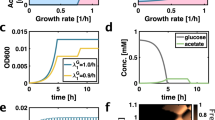
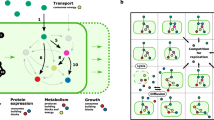
Abstract
The particular importance of evolutionary studies in microbial experimental systems is that starting from the level of the metabolism of individual cells, the adaptive dynamics can be followed step by step by biochemical, genetic, and population dynamical tools. Moreover, the coincidence of evolutionary and ecological time scales helps to clarify the mutual role of ecological and evolutionary principles in predicting adaptive dynamics in general. Ecological principles define the ecological conditions under which adaptive branching can occur. This paper overviews and interprets the results of empirical and modeling studies of the evolution of metabolic cross-feeding in glucose-limited E.coli chemostats and batch cultures in the context of theories of robust coexistence and adaptive dynamics. Empirical results consistently demonstrate that the interactions between cells are mediated by the changing metabolite concentrations in the cultures and modeling confirms that these changes may control the adaptive dynamics of the clones. In consequence, the potential results of evolution can be predicted at the functional level by evolutionary flux balance analysis (evoFBA), while the genetic changes are more contingent. evoFBA follows the scheme of adaptive dynamics theory by calculating the feedback environment that changes during the evolutionary process and provides a promising tool to further investigate adaptive divergence in small microbial communities. Three general conclusions close the paper.
Similar content being viewed by others
Availability of data and material
Not applicable.
Code availability
Not applicable.
References
Armstrong RA, McGehee R (1976) Coexistence of species competing for shared resources. Theor Popul Biol 9:317–328
Armstrong RA, McGehee R (1980) Competitive exclusion. Am Nat 115:151–170
Bajić D, Vila JCC, Blount ZD, Sánchez A (2018) On the deformability of an empirical fitness landscape by microbial evolution. Proc Nat Acad Sci 115:11286–11291
Barabás G, Meszéna G, Ostling A (2012) Community robustness and limiting similarity in periodic environments. Theor Ecol 5:265–282
Barrick JE, Lenski RE (2013) Genome dynamics during experimental evolution. Nat Rev Gen 14:827–839
Blount ZD, Borland CZ, Lenski RE (2008) Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Nat Acad Sci 105:7899–7906
Blount ZD, Barrick JE, Davidson CJ, Lenski RE (2012) Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 489:513–518
Brown JS (2016) Why Darwin would have loved evolutionary game theory. Proc R Soc B-Biol Sci 283:20160847
Csaba G (ed) (1978) A biológiai szabályozás (in Hungarian). Medicina Könyvkiadó, Budapest
Day T (2005) Modelling the ecological context of evolutionary change: deja vu or something new? In: Cuddington K, Beisner B (eds) Ecological paradigms lost routes of theory change. Elsevier, Amsterdam, pp 273–310
De Vos MG, Schoustra SE, De Visser JAG (2018) Ecology dictates evolution? About the importance of genetic and ecological constraints in adaptation (a). EPL (Europhys Lett) 122:58002
Doebeli M (2002) A model for the evolutionary dynamics of cross-feeding polymorphisms in microorganisms. Pop Ecol 44:59–70
D’Souza G, Shitut S, Preussger D, Yousif G, Waschina S, Kost C (2018) Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat Prod Rep 35:455–488
Dykhuizen DE (1998) Santa Rosalia revisited: Why are there so many species of bacteria? Ant Leeuw 73:25–33
Dykhuizen DE, Dean AM (2004) Evolution of specialists in an experimental microcosm. Genetics 167:2015–2026
Friesen ML, Saxer G, Travisano M, Doebeli M (2004) Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution 58:245–260
Geritz SAH, Metz JAJ, Kisdi É, Meszéna G (1997) Dynamics of adaptation and evolutionary branching. Phys Rev Lett 78:2024–2027
Geritz SAH, Kisdi É, Meszéna G, Metz JAJ (1998) Evolutionarily singular strategies and the adaptive growth and branching of the evolutionary tree. Evol Ecol 12:35–57
Good BH, McDonald MJ, Barrick JE, Lenski RE, Desai MM (2017) The dynamics of molecular evolution over 60,000 generations. Nature 551:45–50
Grainger TN, Levine JM, Gilbert B (2019) The invasion criterion: a common currency for ecological research. Trends Ecol Evol 34:925–935
Grosskopf T, Consuegra J, Gaffe J, Willison JC, Lenski RE, Soyer OS, Schneider D (2016) Metabolic modelling in a dynamic evolutionary framework predicts adaptive diversification of bacteria in a long-term evolution experiment. BMC Evol Biol 16:1–15
Gudelj I, Kinnersley M, Rashkov P, Schmidt K, Rosenzweig F (2016) Stability of cross-feeding polymorphisms in microbial communities. PLoS Comput Biol 12:e1005269
Hammerstein P (1996) Streetcar theory and long-term evolution. Science 273:1032–1032
Hans Metz JAJ (2011) Thoughts on the geometry of meso-evolution: collecting mathematical elements for a postmodern synthesis. In: Chalub FACC, Rodrigues JF (eds) The mathematics of Darwin’s legacy. Springer Basel, Basel, pp 193–231. https://doi.org/10.1007/978-3-0348-0122-5_11
Hansen SR, Hubbell SP (1980) Single-nutrient microbial competition: qualitative agreement between experimental and theoretically forecast outcomes. Science 207:1491–1493
Helling RB, Vargas CN, Adams J (1987) Evolution of Escherichia coli during growth in a constant environment. Genetics 116:349–358
Jacob F (1977) Evolution and tinkering. Science 196:1161–1166
Jiang L, Morin PJ (2004) Temperature-dependent interactions explain unexpected responses to environmental warming in communities of competitors. J Anim Ecol 73:569–576
Jiang L, Morin PJ (2007) Temperature fluctuation facilitates coexistence of competing species in experimental microbial communities. J Anim Ecol 76:660–668
Juhász-Nagy P, Vida G (1978) Szupraindividuális organizáció (in Hungarian). In: Csaba G (ed) A biológiai szabályozás. Medicina Könyvkiadó, Budapest, pp 337–406
Kauffman SA, Johnsen S (1991) Coevolution to the edge of chaos: coupled fitness landscapes poised states, and coevolutionary avalanches. J Theor Biol 149:467–505
Kinnersley MA, Holben WE, Rosenzweig F (2009) E Unibus Plurum: genomic analysis of an experimentally evolved polymorphism in Escherichia coli. PLoS Genet 5:e1000713
Kinnersley MA, Wenger J, Kroll E, Adams J, Sherlock G, Rosenzweig F (2014) Ex uno plures: clonal reinforcement drives evolution of a simple microbial community. PLoS Genet 10:e1004430
Kinnersley MA, Schwartz K, Yang DD, Sherlock G, Rosenzweig F (2021) Evolutionary dynamics and structural consequences of de novo beneficial mutations and mutant lineages arising in a constant environment. BMC Biol 19:1–21
Kisdi É, Meszéna G (1993) Density dependent life history evolution in fluctuating environments. In: Yoshimura J, Clark C (eds) Adaptation in stochastic environments. Springer, Berlin Heidelberg, pp 26–62
Laland KN, Odling-Smee J, Hoppitt W, Uller T (2013) More on how and why: cause and effect in biology revisited. Biol Phil 28:719–745
Le Gac M, Plucain J, Hindré T, Lenski RE, Schneider D (2012) Ecological and evolutionary dynamics of coexisting lineages during a long-term experiment with Escherichia coli. Proc Nat Acad Sci 109:9487–9492
Lewis NE, Hixson KK, Conrad TM, Lerman JA, Charusanti P, Polpitiya AD, Adkins JN, Schramm G, Purvine SO, Lopez-Ferrer D (2010) Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol Syst Biol 6:390. https://doi.org/10.1038/msb.2010.47
Lewontin RC (2010) Not so natural selection, the New York review of books. New York Review Books, New York
Lunzer M, Natarajan A, Dykhuizen DE, Dean AM (2002) Enzyme kinetics, substitutable resources and competition: from biochemistry to frequency-dependent selection in lac. Genetics 162:485–499
Meszéna G (2005) Adaptive dynamics: the continuity argument. J Evol Biol 18:1182–1185
Meszéna G, Gyllenberg M, Pásztor L, Metz JAJ (2006) Competitive exclusion and limiting similarity: a unified theory. Theor Popul Biol 69:68–87
Odling-Smee J, Laland KN, Feldman MW (2003) Niche construction: the neglected process in evolution. Princeton University Press, Princeton
Papp B, Notebaart RA, Pál C (2011) Systems-biology approaches for predicting genomic evolution. Nat Rev Gen 12:591–602
Pásztor E (1988) Unexploited dimensions of optimization life history theory. In: de Jong G (ed) Population genetics and evolution. Springer-Verlag, Berlin, Heidelberg, pp 19–32
Pásztor L, Botta-Dukát Z, Magyar G, Czárán T, Meszéna G (2016) Theory-based ecology: a Darwinian approach. Oxford University Press, Oxford
Pásztor L, Barabás G, Meszéna G (2020) Competitive exclusion and evolution: convergence almost never produces ecologically equivalent species: (A comment on mcpeek, “Limiting similarity? The ecological dynamics of natural selection among resources and consumers caused by both apparent and resource competition”). Am Nat 195:E112–E117
Plucain J, Hindré T, Le Gac M, Tenaillon O, Cruveiller S, Médigue C, Leiby N, Harcombe WR, Marx CJ, Lenski RE (2014) Epistasis and allele specificity in the emergence of a stable polymorphism in Escherichia coli. Science 343:1366–1369
Post DM, Palkovacs EP (2009) Eco-evolutionary feedbacks in community and ecosystem ecology: interactions between the ecological theatre and the evolutionary play. Philos Trans R Soc B Biol Sci 364:1629–1640
Rainey PB, Travisano M (1998) Adaptive radiation in a heterogeneous environment. Nature 394:69–72
Rainey PB, Remigi P, Farr AD, Lind PA (2017) Darwin was right: Where now for experimental evolution? Curr Op Gen Dev 47:102–109
Rosenzweig RF, Sharp RR, Treves DS, Adams J (1994) Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics 137:903–917
Rozen DE, Lenski RE (2000) Long-term experimental evolution in Escherichia coli. VIII. Dynamics of a balanced polymorphism. Am Nat 155:24–35
San Roman M, Wagner A (2018) An enormous potential for niche construction through bacterial cross-feeding in a homogeneous environment. PLoS Comp Biol 14:e1006340
San Roman M, Wagner A (2020) Acetate and glycerol are not uniquely suited for the evolution of cross-feeding in E. coli. PLoS Comp Biol 16:e1008433
Smith NW, Shorten PR, Altermann E, Roy NC, McNabb WC (2019) The classification and evolution of bacterial cross-feeding. Front Ecol Evol. https://doi.org/10.3389/fevo.2019.00153
Stewart FM, Levin BR (1973) Partitioning of resources and the outcome of interspecific competition: a model and some general considerations. Am Nat 107:171–198
Tilman D (1977) Resource competition between planktonic algae: an experimental and theoretical approach. Ecology 58:338–348
Tilman D (1982) Resource competition and community structure. Princeton University Press, Princeton
Treves DS, Manning S, Adams J (1998) Repeated evolution of an acetate-crossfeeding polymorphism in long-term populations of Escherichia coli. Mol Biol Evol 15:789–797
Turner PE, Souza V, Lenski RE (1996) Tests of ecological mechanisms promoting the stable coexistence of two bacterial genotypes. Ecology 77:2119–2129
Turner CB, Blount ZD, Lenski RE (2015) replaying evolution to test the cause of extinction of one ecotype in an experimentally evolved population. PLoS One 10:e0142050
Yang DD, Alexander A, Kinnersley M, Cook E, Caudy A, Rosebrock A, Rosenzweig F (2020) Fitness and productivity increase with ecotypic diversity among Escherichia coli strains that coevolved in a simple, constant environment. Appl Environ Microbiol 86:e00051-e120
Yao R, Hirose Y, Sarkar D, Nakahigashi K, Ye Q, Shimizu K (2011) Catabolic regulation analysis of Escherichia coli and its crp, mlc, mgsA, pgi and ptsG mutants. Microb Cell Fact 10:67. https://doi.org/10.1186/1475-2859-10-67
Acknowledgements
I’m grateful to Benjamin Good for raising my attention to the paper of Grosskopf et al. and Franz Rosenzweig for clarifying some points about their work. Géza Meszéna and two anonymous referees helped to improve the manuscript.
Funding
The author’s work was partly supported by NKFI (National Research, Development and Innovation Office) research Grant 123796.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pásztor, L. Population regulation and adaptive dynamics of cross-feeding. BIOLOGIA FUTURA 73, 393–403 (2022). https://doi.org/10.1007/s42977-022-00147-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42977-022-00147-y