves, the expression level increased at 2 h (approximately 2.15-fold higher than the control) and then decreased under this treatment. Under salt treatment (Fig. 3D), the expression of ZmbHLH36 in roots showed an initial increase, followed by a decrease. The highest expression levels were observed at 2 and 5 h, reaching approximately 5.07- and 4.68-fold higher than the control, respectively. Subsequently, ZmbHLH36 expression decreased to its lowest level at 24 h (approximately 0.06-fold of the control). In leaves, ZmbHLH36 expression decreased at 1 h of salt treatment to a level approximately 0.21-fold that of the control and then increased (approximately twofold) at 24 h under this treatment. In summary, these changes in ZmbHLH36 expression indicate that ZmbHLH36 may be involved in plant responses to various abiotic stresses, including osmotic stress, dehydration, low temperatures, and salt stress.
The subcellular localization of ZmbHLH36 in maize protoplasts
To examine the subcellular localization of ZmbHLH36, we fused the coding sequence to the fluorescent marker protein enhanced green fluorescent protein (EGFP) and expressed the resulting fusion from the cauliflower mosaic virus 35S promoter in the construct pYBA1132-ZmbHLH36-EGFP. We introduced pYBA1132-ZmbHLH36-EGFP into protoplasts from etiolated seedlings of maize inbred line Z58 using polyethylene glycol–Ca2+-mediated transfection. The pYBA1132-EGFP empty vector was used as a negative control. After 12 h of cultivation in the dark, protoplasts were observed under a confocal laser-scanning fluorescence microscope (Fig. S1). Protoplasts harboring the pYBA1132-EGFP empty vector exhibited green fluorescence throughout the entire cell, while protoplasts harboring the pYBA1132-ZmbHLH36-EGFP vector showed green fluorescent signals only in the nucleus, indicating that ZmbHLH36 localizes to the nucleus.
Heterologous expression of ZmbHLH36 in transgenic Arabidopsis confers tolerance to salt, mannitol, ABA, and jasmonic acid
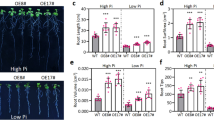
To evaluate the role of ZmbHLH36 in abiotic stress responses, we generated Arabidopsis plants exogenously overexpressing this gene. We grew T3 generation transgenic Arabidopsis plants (L-11, L-17, and L-20) identified by PCR and RT-qPCR alongside control, non-transgenic seedlings on 1/2 MS medium containing various supplements (Fig. S2). The control and transgenic plants showed similar root lengths under normal conditions. However, the roots of the transgenic plants were significantly longer than those of control plants under 150 mM NaCl or 200 mM mannitol treatment. Since bHLH transcription factors are also involved in the crosstalk among plant hormone signaling pathways, we subjected the transgenic Arabidopsis plants to 50 μM ABA or 100 μM jasmonic acid (JA) treatment. The roots of the transgenic plants were significantly longer than those of control plants (Fig. 4). These results suggest that ZmbHLH36 is involved in plant responses to stresses such as salt and osmotic stress and may have a regulatory role in signal transduction pathways of phytohormones such as ABA and JA.
Phenotypic analysis of WT and transgenic Arabidopsis plants under NaCl, mannitol, ABA, and JA treatments. A Transgenic and WT seedlings were cultured on 1/2 MS medium supplemented with 150 mM NaCl, 200 mM mannitol, 50 μM ABA, or 100 μM JA, as well as 1/2 MS medium without any additional additives. B Primary root length of plants on different media; data represent average root length. Data are means ± SD (Student’s t tests, *P < 0.05, **P < 0.01, n = 15, bar = 1.5 cm)
Drought tolerance of ZmbHLH36 transgenic Arabidopsis in soil
Under normal conditions in soil, both ZmbHLH36 transgenic and control Arabidopsis plants exhibited healthy growth with similar vigor. However, when water was withheld from the plants after 2 weeks of growth under normal conditions, the growth of the controls was significantly inhibited, and the control plants were more wilted and had smaller leaves compared to the transgenic plants (Fig. 5A). The transgenic plants exhibited significantly greater stem height, fresh weight, and green leaf ratio than control plants (Fig. 5B-D).
Phenotypic analysis of transgenic Arabidopsis under drought treatment in soil. A Phenotypes of Arabidopsis plants. B Plant height (Student’s t tests, *P < 0.05, n ≥ 12). C Shoot fresh weight (Student’s t tests, **P < 0.01, n ≥ 12). D Green leaf ratio under drought treatment (Student’s t tests, **P < 0.01, n ≥ 12). E Quantification of expression of the marker gene RD29B. F Quantification of expression of the marker gene RD22. G MDA content. H POD activity. Abbreviations are the same as in Fig. 4. Data are means ± SD (Student’s t tests, *P < 0.05, **P < 0.01, n = 3 biological replicates)
To further investigate the regulatory pathway of ZmbHLH36 under abiotic stress, we analyzed the expression patterns of the stress/ABA-responsive genes RESPONSE TO DEHYDRATION 29A (RD29A) (Msanne et al. 2011), RD29B (Nakashima et al. 2006), RD22 (Abe et al. 1997), and RESPONSIVE TO ABA 18 (RAB18) (Tripathy et al. 2021) in WT and ZmbHLH36 transgenic plants by RT-qPCR. Under drought stress, the expression of RD29B and RD22 was promoted in transgenic Arabidopsis plants overexpressing ZmbHLH36 (Fig. 5E and F). Physiological measurements indicated that the transgenic Arabidopsis plants contained reduced malondialdehyde (MDA) contents, indicating reduced oxidative stress, and increased POD activity compared to the non-transgenic control (Fig. 5G and H). These findings demonstrate that heterologously overexpressing ZmbHLH36 enhanced the drought tolerance of Arabidopsis plants.
Salt tolerance of transgenic Arabidopsis expressing ZmbHLH36 in soil
We analyzed the salt tolerance of transgenic Arabidopsis expressing ZmbHLH36 by subjecting soil-grown plants to salt stress (300 mM NaCl) for 1 week. Under salt stress conditions, the non-transgenic control Arabidopsis plants exhibited symptoms such as leaf yellowing and delayed growth (Fig. 6A). By contrast, the transgenic plants showed significantly greater stem height and green leaf ratio compared to the control (Fig. 6B and C). Additionally, the transgenic plants had higher shoot fresh weight than the control (Fig. 6D). RT-qPCR analysis of stress-responsive marker genes revealed that the expression of RD29A, RD22, and RAD18 was promoted in transgenic Arabidopsis plants expressing ZmbHLH36 under salt stress but not under normal conditions (Fig. 6E-G). Physiological measurements indicated that the transgenic Arabidopsis plants had reduced MDA contents and increased POD activity compared to the WT (Fig. 6H and I). Collectively, these results demonstrate that exogenously expressing ZmbHLH36 enhanced the salt tolerance of Arabidopsis plants.
Phenotypic analysis of transgenic Arabidopsis under high-salt treatment in soil. A Phenotypes of Arabidopsis plants. B Plant height (Student’s t tests, **P < 0.01, n ≥ 12). C Green leaf ratio under drought treatment (Student’s t tests, **P < 0.01, n ≥ 12). D Shoot fresh weight (Student’s t tests, **P < 0.01, n ≥ 12). E Quantification of expression of the marker genes RD29A, F RD22, and G RAB18. H MDA content. I POD activity. Abbreviations are the same as in Fig. 4. Data are means ± SD (Student’s t tests, *P < 0.05, **P < 0.01, n = 3 biological replicates)
Discussion
bHLH transcription factors are widely distributed in eukaryotes and participate in the regulation of various biological processes, including cell differentiation, development, metabolism, and stress responses (Sun et al. 2018). In this study, we cloned ZmbHLH36, encoding a maize bHLH transcription factor. Bioinformatic analysis revealed that ZmbHLH36 contains the conserved domain characteristic of the bHLH family. Subcellular localization showed that ZmbHLH36 localizes to the nucleus. We also performed a preliminarily analysis of the biological functions of ZmbHLH36 in plant responses to abiotic stress using transgenic Arabidopsis lines.
Several studies on bHLH transcription factors in other plants have been reported. The maize bHLH transcription factor ZmbHLH91 may be involved in regulating plant tolerance to abiotic stress conditions such as high-salt, drought, and osmotic stress, based on the phenotypes of transgenic Arabidopsis lines expressing ZmbHLH91 (Yue et al. 2022). The Tamarix hispida (Tamarix chinensis) transcription factor ThbHLH1 enhances abiotic stress tolerance by increasing osmotic potential and reducing ROS accumulation (Ji et al. 2016). In this study, our RT-qPCR analysis showed that ZmbHLH36 is expressed in multiple maize tissues, with higher expression in roots at the three-leaf stage vs. the tasseling stage. This finding, combined with the results of previous transcriptome analysis, suggests that ZmbHLH36 might be involved in regulating the growth and development of maize roots (Zhang et al. 2020). Moreover, abiotic stresses such as low-temperature, drought, and salinity, finding that they induced ZmbHLH36 expression. The highest expression level in response to mannitol was observed in roots at 5 h. In response to cold and salt treatments, the expression levels were highest in roots at 2 h. For the dehydration treatment, the expression level of ZmbHLH36 was highest in leaves at 24 h. These results suggest that ZmbHLH36 functions in plant responses to abiotic stress. To test this hypothesis, we generated genetically modified Arabidopsis seedlings exogenously expressing ZmbHLH36 and subjected them to high-salt, osmotic, and other abiotic stress factors. The seedlings expressing ZmbHLH36 had longer roots than WT seedlings under abiotic stress, indicating greater tolerance of the stresses.
We also investigated the possible links between ZmbHLH36 and the signaling pathways of two plant hormones: JA, because bHLH transcription factors play important, generally conserved roles in the signaling cascade triggered by this hormone (Goossens et al. 2017); and ABA, because it plays a crucial role in regulating plant responses to drought stress. AtAIB is a bHLH transcriptional activator involved in regulating ABA signaling in Arabidopsis (Li et al. 2007). The downregulation of AtAIB in Arabidopsis led to reduced sensitivity to ABA, while overexpressing AtAIB had the opposite effect. In the current study, we simulated phytohormone-mediated stress responses by growing Arabidopsis plants on medium supplemented with ABA and JA. Under these hormone treatments, transgenic Arabidopsis plants harboring ZmbHLH36 had significantly longer roots compared to the control plants. We also subjected soil-grown plants to drought and high-salt stress and found that ZmbHLH36 expression enabled Arabidopsis to withstand these stresses, resulting in greener leaves and healthier plants compared to the control. The increased stress tolerance of transgenic Arabidopsis expressing ZmbHLH36 was further supported by the increased expression of the marker genes, reduced MDA content, and increased POD activity under stress conditions.
Our results indicate that ZmbHLH36 acts as a positive regulator of plant responses to high-salt, drought, osmotic, and phytohormone stress. Under stress conditions, ZmbHLH36 may regulate root growth and development, modulate membrane lipid peroxidation, stabilize the cell membrane system, and enhance antioxidant enzyme activity to alleviate the damage caused by drought and high-salt stress.
Materials and methods
Plant materials and growth conditions
Maize inbred line B73 was cultivated at the experimental station of Beijing Academy of Agriculture and Forestry Sciences (39°56′38.41″N, 116°16′57.18″E). Roots and young leaves from plants at the three-leaf stage, as well as roots, stems, leaves, tassels, and silks from plants at the tasseling stage, were collected from inbred line B73. Young embryos, harvested 1 week after pollination, were utilized for tissue-specific differential expression analysis. Root tissues from plants at different stages of growth (V6, V12, VT, and R3) were collected for RT-qPCR. For abiotic stress treatment, B73 maize seeds were placed in a growth chamber at 25 °C and a relative humidity of 70%. The seedlings were transferred to hydroponic containers and grown until the three-leaf stage. Subsequently, the seedlings were treated with a high salt concentration (placed in hydroponic containers containing 0.2 mol L−1 NaCl), osmotic stress (placed in hydroponic containers containing 0.2 mol L−1 mannitol), cold stress (at 4 °C), and dehydration stress (placed on absorbent paper for natural dehydration). The aboveground parts and roots were collected at 0, 1, 2, 5, 10, and 24 h of each treatment, rapidly frozen in liquid nitrogen, and stored at −80 °C.
Bioinformatics analysis
The CDS of the transcription factor gene ZmbHLH36 and the 2-kb sequence upstream of the transcription start codon (ATG) were obtained from the maize database (Ensembl Plants). The online tools SOPMA, THMMM, and Plant CARE were used to perform preliminary bioinformatics analysis of ZmbHLH36.
RNA extraction and RT-qPCR
RNA extraction was performed using a FastPure Universal Plant Total RNA Isolation Kit (Vazyme Biotech Co., Ltd., Beijing) according to the manufacturer’s instructions. cDNA synthesis was carried out using a HiScript III RT SuperMix for qPCR (+ gDNA wiper) kit (Vazyme Biotech Co., Ltd., Beijing). Specific primers, including pZmbHLH36 RT-F, pZmbHLH36 RT-R for the target gene ZmbHLH36, and pGAPDH RT-F, pGAPDH RT-R for the maize reference gene GAPDH, were designed using the Primer-BLAST tool on the NCBI website and utilized for tissue-specific differential expression analysis (primer information can be found in Table S1). Real-time fluorescence amplification was performed using the BIO-RAD CFX96 Real-Time System qPCR instrument (Table S1). Each tissue sample included three biological replicates (separate experiments), and each biological replicate included three technical replicates (replicate samples). The data are presented as the mean ± standard error of three biological replicates. The 2−∆∆CT method was used to obtain relative gene expression levels.
Vector construction and transformation of Arabidopsis
The CDS of ZmbHLH36 without the terminator sequence was amplified using the homologous arm primers pZmbHLH36 OE-F and pZmbHLH36 OE-R (primer information can be found in Table S1). The amplified CDS fragment was recombined into the expression vector pYBA1132 (pYBA1132 was kindly provided by Professor Fengping Yang from the Institute of Crop Sciences, Beijing Academy of Agriculture and Forestry Sciences), driven by the cauliflower mosaic virus (CaMV) E35S promoter, at the N-terminal region of the EGFP (enhanced green fluorescent protein) element, using the seamless cloning kit ClonExpress Ultra One Step Cloning Kit V2 from Vazyme Biotech Co., Ltd., Beijing. Transformed Escherichia coli DH5α competent cells were selected and propagated overnight in 5 mL liquid medium containing kanamycin (Kan+). The bacterial culture was then used for plasmid isolation, followed by digestion with restriction enzymes for verification. Correct plasmids were selected and sent to Sangon Biotech (Shanghai) Co., Ltd. for sequencing.
The sequenced overexpression vector was transformed into Agrobacterium tumefaciens strain GV3101. Single clones were selected for colony PCR and sequencing for verification. Arabidopsis (WT) inflorescences at the flowering stage were transformed with the vector using the floral dip method (Krenek et al. 2015). T1 generation seeds were harvested and air-dried for 1 week, followed by cold stratification for future use. T1 generation seeds were sterilized in 0.3% sodium hypochlorite solution for 15 min, rinsed with sterile water five to eight times, and sown on medium containing 0.02% Basta. After 8–10 days, surviving positive seedlings were transplanted to nutrient soil to obtain single T1 transgenic Arabidopsis plants. Genomic DNA was extracted from the leaves of T1 plants as a template for PCR analysis using the primers pZmbHLH36-F and pZmbHLH36-R. RNA was extracted from the leaves of T1 plants and reverse transcribed into cDNA as a template for RT-qPCR analysis. The expression levels were detected using the primers pZmbHLH36 RT-F, pZmbHLH36 RT-R, pActin1RT-F, and pActin1RT-R as internal controls. T3 generation homozygous plants were obtained from single plants with high expression and tested for subsequent experiments (Table S1).
Root growth assay
To study the root growth of Arabidopsis, the seeds of transgenic and WT Arabidopsis were cultured on 1/2 MS agar plates for 7 days. Five germinated seedlings per line with the same growth status were selected and gently transferred to 1/2 MS agar plates supplemented with 150 mM NaCl, 200 mM mannitol, 50 μM ABA, or 100 μM JA. After 8 days of vertical growth, the root length of the seedlings was measured using ImageJ software (NIH, Bethesda, MD, USA).
Drought and NaCl treatment of transgenic Arabidopsis
WT and transgenic Arabidopsis seeds were sterilized in 0.3% sodium hypochlorite solution for 10 min and sown on 1/2 MS medium. The plates were placed in the Arabidopsis growth chamber and incubated for 6 days. WT and transgenic Arabidopsis seedlings with similar root lengths and growth statuses were selected and transplanted into soil. Four seedlings were transplanted to each pot, with a total of nine pots per group. For the control group, the seedlings were watered normally. For the high-salt treatment, the seedlings were grown in soil for 25–30 days and irrigated with 300 mmol L−1 NaCl solution. Phenotypic data and physiological indicators were measured after 7–10 days. For drought treatment, the seedlings were grown in soil for 2 weeks, then deprived of water for 20–25 days. During this period, the stem height and fresh weight of Arabidopsis were measured using a ruler and analytical balance, respectively. The chlorophyll content of Arabidopsis was observed and analyzed, and the MDA content and POD activity were measured using physiological indicator kits.
Subcellular localization of proteins
The subcellular localization vector was constructed as described above (for plant expression vectors). The pYBA1132-EGFP empty vector lacking the target gene was used as a negative control. The subcellular localization vector and empty vector, each at a concentration of 20 μg, were separately transformed into maize protoplasts derived from yellow maize seedlings. Following incubation at 25 °C under greenhouse conditions for 12 h, the green fluorescent signals were detected under a confocal laser-scanning fluorescence microscope (Nikon).
Statistical analysis
The experiments were repeated three times, and the data are presented as the mean ± standard error of the mean (SEM). The experimental data were analyzed using GraphPad Prism software. In all analyses, P < 0.05 was taken to indicate statistical significance.
References
Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9:1859–1868. https://doi.org/10.1105/tpc.9.10.1859
Van Aken O, Zhang B, Law S, Narsai R, Whelan J (2013) AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol 162:254–271. https://doi.org/10.1104/pp.113.215996
Baillo EH, Kimotho RN, Zhang Z, Xu P (2019) Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes (basel) 10:771. https://doi.org/10.3390/genes10100771
Baldoni E, Genga A, Cominelli E (2015) Plant MYB transcription factors: their role in drought response mechanisms. Int J Mol Sci 16:15811–15851. https://doi.org/10.3390/ijms160715811
Chen Y, Li F, Ma Y, Chong K, Xu Y (2013) Overexpression of OrbHLH001, a putative helix-loop-helix transcription factor, causes increased expression of AKT1 and maintains ionic balance under salt stress in rice. J Plant Physiol 170:93–100. https://doi.org/10.1016/j.jplph.2012.08.019
Ding W, Yu Z, Tong Y, Huang W, Chen H, Wu P (2009) A transcription factor with a bHLH domain regulates root hair development in rice. Cell Res 19:1309–1311. https://doi.org/10.1038/cr.2009.109
Erpen L, Devi HS, Grosser JW, Dutt M (2018) Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tissue Organ Cult 132:1–25. https://doi.org/10.1007/s11240-017-1320-6
Gong Z, Xiong L, Shi H, Yang S, Herrera-Estrella LR, Xu G, Chao DY, Li J, Wang PY, Qin F, Li J, Ding Y, Shi Y, Wang Y, Yang Y, Guo Y, Zhu JK (2020) Plant abiotic stress response and nutrient use efficiency. Sci China Life Sci 63:635–674. https://doi.org/10.1007/s11427-020-1683-x
Goossens J, Mertens J, Goossens A (2017) Role and functioning of bHLH transcription factors in jasmonate signalling. J Exp Bot 68:1333–1347
Guo J, Sun B, He H, Zhang Y, Tian H, Wang B (2021) Current understanding of bHLH transcription factors in plant abiotic stress tolerance. Int J Mol Sci 22:4921. https://doi.org/10.3390/ijms22094921
Hao Y, Zong X, Ren P, Qian Y, Fu A (2021) Basic helix-loop-helix (bHLH) transcription factors regulate a wide range of functions in Arabidopsis. Int J Mol Sci 22:7152. https://doi.org/10.3390/ijms22137152
Ito S, Song YH, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG, Imaizumi T (2012) FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc Natl Acad Sci U S A 109:3582–3587. https://doi.org/10.1073/pnas.1118876109
Ji X, Nie X, Liu Y, Zheng L, Zhao H, Zhang B, Huo L, Wang Y, Tsai C (2016) A bHLH gene from tamarix hispida improves abiotic stress tolerance by enhancing osmotic potential and decreasing reactive oxygen species accumulation. Tree Physiol 36:193–207. https://doi.org/10.1093/treephys/tpv139
Jiang H, Shi Y, Liu J, Li Z, Fu D, Wu S, Li M, Yang Z, Shi Y, Lai J, Yang X, Gong Z, Hua J, Yang S (2022) Natural polymorphism of ZmICE1 contributes to amino acid metabolism that impacts cold tolerance in maize. Nat Plants 8:1176–1190. https://doi.org/10.1038/s41477-022-01254-3
Krenek P, Samajova O, Luptovciak I, Doskocilova A, Komis G, Samaj J (2015) Transient plant transformation mediated by agrobacterium tumefaciens: principles, methods and applications. Biotechnol Adv 33:1024–1042. https://doi.org/10.1016/j.biotechadv.2015.03.012
Li F, Guo S, Zhao Y, Chen D, Chong K, Xu Y (2010) Overexpression of a homopeptide repeat-containing bHLH protein gene (OrbHLH001) from dongxiang wild rice confers freezing and salt tolerance in transgenic Arabidopsis. Plant Cell Rep 29:977–986. https://doi.org/10.1007/s00299-010-0883-z
Li HM, Sun JQ, Xu YX, Jiang HL, Wu XY, Li CY (2007) The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol Biol 65:655–665. https://doi.org/10.1007/s11103-007-9230-3
Li Z, Liu C, Zhang Y, Wang B, Ran Q, Zhang J (2019) The bHLH family member ZmPTF1 regulates drought tolerance in maize by promoting root development and abscisic acid synthesis. J Exp Bot 70:5471–5486. https://doi.org/10.1093/jxb/erz307
Liu F, Zhang H, Ding L, Soppe W, Xiang Y (2020) REVERSAL OF RDO5 1, a homolog of rice seed dormancy4, interacts with bHLH57 and controls ABA biosynthesis and seed dormancy in Arabidopsis. Plant Cell 32:1933–1948. https://doi.org/10.1105/tpc.20.00026
Liu J, Liu J, He M, Zhang C, Liu Y, Li X, Wang Z, Jin X, Sui J, Zhou W, Bu Q, Tian X (2023) OsMAPK6 positively regulates rice cold tolerance at seedling stage via phosphorylating and stabilizing OsICE1 and OsIPA1. Theor Appl Genet 137:10. https://doi.org/10.1007/s00122-023-04506-8
Liu W, Tai H, Li S, Gao W, Zhao M, Xie C, Li W (2014) BHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol 201:1192–1204. https://doi.org/10.1111/nph.12607
Msanne J, Lin J, Stone JM, Awada T (2011) Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234:97–107. https://doi.org/10.1007/s00425-011-1387-y
Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Transcriptional regulation of ABI3 and ABA responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol Biol 60:51–68. https://doi.org/10.1007/s11103-005-2418-5
Pham VN, Kathare PK, Huq E (2018) Phytochromes and phytochrome interacting factors. Plant Physiol 176:1025–1038. https://doi.org/10.1104/pp.17.01384
Song Y, Li S, Sui Y, Zheng H, Han G, Sun X, Yang W, Wang H, Zhuang K, Kong F, Meng Q, Sui N (2022) SbbHLH85, a bHLH member, modulates resilience to salt stress by regulating root hair growth in sorghum. Theor Appl Genet 135:201–216. https://doi.org/10.1007/s00122-021-03960-6
Sun X, Wang Y, Sui N (2018) Transcriptional regulation of bHLH during plant response to stress. Biochem Biophys Res Commun 503:397–401. https://doi.org/10.1016/j.bbrc.2018.07.123
Tran LP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481–2498. https://doi.org/10.1105/tpc.104.022699
Tripathy MK, Deswal R, Sopory SK (2021) Plant RABs: role in development and in abiotic and biotic stress responses. Curr Genomics 22:26–40. https://doi.org/10.2174/1389202922666210114102743
Xie ZL, Nolan TM, Jiang H, Yin YH (2019) AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front Plant Sci 10:228. https://doi.org/10.3389/fpls.2019.00228
Yan Z, Li K, Li Y, Wang W, Leng B, Yao G, Zhang F, Mu C, Liu X (2023) The ZmbHLH32-ZmIAA9-ZmARF1 module regulates salt tolerance in maize. Int J Biol Macromol 253:126978. https://doi.org/10.1016/j.ijbiomac.2023.126978
Yang T, Yao S, Hao L, Zhao Y, Lu W, Xiao K (2016) Wheat bHLH-type transcription factor gene TabHLH1 is crucial in mediating osmotic stresses tolerance through modulating largely the ABA-associated pathway. Plant Cell Rep 35:2309–2323. https://doi.org/10.1007/s00299-016-2036-5
Yue MF, Zhang C, Zheng DY, Zou HW, Wu ZY (2022) Response of maize transcriptional factor ZmbHLH91 to abiotic stress. Acta Agron Sin 48:3004–3017
Zhang C, Li X, Wang Z, Zhang Z, Wu Z (2020) Identifying key regulatory genes of maize root growth and development by RNA sequencing. Genomics 112:5157–5169. https://doi.org/10.1016/j.ygeno.2020.09.030
Zhang H, Liu S, Ren T, Niu M, Liu X, Liu C, Wang H, Yin W, Xia X (2023) Crucial abiotic stress regulatory network of NF-Y transcription factor in plants. Int J Mol Sci 24:4426. https://doi.org/10.3390/ijms24054426
Acknowledgements
We would like to express our gratitude to Liu Chen-Ming from our laboratory for his assistance during the course of this study, as well as his careful review and valuable comments during the writing of the first draft of this paper. This work was supported by the National Natural Science Foundation of China (Grant Nos. 32001430, 32171952, 31971839).
Author information
Authors and Affiliations
Contributions
ZW and CZ conceived the original idea, designed the experiments, and provided overall supervision of the work; ZD, KZ, DZ, and SG performed the experiments; ZD, HZ, SG, and CZ analyzed and interpreted the data; ZD and CZ wrote the manuscript. All authors read and approved the final manuscript. ZD, KZ, and DZ contributed equally to this work.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Data availability
All data are available from the corresponding author upon reasonable request.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dai, Z., Zhao, K., Zheng, D. et al. Heterologous expression of the maize transcription factor ZmbHLH36 enhances abiotic stress tolerance in Arabidopsis. aBIOTECH (2024). https://doi.org/10.1007/s42994-024-00159-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42994-024-00159-3