Abstract
Flowering time (or heading date) is an important agronomic trait that determines the environmental adaptability and yield of many crops, including rice (Oryza sativa L.). Hd3a BINDING REPRESSOR FACTOR 1 (HBF1), a basic leucine zipper transcription factor, delays flowering by decreasing the expression of Early heading date 1 (Ehd1), Heading date 3a (Hd3a), and RICE FLOWERING LOCUS T 1 (RFT1), but the underlying molecular mechanisms have not been fully elucidated. Here, we employed the hybrid transcriptional factor (HTF) strategy to enhance the transcriptional activity of HBF1 by fusing it to four copies of the activation domain from Herpes simplex virus VP16. We discovered that transgenic rice lines overexpressing HBF1-VP64 (HBF1V) show significant delays in time to flower, compared to lines overexpressing HBF1-MYC or wild-type plants, via the Ehd1–Hd3a/RFT1 pathway, under both long-day and short-day conditions. Transcriptome deep sequencing analysis indicated that 19 WRKY family genes are upregulated in the HBF1V overexpression line. We demonstrate that the previously unknown gene, OsWRKY64, is a direct downstream target of HBF1 and represses flowering in rice, whereas three known flowering repressor genes, Days to heading 7 (DTH7), CONSTANS 3 (OsCO3), and OsWRKY104, are also direct target genes of HBF1 in flowering regulation. Taking these results together, we propose detailed molecular mechanisms by which HBF1 regulates the time to flower in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The timing of flowering is coordinately controlled by endogenous and environmental factors, such as photoperiod, temperature, nutrient availability, phytohormones, and plant age (Quiroz et al. 2021). As rice is a facultative short-day (SD) plant, photoperiod-mediated flowering (Oryza sativa L.) is critical for its regional adaptation and yield. Transcriptional and post-transcriptional components regulating rice photoperiodic flowering have been extensively investigated. Two major pathways with cross-connections have been identified: the evolutionarily conserved OsGIGANTEA (OsGI)–Heading date 1 (Hd1)–Heading date 3a (Hd3a)–RICE FLOWERING LOCUS T 1 (RFT1) pathway, analogous to Arabidopsis (Arabidopsis thaliana) GI–CONSTANS (CO)–FLOWERING LOCUS T (FT) pathway; and the rice-specific Grain number, plant height, and heading date 7 (Ghd7)–Early heading date 1 (Ehd1)–Hd3a/RFT1 pathway (Zhou et al. 2021). The two pathways are integrated at the level of the florigen genes Hd3a and RFT1 to modulate flowering. Hd3a and RFT1, from the phosphatidylethanolamine-binding protein (PEBP) family, are close homologs of Arabidopsis FT (Kojima et al. 2002). Although Hd3a and RFT1 are expressed in leaves, their protein products are delivered to the shoot apical meristem (SAM) through the phloem, where they activate the transcription of downstream floral identity genes and trigger the transition to flowering (Tamaki et al. 2007). They exhibit distinct functions, with Hd3a inducing flowering under SD conditions, while RFT1 promotes flowering under long-day (LD) conditions (Komiya et al. 2009).
Most flowering-related factors act upstream of Hd3a or RFT1, influencing flowering time by regulating Hd3a or RFT1 expression. OsGI encodes an ortholog of Arabidopsis GI that activates Hd1 transcription in the conserved pathway (Huang et al. 2022). The loss of OsGI function results in a late-flowering phenotype under SD conditions, but the effect is weak under LD conditions (Lee et al. 2016). Hd1 exerts a dual function in controlling rice flowering, suppressing flowering under LD conditions but promoting it under SD conditions by regulating Hd3a expression (Turck et al. 2008). Ghd7 and Ehd1 are specific to monocots and have no clear orthologs in Arabidopsis (Zhang et al. 2021). Ehd1 functions as an activator of flowering under both LD and SD conditions and acts upstream of Hd3a and RFT1 independently of Hd1 (Zhou et al. 2021). Ghd7 suppresses Ehd1 expression, thereby repressing the floral transition (Xue et al. 2008). Furthermore, Ghd7 interacts with Hd1 to form a complex that can repress Ehd1 transcription through binding to cis-regulatory sequences in the Ehd1 promoter (Zhang et al. 2017b). As these genes are at the core of flowering regulation in rice, their transcript levels offer convenient tools to monitor the state of the flowering pathways in this crop when dissecting its multiple underlying regulatory layers.
In addition to these key flowering genes, multiple flowering regulators have been identified in rice. Days to heading 7 (DTH7, also named PSEUDO-RESPONSE REGULATOR 37, OsPRR37) encodes a pseudo-response regulator that downregulates Ehd1 and Hd3a expression and results in a late-flowering phenotype under LD conditions. Many European and Asian rice cultivars from higher latitudes harbor nonfunctional DTH7 alleles and have an early-flowering phenotype (Koo et al. 2013). Independently of the Ehd1 pathway, another flowering repressor, the CONSTANS-LIKE (COL) protein OsCO3, controls flowering time under SD conditions by negatively regulating the expression of Hd3a (Kim et al. 2008). Many transcription factors are also involved in rice flowering regulation. OsWRKY11 (also named semi-dwarf and late flowering 1, Dlf1) acts as a transcriptional activator and regulates flowering by downregulating Ehd2 expression (Cai et al. 2014). Similarly, OsWRKY104 suppresses Ehd1 expression and confers a late-flowering phenotype in rice (Zhang et al. 2016).
The basic leucine zipper (bZIP) family of transcription factors (TF) play diverse roles in rice development, including abiotic stress responses, light signal transduction, flower development, pathogen defense, and seed maturation. Little is known, however, about their roles in rice flowering (Zong et al. 2020). OsFD1, the counterpart of Arabidopsis FD in rice, forms a florigen activation complex (FAC) with Hd3a and 14-3-3 proteins in the SAM, thus promoting flowering by inducing the expression of downstream MADS-box transcription factor genes (Taoka et al. 2011). Many other bZIP TFs can form alternative FACs by replacing OsFD1 in the complex and regulate flowering. For instance, the bZIP TF Hd3a BINDING REPRESSOR FACTOR 1 (HBF1, also called bZIP42) forms a FAC by substituting for OsFD1, acting as a suppressor of rice flowering (Brambilla et al. 2017). HBF1 physically interacts with Hd3a and HBF1 overexpression decreased the expression of Ehd1, Hd3a and RFT1 in leaves, causing delayed flowering (Brambilla et al. 2017). These studies indicate that Hd3a-mediated transcriptional activation or repression complexes can regulate rice flowering via other bZIP TFs. Our previous study revealed that the bZIP family members ABA RESPONSIVE ELEMENT BINDING FACTOR 1 (OsABF1) and its closest homolog OsbZIP40 suppress the floral transition by activating OsWRKY104 transcription in a photoperiod-independent manner (Zhang et al. 2016). Recently, bZIP71 was demonstrated to delay flowering by suppressing Ehd1 expression in rice (Li et al. 2022).
In this study, we explored the molecular mechanisms underlying HBF1-mediated regulation of time to flower in rice. Transcriptome deep sequencing (RNA-seq) analysis showed that many WRKY family genes are upregulated upon overexpression of HBF1V, encoding a fusion between HBF1 and four copies of the VP16 activation domain. Among them, we focused here on the flowering repressor OsWRKY64 as a direct downstream target of HBF1. In addition, we demonstrate that HBF1 can directly activate the transcription of multiple other flowering repressor genes, including DTH7, OsCO3, and OsWRKY104, to inhibit rice flowering through both Ehd1-dependent and -independent pathways.
Results
Hybrid transcription factor HBF1-VP64 delays flowering time in rice
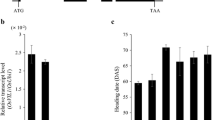
In the hybrid transcription factor (HTF) system, the sequence encoding VP64 (tetrameric repeats of the VP16 activation domain from Herpes simplex virus) is cloned in-frame with the coding sequence of the TF gene of interest. This method has been demonstrated to be effective for the study of TFs in rice and other species (Zhao et al. 2015). In our study, we obtained eight transgenic lines constitutively expressing HBF1-VP64 in rice under the control of the maize (Zea mays) UBIQUITIN1 promoter (Ubipro) through Agrobacterium (Agrobacterium tumefaciens)-mediated transformation (Fig. 1A, upper panel); these Ubipro:HBF1-VP64 lines will be referred to as HBF1V thereafter. We established that all positive HBF1V lines delay flowering time under natural day (ND, summer in Beijing) conditions, in particular lines HBF1V-1 and HBF1V-2, which we chose for characterization. Reverse-transcription quantitative PCR (RT-qPCR) analysis determined that HBF1 expression in the HBF1V-1 and HBF1V-2 lines is higher than that in wild type (WT, Kita-ake) (Fig. 1B). To precisely assess their flowering time, we grew WT, HBF1V-1, and HBF1V-2 plants under long-day (LD, 14 h light/ 10 h dark), short-day (SD, 10 h light/14 h dark), and ND conditions and counted the number of days from sowing to heading as flowering time. We determined that the HBF1V-1 and HBF1V-2 lines both flower significantly later than WT under LD, SD, and ND conditions (Fig. 1C and D).
Overexpression of HBF1V or HBF1M results in the late flowering phenotype. A Diagram of the HBF1V and HBF1M constructs. HBF1V, Ubipro:HBF1-VP64; HBF1M, 35Spro:HBF1-MYC. B Expression of HBF1 in indicated lines by reverse-transcription quantitative PCR (RT-qPCR). C Representative flowering image of indicated genotypes under natural day (ND) conditions in Beijing. D Flowering time of each genotype under LD or SD conditions. LD, long-day (14 h light/10 h dark), SD, short day (10 h light/14 h dark). Data were means ± s.d. (standard deviations, n = 20). The data of each genotype were compared with wild type (WT, Kita-ake) (Student’s t tests, **P < 0.01, n = 20)
To elucidate the function of HBF1 in regulating time to flower, we generated the construct 35Spro:HBF1-MYC (HBF1M) and obtained multiple transgenic lines through Agrobacterium-mediated transformation (Fig. 1A, lower panel). We confirmed the high expression of HBF1 in these transgenic plants by RT-qPCR; we chose the two independent HBF1M overexpression lines HBF1M-1 and HBF1M-2, with similar HBF1 expression levels to HBF1V lines (Fig. 1B). Although the overexpression of HBF1M significantly delayed flowering time, this effect was less pronounced than that seen for HBF1V lines under LD, SD, and ND conditions, indicating that HBF1V exerts a stronger flowering repressor activity than HBF1M in these transgenic plants (Fig. 1C and D). We conclude that HBF1 is a negative regulator of rice flowering time, with HBF1V exerting a stronger delaying effect on flowering than HBF1M.
To explore the quantitative difference in flowering time shown by the HBF1V and HBF1M lines, we examined the transcriptional activity of HBF1M and HBF1V proteins in a yeast (Saccharomyces cerevisiae) transcriptional activation assay. To this end, we fused HBF1V or HBF1M to the DNA-binding domain of yeast GAL4 (BD) and transformed the encoding constructs individually into a yeast strain. We observed that HBF1 exhibits transcriptional activation activity, whereas the negative control (the GAL4 BD alone) did not. Moreover, HBF1V had stronger transcriptional activation activity than HBF1M (Fig. 2), indicating that the addition of the VP64 domain markedly increased the transcriptional activation activity of HBF1. Combined with the flowering phenotype reporter above, these results indicate that the stronger transcriptional activation activity of HBF1V is likely responsible for the later-flowering phenotype of HBF1V transgenic lines relative to HBF1M lines.
Yeast transcriptional activation activity assay of HBF1 protein. (Left panel) Diagrams of yeast bait constructs containing HBF1-MYC (HBF1M) and HBF1-VP64 (HBF1V). DNA-binding domain of yeast GAL4 (BD) and BD fused with DST were used as negative and positive controls respectively. (Middle panel) Plate auxotroph assays showed transcriptional activation activity of each protein. (Right panel) Quantitative the transcriptional activation activity of each protein. β-galactosidase activity data were means of ± s.d. (Student’s t tests, **P < 0.01, n = 3)
HBF1 delays flowering partially through the Ehd1 pathway
A previous study showed that HBF1 transcripts are highly abundant in the SAM and leaves (Brambilla et al. 2017). We monitored HBF1 expression in various tissues of Kita-ake using RT-qPCR, which revealed that HBF1 is expressed in all tissues, with high expression in leaves and sheaths (Fig. S1A). Consistent with these results, we detected strong β-glucuronidase (GUS) staining in the leaves, sheaths, stems, roots and panicles of transgenic plants harboring the HBF1pro:GUS reporter construct (Fig. S1B). As the diurnal expression pattern of flowering genes is critical to their role, we measured the expression of four key flowering genes (Ehd1, RFT1, Hd3a, and Hd1) in leaves over one diurnal cycle by collecting samples every 4 h. We determined that the expression of Ehd1, RFT1, and Hd3a is significantly downregulated in the HBF1V-2 line compared to WT under LD and SD conditions, while Hd1 expression showed little difference from WT (Fig. 3A and B). We examined the expression of another 12 flowering-time-related genes in rice, but detected no significant differences between WT and HBF1V-2 under either LD or SD conditions (Fig. S2 and S3). These results demonstrate that HBF1V delays flowering time through lowering the expression of Ehd1, Hd3a, and RFT1 in leaves.
HBF1 delays floral initiation incompletely dependent on Ehd1 pathway. A, B Expression analysis of Ehd1, Hd3a, RFT1 and Hd1 in WT and HBF1V-2 plants. Plants were grown under LD (A) or SD (B) conditions for 4 weeks, newly expended leaves were collected every 4 h from the beginning of the light period for RNA extraction. Three biological replicates were performed, and Ubiquitin (UBQ) was used as internal control. Data were means ± s.d. (n = 3). C Phenotypes of indicated genotypes at heading stage grown under ND conditions. Ehd1-1 and Ehd1-2 were generated by transformation of the Ubipro:Ehd1-FLAG plasmids into wild-type. HBF1V Ehd1-1 and HBF1V Ehd1-2 were two independent T1 lines generated by transformation of Ubipro:Ehd1-FLAG construct into the HBF1V-2 homozygotes line. D Statistical analysis of flowering time in indicated genotypes. Data of each genotype were compared with WT (Student’s t tests, **P < 0.01, n = 20). E Western blot detects HBF1V or Ehd1-FLAG in each genotype by using anti-VP16 or anti-FLAG antibody respectively. The ponceau S staining band of Rubisco large subunit was used as loading control
To analyze the genetic relationship between HBF1 and Ehd1, we generated the Ubipro:Ehd1-FLAG construct and transformed it into WT and HBF1V-2. We obtained several transgenic lines, from which we chose four overexpression lines, named Ehd1-1, Ehd1-2, HBF1V Ehd1-1, and HBF1V Ehd1-2, for characterization. We observed a late-flowering phenotype for HBF1V Ehd1 overexpression lines and WT compared to Ehd1 overexpression lines under LD conditions (Fig. 3C and D). We confirmed the accumulation of the respective tagged proteins in each transgenic line by immunoblotting with anti-FLAG and anti-VP64 antibodies (Fig. 3E). Thus, molecular and genetic lines of evidence indicate that HBF1 delays flowering in a manner partially dependent on the Ehd1 pathway.
HBF1 inhibits rice flowering by directly activating OsWRKY64 transcription
Although HBF1 was previously shown to bind to the Ehd1 promoter (Brambilla et al. 2017), Ehd1 was expressed at lower levels in the HBF1 overexpression lines relative to WT (Fig. 3A). Since HBF1 is proposed to work as a transcriptional activator, we suspected that other downstream genes of HBF1 may function upstream of Ehd1. To uncover the downstream targets of HBF1, we performed RNA-seq using leaves of 4-week-old WT and HBF1V-2 seedlings. We identified differentially expressed genes (DEGs) in HBF1V seedlings relative to WT with cutoffs of fold-change ± 2, P-value < 0.01, and false discovery rate (FDR) < 0.01. A volcano plot of these DEGs indicated that 1292 genes are upregulated and 797 genes are downregulated in HBF1V seedlings compared to WT (Fig. 4A and Table S2). We randomly selected 20 upregulated DEGs for validation by RT-qPCR (Fig. S4). A careful inspection of the RNA-seq data revealed that 19 out of 129 WRKY family genes in japonica rice are upregulated in HBF1V-2 seedlings (Table S3). A phylogenetic analysis of the proteins encoded by all upregulated WRKY genes in HBF1V-2 using MEGA5.2 showed that OsWRKY64 is the closest homolog to OsWRKY104 (Fig. 4B). Although OsWRKY104 has been identified as a direct target of OsABF1 (Zhang et al. 2016), the function of e genes in flowering regulation remains elusive due to the lack of the loss-of-function mutants of these genes, which prompted us to investigate the role of OsWRKY64 in the regulation of flowering.
The 19 WRKY genes are up-regulated significantly in RNA-seq data. A Volcano plot displays differentially expressed genes between WT and HBF1V-2 (Fold Change (FC) > 2, P value < 0.01, and False Discovery Rate (FDR) < 0.01). WT and HBF1V-2 transgene lines were grown under continuous light in plant growth chamber at 28 °C for 4 weeks. Ten newly expended leaves were collected to extract total RNA and three biological replicates were performed to verify the RNA-sequence results. B Phylogenetic tree of the 19 WRKY genes that up-regulated in HBF1V-2 (Left panel). Protein sequences were downloaded from the MSU Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/analyses_search_locus.shtml) databases and then used for Neighbor-joining phylogenetic analysis (MEGA5.2). Fold change analysis of 19 WRKY genes in HBF1V compared with those in WT (Right panel)
RT-qPCR analysis verified that OsWRKY64 transcript abundance is higher in HBF1V-2 seedlings than in WT under both LD and SD conditions (Fig. 5A and B). Consistent with this finding, we identified three ABRE (ABA-response element, ACGT-containing sequences) cis-elements, known binding elements for bZIP TFs, within the OsWRKY64 promoter sequence. We therefore investigated whether HBF1 might bind to the OsWRKY64 promoter by performing an in vivo chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) experiment using WT Kita-ake seedlings and an anti-HBF1 antibody (Fig. 5C). Indeed, we observed robust binding of HBF1 to the b and c sites in the OsWRKY64 promoter (Fig. 5C). Transient expression assays in Nicotiana benthamiana leaves revealed that HBF1 can increase firefly luciferase (LUC) activity derived from a OsWRKY64pro:LUC reporter construct (Fig. S5), suggesting that HBF1 positively regulates OsWRKY64 transcription. To explore the function of OsWRKY64 in rice, we generated OsWRKY64 overexpression and RNA interference (RNAi) lines. We chose the two independent transgenic Ubipro:OsWRKY64-FLAG-OX (OX#1 and OX#2) lines with increased accumulation of OsWKRY64-FLAG protein, as well as two OsWRKY64-RNAi lines with lower OsWRKY64 transcript levels, for further analysis (Fig. 5D and E). The two OsWRKY64 overexpression lines flowered late, while the two RNAi lines flowered early, compared to WT when grown under LD, SD, or ND conditions (Fig. 5F and G). These results suggest that OsWRKY64 is a direct target gene of HBF1 and inhibits flowering in rice.
OsWRKY64 is direct target of HBF1. A, B Expression analysis of OsWRKY64 in each genotype under LD (A) or SD (B) conditions using RT-qPCR. Three biological replicates were performed and the UBQ were used as internal control. Data were means ± s.d. (Student’s t tests, **P < 0.01, n = 3). C Verification of the binding sites of HBF1 in OsWRKY64 promoter by ChIP-qPCR. ChIP samples were collected from Kita-ake plants and precipitated with anti-HBF1 antibody. qPCR data were normalized to the input signal. The binding of HBF1 to 25S rDNA was used as negative control. Data were means ± s.d. (n = 3, Student’s t tests, **P < 0.01). The promoter diagram of OsWRKY64 is shown in the boxes. The short lines above a, b or c represent the distribution of PCR fragments on promoter region. The dots indicate the position of ACGT core sequence, triangle indicates the position of translation start site (TSS), + 1 represents the position of start codon ATG. D Western blot analysis of OsWRKY64-FLAG in indicated genotypes by using anti-FLAG antibody. The ponceau S staining bands of Rubisco large subunit was used as loading control. E RT-qPCR analysis of OsWRKY64 expression in indicated lines. Data were means ± s.d. (Student’s t tests, **P < 0.01, n = 3). F Flowering image of each genotype under ND conditions in Beijing. OX, Ubipro:OsWRKY64-FLAG; RNAi, OsWRKY64 RNAi. G Statistical analysis of flowering time in each genotype under LD, SD, and ND conditions, data were means ± s.d. (Student’s t tests, **P < 0.01, n ≥ 12)
HBF1 is a transcriptional activator of DTH7, OsCO3, and OsWRKY104
From the RNA-seq dataset, we noticed that three flowering repressors, DTH7, OsCO3, and OsWRKY104, were upregulated in the HBF1V-2 line (Table S2). To explore this observation, we conducted an RT-qPCR analysis over a diurnal time course under LD and SD conditions. The expression of OsCO3 and OsWRKY104 was upregulated in HBF1V-2 compared to WT during the daytime under both LD and SD conditions, whereas DTH7 was upregulated in LD only but not in SD, suggesting that HBF1 might act as a direct activator of DTH7, OsCO3, and OsWRKY104 to regulate flowering time in rice (Fig. 6A–C). To test this hypothesis, we inspected the promoter sequences of DTH7, OsCO3, and OsWRKY104, which revealed ABRE (ACGT-containing sequences) cis-elements (Fig. 6D–F, insets). We performed ChIP-qPCR to test the direct interactions between HBF1 and these three promoters. We established that HBF1 can specifically bind to all three promoters, but not to the 25S rDNA locus, which was used as negative control (Fig. 6D–F). Transient expression assays in N. benthamiana leaves revealed that the presence of HBF1 results in higher LUC activity from the DTH7pro:LUC, OsCO3pro:LUC, and OsWRKY104pro:LUC reporter constructs (Fig. S5), suggesting that HBF1 can positively regulate their transcription. These results support the notion that HBF1 directly activates the transcription of DTH7, OsCO3, and OsWRKY104 in the context of flowering regulation.
HBF1 is transcriptional activator of DTH7, OsCO3, and OsWRKY104. A–C, RT-qPCR analysis of the dynamic expression of DTH7 (A), OsCO3 (B), and OsWRKY104 (C) in each genotype under LD or SD conditions. Three biological replicates were performed and UBQ was used as internal control. D–F Verification of HBF1 binding sites in DTH7 (D), OsCO3 (E), and OsWRKY104 (F) promoters by ChIP-qPCR. ChIP samples were collected from Kita-ake plants and precipitated with anti-HBF1 antibody. qPCR data were normalized to the input signal. The binding of HBF1 to 25S rDNA was used as negative control. Data were means ± s.d. (n = 3, Student’s t tests, **P < 0.01). The promoter diagrams of DTH7, OsCO3, and OsWRKY104 were shown in the boxes. The short lines under or above a, b or c represent the distribution of PCR fragments, and the ACGT core sequence were indicated by black dots. Triangle indicates the position of TSS, + 1 represents the first nucleotide of start codon ATG
Discussion
HBF1V confers an enhanced function to HBF1 in rice flowering
Although bZIP TFs are important in various biological programs, especially in abiotic stress tolerance, in rice, little is known about their involvement in flowering. HBF1/OsbZIP42 was reported to form transcriptional repressive complexes with Hd3a to regulate the expression of Ehd1, Hd3a, and RFT1 in leaves (Brambilla et al. 2017). However, few direct target genes of HBF1 have been identified in rice beside Ehd1. In this study, we demonstrated using the HTF strategy that HBF1 represses flowering in rice (Zhao et al. 2015). The VP64 fragment fused to HBF1 enhanced its transcriptional activity; HBF1V lines exhibited a greater delay in their flowering compared to HBF1M and WT, indicating that the HTF strategy was effective in enhancing the function of HBF1 in flowering regulation of rice. Our study also confirmed that the HTF strategy is a very effective method to study TFs (Zhang et al. 2016).
HBF1 regulates flowering through Ehd1-dependent and -independent pathways
Ehd1 encodes a B-type response regulator that promotes flowering by inducing the expression of Hd3a and RFT1. In this study, molecular and genetic evidence support the idea that HBF1 partially depends on the Ehd1 pathway to repress flowering time, which is consistent with a previous study (Brambilla et al. 2017). Indeed, we showed two flowering repressor genes, DTH7 and OsCO3, to be direct targets of HBF1 (Fig. 7). DTH7 mainly acts as a suppressor of Ehd1 in an LD-dependent manner (Gao et al. 2014). The wild-type background used in our study, Kita-ake, is thought to harbor nonfunctional alleles of several flowering genes, including DTH7, with the polymorphisms in DHT7 causing changes in conserved amino acids that may affect DTH7 function. Notably, DTH7 transcript levels in Kita-ake were not lower than those of cultivars carrying a fully functional DTH7 allele (Gao et al. 2014), indicating that Kita-ake may carry a weak DTH7 allele rather than a loss-of-function allele. Therefore, our study raises the possibility that DTH7 is a direct target of HBF1 in rice to regulate flowering. OsCO3 regulates the expression of Hd3a but not Ehd1, to delay flowering under SD conditions (Kim et al. 2008), offering evidence that HBF1 functions in rice flowering independently of the Ehd1 pathway. Additionally, our study suggests that HBF1 regulates rice flowering through distinct pathways under LD and SD conditions.
WRKY genes mediate the regulation of flowering time in rice
In rice, few WRKY TFs have been reported to participate in flowering time, with the exceptions of OsWRKY11 and OsWRKY104. However, the function of WRKY genes in flowering remains elusive due to the lack of the loss-of-function mutants. In our study, RNA-seq analysis indicated that 19 WRKY family genes were upregulated in HBF1V-2 plants. Among these genes, we established that OsWRKY64 transcription is directly activated by HBF1 and represses rice flowering. Knockdown of OsWRKY64 by RNA interference significantly promoted flowering time in rice. Therefore, our results provide evidence in support for the important roles of WRKY members in rice flowering.
The relationship between HBF1 and OsABF1 in rice flowering
bZIP TFs can form homo- or heterodimers to bind to their cognate DNA sequence. In our previous work, we showed that HBF1 interacted with OsABF1 in Arabidopsis protoplasts (Fig. S6A), indicating that HBF1 and OsABF1 may form a heterodimer to target their candidate genes. In the RNA-seq dataset generated here, 45% of all genes upregulated in HBF1V were also more highly expressed in OsABF1V than in WT (Fig. S6B, Table S4), suggesting that HBF1 and OsABF1 might regulate the expression of the same genes during plant development. During rice flowering, both HBF1 and OsABF1 bound to the promoters of OsCO3, DTH7, OsWRKY64, and OsWRKY104 (Fig. S6C). The transcript level of OsWRKY64 was upregulated in OsABF1V under SD but not LD conditions (Fig. S6D). However, the expression of DTH7 and OsCO3 was unchanged in OsABF1V transgenic lines (Fig. S6E and F), suggesting that HBF1 may target specific flowering-related genes, or share the same set of target genes as OsABF1 to regulate flowering time in rice.
Materials and methods
Generation of transgenic rice
To generate HBF1V overexpression lines, HBF1 cDNA was inserted into pBCV (constructed by our lab) expression vector using the Gateway cloning system (Zhao et al. 2015). For HBF1M overexpression lines, the HBF1 cDNA driven by 35S promoter was cloned into binary vector pCAMBIA1390 (reconstructed by our lab) using the Gateway cloning system. To generate Ehd1 and OsWRKY64 overexpression lines, their cDNA was inserted into the pHCF (constructed by our lab) vector at PstI site using the Infusion system (Clontech) (Zhang et al. 2016). To generate OsWRKY64-RNAi plants, a 266-bp fragment of the OsWRKY64 gene was inserted into the pANDA vector using the Gateway cloning system. The constructs were introduced into Kita-ake rice (Oryza sativa japonica) or indicated background by Agrobacterium tumefaciens-mediated transformation (Hiei et al. 1994).
Growth conditions
For RT-qPCR, RNA-seq, and ChIP assays, seeds of wild-type rice and transgenic lines were germinated for 2 d on wet filter paper in petri dishes at 37 °C. The uniformly germinated seeds were picked up and sown in bottomless 96-well plates and hydroponically grown (distiller water with 1/10 Murashige and Skoog). To investigate the flowering phenotypes, all plants were grown under natural day (ND) conditions in Beijing (39°54′N, 116°23′E), China, or under long-day (LD) (14 h light, 28 °C; 10 h dark, 24 °C) conditions or short-day (SD) (10 h light, 28 °C; 14 h dark, 24 °C) conditions in plant growth chambers.
Yeast transcriptional activation activity assay
To test the transcriptional activation activity, the indicated CDS was fused with GAL4 DNA-binding domain in the pGBKT7 vectors using Infusion system (Clotech) and transformed into the yeast strain AH109. The empty vector (BD) and BD-DST vector were used as negative and positive controls, respectively. Measurement of the β-galactosidase activity and the colony-lift filter assay were performed according to the Yeast Protocols Handbook (Clontech) using chlorophenol red-β-d-galactopyranoside (CPRG, Roche Biochemical) or X-gal (Inalco, Cat.# 1758-0300) as the substrate.
Chromatin immunoprecipitation (ChIP) assay
The HBF1V-2 or WT plants under continuous light (CL) were used for ChIP assays. ChIP was performed as described previously (Zhang et al. 2016). Briefly, 3 g of leaves from 4-week-old seedlings were cross-linked by 1% formaldehyde under vacuum for 15 min twice. Then the samples were ground to powder in liquid nitrogen prior to isolating chromatin. After sonicated, the chromatin complexes were incubated with anti-VP16 or anti-HBF1 antibody as described. The precipitated DNA was recovered in water for quantitative real-time PCR. The enrichment value was normalized to that of input DNA (% of input).
RNA-seq and data analysis
WT and HBF1V-2 lines were cultivated under continuous light at 28 °C for 4 weeks in plant growth chamber. Ten latest fully expended leaves of each genotype were collected for total RNA extraction and three biological replicates were performed. The sequencing library was constructed following the manufacturer`s instructions, and then sequenced with Illumina HiSeq 2000 at ANOROAD company. Clean reads were mapped to the O. ssp. japonica genome reference by TopHat. The differentially expressed genes were analyzed by Cuffdiff (q < 0.05) based on FPKM (fragments per kilobase of exon model per million mapped fragments). Differentially expressed genes were defined as those with fold changes ≥ 2 or ≤ 0.5. The volcano plot and venn diagram were analyzed through Omicshare platflom from Gene Denovo Biotechnology Co. (https://www.omicshare.com/).
Gene expression analyses
To test the mRNA expression of flowering-associated genes in a time course manner under LDs or SDs, plants were grown for 4 weeks and samples were collected every 4 h from the beginning of the light period. RNA was isolated using TRIZOL (Invitrogen) and treated with DNase I (Invitrogen). The cDNA was synthesized from 3.0 μg total RNA using TransScript® II One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech). LightCycler 480 SYBR Green I Master (Roche) was used for the quantitative PCR reaction.
Immunoblot analyses
The anti-HBF1 polyclonal antibody was generated by inoculating rabbits with TF-His- HBF1 recombination protein (Bio-med). The anti-VP16 and anti-OsABF1 polyclonal antibodies were generated in previous study (Zhang et al. 2016). To extract the total protein for immunoblot, the young leaves were ground in liquid nitrogen and mixed with 5 × SDS-PAGE loading buffer [250 mM Tris (pH 6.8), 10% (w/v) SDS, 0.5% (w/v) bromphenol blue, 50% (v/v) glycerol, 5% (v/v) 2-mercaptoethanol], boiled for 5 min, and spun at 12,000 rpm for 5 min at room temperature. The supernatants were fractioned by 10% SDS-PAGE, and the membrane was probed with the indicated antibody.
GUS histochemical staining
To obtain HBF1pro:GUS transgenic plants, a 2221 bp promoter region of HBF1 was amplified from the genome of Nipponbare, and inserted into the HindIII and BamHI sites of the pCAMBIA3301-GUS vector. GUS histochemical staining assays were performed according to the previous method (Zou et al. 2015).
BiFC assays
The HBF1 or OsABF1 coding sequences were cloned into the pSPYNE (R) or the pSPYCE (MR) vector (Zou et al. 2015). The vectors were co-transformed into Arabidopsis mesophyll protoplasts and incubated overnight before observation. Fluorescence signals were visualized using a Leica TCS-SP4 confocal microscope.
Transient expression assay
To generate the OsWRKY64pro:LUC, DTH7pro:LUC, OsCO3pro:LUC, and OsWRKY64pro:LUC reporter constructs, ~ 2 kb promoters of these genes were cloned into the pGreenII 0800-LUC vector. The Renilla Luciferase (REN) gene under the control of 35S promoter in the pGreenII 0800-LUC vector was used as the internal control. The coding region of HBF1 was cloned into pGreen-35S:GFP vector to produce 35Spro: HBF1-GFP construct and used as an effector. These effector and reporter or the control was transformed individually into Agrobacterium tumefaciens strain GV3101. GV3101 cells harboring the indicated constructs were mixed at a ratio of 1:1 and introduced into N. benthamiana leaves. The LUC and REN activities were measured using the Dual-Luciferase Reporter Assay System under the manufacturers’ instructions. The LUC/REN ratio was presented with three biological replicates.
Primers and accession numbers
All the primers in this study were listed in Table S1. Sequence data from this article can be found in the MSU Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/analyses_search_locus.shtml) databases (Kawahara et al. 2013) under the following accession numbers: HBF1 (LOC_Os05g41070), OsABF1 (LOC_Os01g64730), Ehd1 (LOC_Os10g32600), Hd1 (LOC_Os06g16370), Hd3a (LOC_Os06g06320), RFT1 (LOC_Os06g06300), DTH7 (LOC_Os07g49460), OsCO3 (LOC_Os09g06464), OsWRKY104 (LOC_Os11g02520), and OsWRKY64 (LOC_Os12g02450).
Data availability
All relevant data are within the manuscript and its Supplementary files.
References
Brambilla V et al (2017) Antagonistic transcription factor complexes modulate the floral transition in rice. Plant Cell 29:2801–2816. https://doi.org/10.1105/tpc.17.00645
Cai Y et al (2014) Dlf1, a WRKY transcription factor, is involved in the control of flowering time and plant height in rice. PLoS ONE 9:e102529. https://doi.org/10.1371/journal.pone.0102529
Gao H et al (2014) Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc Natl Acad Sci U S A 111:16337–16342. https://doi.org/10.1073/pnas.1418204111
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282. https://doi.org/10.1046/j.1365-313x.1994.6020271.x
Huang X, Huang L, Zhao X, Jia J, Zhang G, Zhang M, Jiang M (2022) A J-Protein OsDjC46 interacts with ZFP36 to participate in ABA-mediated antioxidant defense in rice. Antioxidants. https://doi.org/10.3390/antiox11020207
Kawahara Y et al (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4. https://doi.org/10.1186/1939-8433-6-4
Kim SK, Yun CH, Lee JH, Jang YH, Park HY, Kim JK (2008) OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta 228:355–365. https://doi.org/10.1007/s00425-008-0742-0
Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43:1096–1105. https://doi.org/10.1093/pcp/pcf156
Komiya R, Yokoi S, Shimamoto K (2009) A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136:3443–3450. https://doi.org/10.1242/dev.040170
Koo BH et al (2013) Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol Plant 6:1877–1888. https://doi.org/10.1093/mp/sst088
Lee YS, Yi J, An G (2016) OsPhyA modulates rice flowering time mainly through OsGI under short days and Ghd7 under long days in the absence of phytochrome B. Plant Mol Biol 91:413–427. https://doi.org/10.1007/s11103-016-0474-7
Li X et al (2022) bZIP71 delays flowering by suppressing Ehd1 expression in rice. J Integr Plant Biol 64:1352–1363. https://doi.org/10.1111/jipb.13275
Quiroz S et al (2021) Beyond the genetic pathways, flowering regulation complexity in Arabidopsis thaliana. Int J Mol Sci. https://doi.org/10.3390/ijms22115716
Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316:1033–1036. https://doi.org/10.1126/science.1141753
Taoka K et al (2011) 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476:332–335. https://doi.org/10.1038/nature10272
Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59:573–594. https://doi.org/10.1146/annurev.arplant.59.032607.092755
Xue W et al (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genetics 40:761–767. https://doi.org/10.1038/ng.143
Zhang C et al (2016) A drought-inducible transcription factor delays reproductive timing in rice. Plant Physiol 171:334–343. https://doi.org/10.1104/pp.16.01691
Zhang Z et al (2017b) Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions. Sci Rep 7:5388. https://doi.org/10.1038/s41598-017-05873-1
Zhang J, Fan X, Hu Y, Zhou X, He Q, Liang L, Xing Y (2021) Global analysis of CCT family knockout mutants identifies four genes involved in regulating heading date in rice. J Integr Plant Biol 63:913–923. https://doi.org/10.1111/jipb.13013
Zhao T et al (2015) Using hybrid transcription factors to study gene function in rice. Sci China Life Sci 58:1160–1162. https://doi.org/10.1007/s11427-015-4937-x
Zhou S et al (2021) Transcriptional and post-transcriptional regulation of heading date in rice. New Phytol 230:943–956. https://doi.org/10.1111/nph.17158
Zong W, Yang J, Fu J, Xiong L (2020) Synergistic regulation of drought-responsive genes by transcription factor OsbZIP23 and histone modification in rice. J Integr Plant Biol 62:723–729. https://doi.org/10.1111/jipb.12850
Zou X et al (2015) Over-expression of an S-domain receptor-like kinase extracellular domain improves panicle architecture and grain yield in rice. J Exp Bot 66:7197–7209. https://doi.org/10.1093/jxb/erv417
Acknowledgements
This research was supported by grants from the National Natural Science Foundation of China (No. 31771758), and the National Transgenic Major Project of China (No. 2018ZX0800925B).
Author information
Authors and Affiliations
Contributions
CZ and JL conceived of and designed experiments and edited the manuscript; CL and LZ performed the experiments; XW, CY, BL, TZ and HL helped in performing the experiments and writing this manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, C., Zhang, L., Wang, X. et al. The transcription factor HBF1 directly activates expression of multiple flowering time repressors to delay rice flowering. aBIOTECH 4, 213–223 (2023). https://doi.org/10.1007/s42994-023-00107-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42994-023-00107-7