Abstract
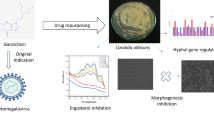
Candida albicans represents an interesting microorganism to study complex host-pathogen interactions and for the development of effective antifungals. Our goal was to assess the efficacy of 4-chloro-3-nitrophenyldifluoroiodomethyl sulfone (named Sulfone) against the C. albicans infections in the Galleria mellonella host model. We assessed invasiveness of CAI4 parental strain and mutants: kex2Δ/KEX2 and kex2Δ/kex2Δ in G. mellonella treated with Sulfone. We determined that KEX2 expression was altered following Sulfone treatment in G. mellonella-C. albicans infection model. Infection with kex2Δ/kex2Δ induced decreased inflammation and minimal fault in fitness of larvae vs CAI4. Fifty percent of larvae died within 4–5 days (P value < 0.0001) when infected with CAI4 and kex2Δ/KEX2 at 109 CFU/mL; survival reached 100% in those injected with kex2Δ/kex2Δ. Larvae treated with Sulfone at 0.01 mg/kg 30 min before infection with all C. albicans tested survived infection at 90–100% vs C. albicans infected-PBS-treated larvae. Hypersensitive to Sulfone, kex2Δ/kex2Δ reduced virulence in survival. KEX2 was down-regulated when larvae were treated with Sulfone: 30 min before and 2 h post-SC5314-wild-type infection respectively. kex2Δ/kex2Δ was able to infect larvae, but failed to kill host when treated with Sulfone. Sulfone can be used to prevent or treat candidiasis. G. mellonella facilitates studding of host-pathogen interactions, i.e., testing host vs panel of C. albicans mutants when antifungal is dosed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since incidences and frequency of the Candida albicans infections dominate over non-albicans species, there is still need to study the virulence of this pathogen, which represents an interesting window into the evolution of complex host-pathogen interactions and for the development of effective antifungal treatments [1,2,3]. The serine protease encoded by KEX2 is among the C. albicans’ virulence factors that mediate its success as a pathogen. It is a representative of subtilisin family of proteins, processes enzymes, and its critical role in virulence was demonstrated previously [4,5,6,7,8]. As this approach still needs further analyses, here we continue studies assessing the Galleria mellonella-C. albicans infection model and the efficiency of novel antimicrobial agent. Recent studies [9,10,11,12,13,14,15,16,17,18,19,20] increased interest in the G. mellonella larvae as an alternative in vivo model due to the immunological and developmental similarities between insects and mammals. Therefore, the results obtained using these insects can serve as a starting point to study the C. albicans pathogenesis and generate hypotheses to be further tested in vertebrate models.
Since candidiasis is difficult to eradicate with the existing antimycotics [21], a new antifungal compound overcomes these deficiencies. We found [8] that the 4-chloro-3-nitrophenyldifluoroiodomethyl sulfone (named below Sulfone) treatment reduced the C. albicans pathogenicity in G. mellonella. It was shown [8] that the Sulfone displayed the minimal fungicidal concentration (MFC) against C. albicans at 0.25 μg/mL in in vitro studies and it was non-toxic against G. mellonella (lethal dose > 16 μg/mL). However, the impact of the Sulfone on metabolic pathways’ inhibition in C. albicans is not well understood yet. Our goals were to assess the efficacy of the Sulfone against the C. albicans infections in the G. mellonella host model and to compare the KEX2 mutants’ susceptibility with the Sulfone in vivo. Additionally, we investigated whether the KEX2 expression plays a role in the virulence of C. albicans in G. mellonella. The role of KEX2 in virulence was tested by screening for attenuation in the C. albicans mutants. We determined in histopathological examinations whether the KEX2 mutations affected the C. albicans’ tissue invasion capabilities. We examined if the KEX2 expression is altered following the Sulfone treatment in the G. mellonella-C. albicans infection model.
Material and methods
4-Chloro-3-nitrophenyldifluoroiodomethyl Sulfone’s synthesis
4-Chloro-3-nitrophenyldifluoroiodomethyl sulfone was synthesized according to the scheme in Fig. 1, starting with the commercial 4-chlorophenyldifluoromethyl sulfone, which was iodinated through the reaction with iodine bromide, carried out in carbon tetrachloride with potassium hydroxide as a base [8]. The next step was nitration by fuming nitric acid and concentrated sulfuric acid.
Strains culture, reagents, and growth conditions
Candida albicans strains used in the study are listed in Table 1. C. albicans cultures were grown in YEPD medium [24] at 30 °C overnight. Cultures were washed in sterile PBS and adjusted to the required cell density. YEPD medium supplemented with uridine at the final concentration of 50 μg/mL [25] was used when required. Transformants were selected on the MMD (0.7% wt/vol yeast nitrogen base without amino acids, 2% wt/vol glucose, 770 μg/mL Complete Supplement CSM-URA) liquid and agar (2%) media. The 5-FOA medium (4% wt/vol glucose, 0.2% wt/vol 5-fluoroorotic acid, 1.34% wt/vol yeast nitrogen base with amino acids, 4% wt/vol agar) was used when required.
Generation of the C. albicans KEX2 mutant strains using the mini-ura-blaster technique [23]
The C. albicans strains attenuated in KEX2 (Table 1) were constructed as follows: the two KEX2 alleles in C. albicans CAI4 [23] were disrupted using the URA3-dpl200 disruption cassette, that was PCR amplified from plasmid pDDB57 [25] using the specific primer pairs, containing complementary sequences to the 5′- and 3′-regions of the target ORF 19.4755 (Table 2). The transformants were generated in two rounds using the lithium acetate/single-stranded carrier DNA/PEG method as described in [25, 26] and the URA3 marker was recycled by growing transformants on the 5-FOA medium. Briefly, in the first round of the transformation, we used primers (Table 2) amplifying plasmid DNA (URA3-dp1200 disruption cassette) in PCR with 70 bp of KEX2 flanking homology on either side and then transformed the PCR product into CAI4. The URA+ transformants grown on the MMD medium were screened with PCR that had undergone homologous integration at KEX2. Following this, the transformants were placed on 5-FOA to recycle the selection marker and use this strain in the second round of gene disruption. Due to high transformation frequencies (37 ± 5.7 CAI4 transformants/μg DNA [27]), the Candida integrity plasmid CIP10 was used as the positive control in the lithium acetate procedure. Consistent with this, CIP10 transformed C. albicans CAI4 [23] about 20 times more efficiently than the control plasmid (YPB–ADHpt).
Galleria mellonella collection and treatment
The last instar larvae weighing between 180 and 250 mg were selected for the study. For killing assays, the C. albicans inoculum of 1 × 109 cells per 10 μL of PBS was tested per larva. Larvae were incubated at 28 °C in 9-cm Petri dishes without food for up to 96 h post-infection and inspected every 24 h for survival. Since larvae deprived of nutrition demonstrated increased susceptibility to infection with the fungal pathogen C. albicans [28], we used the same conditions to show unequivocally the Sulfone activity/toxicity using the C. albicans-G. mellonella model. It is worthy to note that despite slight differences in the level of hemocytes and antimicrobial peptides between non- and starved larvae, their effectivity in killing C. albicans is not affected [28]. Since it was found that prophylaxis test is more likely to be successful because a compound is administered before infection, here we tested the Sulfone for either its prophylaxis or post-exposure activity [29]. Thus, we chose four time points to test the Sulfone dosing, e.g., 30 min and 1 h before infection (b. i.) with C. albicans as well as 30 min and 1 h post-infection (p. i.). In one experiment, 10 larvae per group were tested, where each one was injected with 10 μL of the Sulfone (at 0.01 mg/kg of larva) and with the C. albicans inoculum of 1 × 109 cells per 10 μL of PBS (treated). The Sulfone and C. albicans inoculum were administered by injection into a different pro-leg using a Hamilton syringe. The Sulfone dose (0.01 mg/kg) was selected based on the previous studies [8] which proved that this dose is fungicidal against the C. albicans wild type and mutants (KEX2/kex2∆ and kex2∆/kex2∆) in in vitro studies. Syringes were changed between treatments with different strains. Ten larvae without the C. albicans inoculum and 10 larvae inoculated with RPMI or PBS were included for control purposes. The larvae were incubated for 96 h in the above-described conditions. Host health index [30] of the treated larvae was assessed to rank the virulence of the C. albicans strains against G. mellonella [30]. Thus, larvae were monitored daily for the following attributes: activity and survival. Larvae were considered dead if they did not move after stimulation. Randomly selected live, non-melanized larvae, and treated larvae (each in triplicate) were suspended in 0.5 mL of PBS and mechanically disrupted (homogenized), then centrifuged at 100,000g for 1 h. Briefly, 100 μL of aliquots from the serial dilution (1/10,000,000) of supernatants were placed onto YEPD agar medium with penicillin (100 μg/mL) and incubated at 37 °C for 48 h. Thus, we confirmed the Sulfone’s fungicidal activity (MFC = 0.25 μg/mL) in vivo by comparison of the number of CFU of the C. albicans recovered from the larvae treated with the Sulfone and with the starting inoculum. The Sulfone’s antifungal activity was calculated using the formula: log reduction R = log CFU/mL control Candida, log CFU/mL Candida treated with the Sulfone, where R means the relative number of live fungal cells eliminated by the antifungal agent. To interpret the Sulfone activity in vivo, we adopted the criteria described by Majoros et al. [31], where fungicidal agent is that causing a ≥ 3 log reduction; i.e., after the treatment with the agent, the number of fungal cells is 1000 times smaller than the initial (control) number of fungal cells.
Fungal cell staining with hematoxylin and eosin and periodic acid Schiff
Larvae were infected as described above. After 96 h of the maintenance of the larvae at 28 °C (see above), the surviving larvae from each group such as controls (PBS, RPMI, untreated) and those exposed to Candida and the Sulfone were anesthetized by chilling at − 20 °C for 18 h and then fixed, dehydrated, and stained, as described previously [8]. The stained tissues were observed using an Olympus FLUOROVIEW FV1000 confocal laser scanning microscope CLSM (Olympus, USA). Images were assembled using the Photoshop software (Adobe Photoshop CS3 Extended, France). Three larvae from each experimental group were used and the experiment was repeated on 3 independent occasions.
RNA isolation and reverse transcription—quantitative polymerase chain reaction
Larvae were injected as described above. Three surviving larvae originated from each experimental group after 96 h, i.e., the larvae injected with Candida and Sulfone in time intervals as well as the untreated control group; they were frozen in liquid nitrogen and ground to powder with a mortar and pestle. The samples were homogenized and total RNA of C. albicans was isolated as described previously [32]. The total RNA from each sample was used in separate reverse transcription reactions with oligo dT. Total RNA from each sample (5 μL) was reverse transcribed with the Thermo Scientific RevertAid First Strand cDNA Synthesis Kit (K1691, Thermo Fisher Scientific, Waltham, MA USA). Each biological replicate was assayed for the targeted gene KEX2 as well as the endogenous reference gene ACT1 using the Thermo Scientific Maxima SYBER Green qPCR Master Mix (K0221, Thermo Fisher Scientific, Waltham, MA USA). The control groups were introduced to assess the influence of the RPMI Sulfone solvent and the lack of treatment on the KEX2 expression. qPCR thermal cycling and fluorescent data acquisition were performed with a Light Cycler 96 Instrument (Roche Diagnostics GmbH, Germany/Roche Molecular Systems, Inc.) and Cq values were called using the Light Cycler 96 software. A 2−∆∆Cq method [33] was then used to process these data to calculate the relative gene expression for the KEX2 experiment. The experiment was performed in triplicate.
Statistical analysis
Each experiment was performed at least in three replicates and the data were presented as mean values ± standard deviations (SD). Survival curves were plotted and differences in survival (logrank Mantel-Cox test and Gehan-Breslow-Wilcoxon test) were analyzed with the Kaplan-Meier Method using Graph Pad Prism 7 software (Inc.). P value < 0.05 was considered significant. Additionally, the variance ratio test (F test) was performed using MedCalc Statistical Software version 18.6 (MedCalc Software bvba, Ostend, Belgium; http://www.medcalc.org; 2018).
Results
Disruption of KEX2 alleles reduces fungal injury during the host-fungus interaction
Using the mini-ura-blaster technique, we disrupted KEX2 to study its relevance in the C. albicans virulence in vivo. The electrophoresis results of the CAI4 parental strain and KEX2 disruptants are presented in Fig. 2. The CAI4 strain (Ura−) with two deleted alleles of URA3 was used as the parental strain (Fig. 2, line 1). The KEX2 alleles were replaced by the C. albicans URA3 gene flanked by direct repeats of the hisG sequence from Salmonella typhimurium (Fig. 2, lines 2–6). After CAI4 had been transformed with the URA blaster cassette, the Ura+ transformants were selected on uracil-deficient medium. The cells that had lost URA3 by homologous recombination of the hisG sequences were selected on the medium containing 5-FOA because of its toxicity against the Ura+ cells (Fig. 2, lines 7–8). The larvae were infected with 1 × 109 blastoconidia per larva of each strain: CAI4, KEX2/kex2∆, and kex2∆/kex2∆, and the survival was monitored (Fig. 3). Killing of the larvae depended on the strain injected. The larvae were killed significantly faster (P value< 0.0001) when infected with CAI4 and kex2Δ/KEX2 at the inoculum density tested. Fifty percent of the larvae died within 4–5 days, while the survival still reached 100% in those injected with kex2Δ/kex2Δ, and after 6 days, all the latter larvae survived. The tissue sections were performed on the sixth day after the initiation of the fungal infection (Fig. 4). In the larvae treated with kex2∆/kex2∆, there were only very few infected areas compared with the larvae injected with CAI4 or KEX2/kex2∆. The larvae inoculated with kex2∆/kex2∆ showed smaller nodules; these limited to the peripheral larval tissues. There was a relationship between the larvae death rate (Fig. 3) and progression to pathogenesis (mature nodules entrapping CAI4 or KEX2/kex2∆ appeared, Fig. 3a–d). Therefore, we found evidence of the resistance of the larvae against the infection with kex2∆/kex2∆ (Figs. 3 and 4e, f).
G. mellonella killing by different C. albicans strains depends on the KEX2 gene disruption. The kex2∆/KEX2 and kex2∆/kex2∆ mutants along with the parental strain CAI4 were tested. PBS was used as a control. Groups of larvae from 110 to 150 per each strain were infected, as described in “Material and methods,” and survival was monitored every day. Survival experiment was performed at 28 °C. Larvae treated with PBS showed 100% survival in the tested period (data not presented). Statistics were calculated using Kaplan-Meier Graph Pad Prism 7 (logrank Mantel-Cox test, logrank test for trend, and Gehan-Breslow-Wilcoxon test), asterisks indicate significant differences, P value < 0.0001
C. albicans transformants confirmed by PCR reaction using the screening primer pair (Table 2) amplifying the whole KEX2 locus. Line: MM, marker; 1, CAI4 (KEX2/KEX2); 2, Kex2−kex2::URA3dp1200 transformant; 3–6, KEX2/kex2::URA3dp1200 transformants; 7–8, 5-FOA-resistant segregants
Galleria mellonella response to C. albicans infection. Histological analysis of larvae was performed using hematoxylin-eosin (HE) staining (a, c, e) and periodic acid Schiff (PAS) staining (b, d, f) at 6 days post-infection. a, b Larvae injected with CAI4. Fungal cells were isolated by hemocyte aggregation forming melanized nodules (arrows). c, dC. albicans kex2∆/KEX2 promotes hemocyte aggregation (arrows). e, f Small nodules (arrows) were formed in larvae infected with kex2Δ/kex2Δ. Representative images are shown from histological analysis of 3 larvae per each strain per 3 independent experiments
The efficiency of the sulfone during the infection of larvae with C. albicans
We treated the larvae with the Sulfone at 0.01 mg/kg per larva separately in regiments as follows: 1 h or 30 min b. i. with C. albicans. The survival curves correlated with the strains and the Sulfone time course, with a higher death rate (57%) observed when the Sulfone was administered 30 min b. i. with CAI4 (Fig. 5b, P = 0.0002 vs PBS-treated group). The Sulfone-treated larvae 1 h b. i. with CAI4, KEX2/kex2∆, or kex2∆/kex2∆ separately were protected against the development of candidiasis (Fig. 5b–d; P < 0.0001 vs PBS-treated group). Moreover, we evaluated the preventing effect of the Sulfone from the infection development when it was administered b. i. with C. albicans. Plating of the larval extracts provided evidence of the Sulfone’s antifungal activity in vivo (Table 3). We observed a reduction in the number of CFUs recovered from the Sulfone-treated larvae p. i. with C. albicans compared with the untreated larvae. In the Sulfone-treated larvae, the CFUs did not increase over the 6-day period as compared with the C. albicans control CFUs in the Sulfone-untreated larvae (Table 3). Collectively, the Sulfone increased the larval survival and caused 8.8-log reduction of CFUs when injected 1 h b. i. with CAI4 vs control. In contrast, the Sulfone inhibited the proliferation of C. albicans kex2∆/KEX2 and kex2∆/kex2∆ regardless of its administration (Table 3). Each infected larva was treated with the Sulfone at time course (b. i. and p. i.). The two major attributes such as activity and survival were assessed. The surviving larvae exhibited mobility after 96-h incubation. The wax worms injected with the Sulfone 1 h b. i. and p. i. with C. albicans showed a higher health index score (higher activity and survival) compared with those PBS-treated (Fig. 6, insignificant variation, P > 0.063).
Effect of the Sulfone on C. albicans virulence in vivo Wax worms were injected with a dose of the Sulfone of 0.01 mg/kg in time intervals such as 1 h or 30 min before infection with C. albicans at 109 CFU/mL. Kaplan-Meier survival curves of wax worms injected with a SC5314, b CAI4, cKEX2/kex2∆, and dkex2∆/kex2∆. Logrank (Mantel-Cox) test and Gehan-Breslow-Wilcoxon test: a survival proportion of SC5314 + PBS (− 1 h), SC5314 + SULFONE (− 30 min), and SC5314 + SULFONE (− 1 h) was 90, 75.862, and 100% respectively. Single asterisk indicates the survival curves sig different, P value = 0.0188. b Survival proportion of CAI4 + PBS (− 1 h), CAI4 + SULFONE (− 30 min), and CAI4 + SULFONE (− 1 h) was 90, 42.857, and 90% respectively. Double asterisks indicate the survival curves sig different, P value = 0.0002. c Survival proportion of kex2∆/KEX2 + PBS (− 1 h), kex2∆/KEX2 + SULFONE (− 30 min), and kex2∆/KEX2 + SULFONE (− 1 h) was 90, 96.667, and 100% respectively. The survival curves are not sig different, P value = 0.2585. d Survival proportion of kex2∆/kex2∆ + PBS (− 1 h), kex2∆/kex2∆ + SULFONE (− 30 min), and kex2∆/kex2∆ + SULFONE (− 1 h) was 33.333, 93.333, and 100% respectively. Triple asterisks indicate the survival curves sig different, P value < 0.0001
Effect of the Sulfone on larva health index. Treatment with the Sulfone at dose of 0.01 mg/kg per larva or PBS (control) was performed at time course such as 1 h or 30 min before infection with C. albicans (b. i.), 1 h or 30 min post-infection (p. i.) with C. albicans. a–d Variance ratio test (F test) of health index scores of wax worms was performed after 96-h incubation at 37 °C. In each graph, the differences between two curves (Sulfone vs PBS) were not significant (P value > 0.063)
The sulfone treatment modulates the KEX2 expression in the G. mellonella-C. albicans infection model
KEX2 was quantified as the relative gene expression data from each sample, and was normalized to the endogenous reference ACT1 gene of C. albicans [32]. To evaluate the KEX2 expression in the C. albicans treated with the antifungal Sulfone, we chose fungal inoculum at conc. of 109 CFU/mL of PBS. Using the 2−∆∆Cq method, it was shown that KEX2 was slightly upregulated when the Sulfone at 0.01 mg/kg was dosed 30 min and 1 h p. i. with C. albicans SC5314 compared with the larvae injected only with the fungal inoculum of 109 CFU/mL of PBS. During in vivo infection, KEX2 was down-regulated when the larvae were treated with the Sulfone: 30 min b. i. and 2 h p. i. with SC5314 respectively (Table 4).
Discussion
Besides C. albicans, kexin-like proteinases (Kex2) were identified in Saccharomyces cerevisiae, Pichia pastoris, and C. glabrata [34]. These Authors [34] showed that fungal Kex2 proteinases are similar in their substrate activities which have different functions according to the different biological backgrounds of the investigated fungi, including pathogenicity in humans. The KEX2 gene from C. glabrata showed 51% and 62% identity and high structural similarities to its counterparts in C. albicans and S. cerevisiae. Bader et al. [35] revealed that Kex2 is involved in the processing of the proteins that are essential for cell surface integrity of C. glabrata. In C. albicans, Kex2 plays a role in the cell wall formation and interferes with aspartic proteases (Saps) in the cell wall’s remodeling mechanisms [4,5,6,7]. Moreover, the C. albicans cells attenuated in KEX2 showed a defect in polarized growth and morphology [4,5,6,7]. Our in vivo results (lack of hyphae in histological analysis Fig. 4) are consistent with the in vitro findings of Newport and Agabian [5] that the double KEX2 mutation affects morphogenesis. Furthermore, we are consistent with [5] showing that the KEX2 disruption in C. albicans has a pleiotropic effect such as elevated sensitivity to the Sulfone. Our studies in C. albicans showed that the KEX2 gene acts in its virulence process in vivo. It is a gene whose product processes enzymes which are critical for the virulence and ability of C. albicans to evade detection and destruction by the host’s immune system [4,5,6,7]. Bearing in mind that the understanding of the role of KEX2 needs further investigation, we would like to emphasize below several observations that suggest its role during fungal challenge in the G. mellonella model. In contrast with the parental strain CAI4, among the larvae infected with the kex2Δ/kex2Δ mutant, 100% survived after day 5 (Fig. 3). Notably, the larvae infected with the relevant kex2Δ/KEX2 mutant showed survival closer to the CAI4 parental strain (50% survived after day 4). This could be attributed to the absence of both of the KEX2 alleles which are required for the attenuation the C. albicans virulence trait, while both alleles are not needed to maintain the C. albicans’ viability in vivo as shown here, and in vitro as previously reported [5]. Moreover, this phenomenon was described previously for other gene networks [36]. The larvae challenged with the kex2Δ/kex2Δ mutant displayed a decreased inflammatory response, as indicated by low nodule loads seen in the fat body (Fig. 4). Furthermore, this nodule-defective mutant displayed a minimal fault in fitness of the infected larvae. In contrast, the CAI4 parental cells exhibited abundant nodule formation comparable with KEX2/kex2Δ (Fig. 4). Interestingly, this consideration emphasizes further that the role of KEX2 is more attributable to virulence in vivo. These data and the previous reports [18, 30] ruled out the ability of C. albicans to proliferate within hemocytes, leading to infection, and overwhelming the larval immune response.
On the basis of our previous study [8], we selected the Sulfone as the most promising lead candidate for further characterization in vivo. The findings presented in [8] and the current studies showed that administration of the Sulfone triggered immune response in larvae. We observed elevated hemocyte aggregation (Fig. 4) and fungicidal activity without the Sulfone’s negative influence on survival and pupation (Fig. 6). We included the Sulfone antifungal compound in our screen in order to identify whether it targets KEX2. The kex2Δ/kex2Δ mutant was hypersensitive to the Sulfone, and reduced virulence in the survival assay was noted (Fig. 5). As judged from our results and described for known antifungal drugs elsewhere [37, 38], complementation of other fungal genes can be successfully reduced by antifungal drugs when any one of them is deleted. In our study, this could be explained by the inability of other genes to substitute for the absence of KEX2 in that strain when the Sulfone treatment is performed. Inhibition of other genes by the Sulfone is sufficient to prevent the development of infection.
Our study first involved identifying the expression of the responsive KEX2 gene in vivo when treated with the Sulfone at 0.01 mg/kg per larva. The Sulfone affected the C. albicans cells’ proliferation in vivo in the time course tested, except for the Sulfone injected 30 min b. i. with SC5314 (Table 4). In this condition, the KEX2 expression was almost unchanged, which indicates that the Sulfone both prevents the fungal cells’ proliferation and inhibits the virulence factor, thus it prevents the KEX2 upregulation during infection. Targeting virulence is attractive in terms of antifungal drug development because it expands the extremely limited repertoire of targets in fungi [39]. Since the larvae injected with the CAI4 parental strain displayed lowered health index parameters compared with the relevant Sulfone-treated cells and mutants (Fig. 6), we hypothesized that the Sulfone targets KEX2. Due to the Sulfone’s action mode (inhibition of filamentation, adhesion, and biofilm formation, described in [8]), we decided to use a pretreatment modality to mimic its use in a prophylactic antifungal regimen starting 1 h and 30 min b. i. with C. albicans, thus maximizing the chances to detect any protective effect. The larvae which were administered the Sulfone 30 min b. i. with all the C. albicans tested survived the infection at 90–100%, this compared with the mortality observed in the infected and PBS-treated larvae (placebo control group in Fig. 6). We did not observe a perfect correlation between the two experiments, as measured in survival assays (Fig. 3 vs Fig. 6). For instance, the kex2Δ/kex2Δ mutant was less virulent in the untreated larvae (either with the Sulfone or PBS) than in the PBS-treated ones 1 h b. i. with this mutant, yet the larvae displayed a similar fungal load (Fig. 6). One possible explanation is that for the infected larvae, additional stress related to the injection with PBS or the cuticle piercing can be deadly; the larvae started dying in large proportions already on day 1. Thus, the death rate may be misleading, with an error introduced by the PBS injection: the infected and PBS-treated larvae yielded higher mortality, whereas the infected but PBS-untreated ones survived. Thus, a definitive conclusion will be revealed when this experiment has been repeated much more extensively than in the preliminary studies reported here. As shown by Ignasiak and Maxwell [40], sodium chloride is not toxic against G. mellonella, but the cuticle piercing with a needle as such can be traumatic for the larvae. Thus, in each step in the study, three following control groups: untreated, traumatized, and buffer-injected control ought to be included [40]. These Authors [40] found that although the G. mellonella larvae cannot fully replace the mammalian models, they provide the statistical robustness which animal models lack. Moreover, the antibiotic doses recommended for use in humans can be effective in systemic infections in the larvae, and the acute toxicity of compounds in wax moth larvae correlate to the toxicity in mice and rats.
Collectively, since the C. albicans virulence involves a complex regulatory network of genes, the presence of overlapping genes could mask any detectable phenotype due to an altered expression of other genes in the kex2Δ/kex2Δ cells. However, our results supported the well-established Sulfone’s anti-virulence activity [8].
Our data of the Sulfone revealed striking observations: kex2Δ/KEX2 and kex2Δ/kex2Δ were less virulent when the Sulfone was administered 1 h or 30 min b. i. of the larvae. The C. albicans parental strain’s inoculum of 109 CFU/mL of PBS induced the larval mortality at 50% compared with that those infected with the C. albicans kex2Δ/kex2Δ mutant. This mutant was able to infect the larvae but failed to kill the host cells when treated with the Sulfone. Moreover, treating the larvae with the Sulfone 30 min or 1 h b. i. with SC5314 prevented further C. albicans’ growth and effectively prevented the larvae from death. In conclusion, our novel findings described in this article suggest that the Sulfone can be used in direct therapy to prevent or treat potentially fatal fungal infections. Since no animal or animal-derived model of infection completely replicates human diseases [37], the G. mellonella systemic candidiasis model that we used here facilitates the host-pathogen interactions, i.e., testing a host vs a panel of C. albicans mutants when a new antifungal agent is dosed.
References
Kragelund C (2017) Exploiting new knowledge of Candidal infection for future antifungal combat. Oral Dis 23:543–547. https://doi.org/10.1111/odi.12546
Böhm L, Torsin S, Tint SH, Eckstein MT, Ludwig T, Pérez JC (2017) The yeast form of the fungus Candida albicans promotes persistance in the gut of gnobiotic mice. PLoS Pathog 13(10):e1006699. https://doi.org/10.1371/journalppat1006699 eCollection 2017 Oct.
Romo JA, Pierce CG, Chaturvedi AK, Lazzell AL, McHardy SF, Saville SP, Lopez-Ribot JL (2017) Development of anti-virulence approaches for candidiasis via a novel series of small-molecule inhibitors of Candida albicans filamentation. mBio 8:e01991-17. https://doi.org/10.1128/mBio.01991-17
Bresnahan PA, Leduc R, Thomas L, Thorner J, Gibson HL, Brake AJ, Barr PJ, Thomas G (1990) Human fur gene encodes a yeast KEX2-like endoprotease that cleaves pro-beta NGF in vivo. J Cell Biol 111:2851–2859. https://doi.org/10.1083/jcb.111.6.2851
Newport G, Agabian N (1997) KEX2 influences Candida albicans proteinase secretion and hyphal formation. J Biol Chem 272:28954–28961. https://doi.org/10.1074/jbc.272.46.28954
Newport G, Kuo A, Flattery A, Gill C, Blake JJ, Kurtz MB, Abruzzo GK, Agabian N (2003) Inactivation of Kex2p diminishes the virulence of Candida albicans. J Biol Chem 278:1713–1720. https://doi.org/10.1074/jbc.M209713200
Holyoak T, Kettner CA, Petsko GA, Fuller RS, Ringe D (2004) Structrural basis for differences in substrate selectivity in Kex2 and furin protein convertases. Biochemistry 43:2412–2421. https://doi.org/10.1021/bi035849h
Staniszewska M, Bondaryk M, Kazek M, Gliniewicz A, Braunsdorf C, Schaller M, Mora-Montes HM, Ochal Z (2017) Effect of serine protease KEX2 on Candida albicans virulence under halogenated methyl sulfones. Future Microbiol 12:285–306. https://doi.org/10.2217/fmb-2016-0141
Cotter G, Doyle S, Kavanagh K (2000) Development of an insect model for the in vitro pathogenicity testing of yeasts. FEMS Immunol Med Microbiol 27:163–169
Mowlds P, Kavanagh K (2008) Effect of pre-incubation temperature on susceptibility of Galleria mellonella larvae to infection by Candida albicans. Mycopathologia 165:5–12. https://doi.org/10.1007/s11046-007-9069-9
Fuchs BB, O’Brien E, Khoury JB, Mylonakis E (2010) Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence 1:475–482. https://doi.org/10.4161/viru.1.6.12985
Kelly J, Kavanagh K (2010) Proteomic analysis of proteins released from growth-arrested Candida albicans following exposure to caspofungin. Med Mycol 48:598–605. https://doi.org/10.3109/13693780903405782
Kelly J, Kavanagh K (2011) Caspofungin primes the immune response of the larvae of Galleria mellonella and induces a non-specific antimicrobial response. J Med Microbiol 60:189–196. https://doi.org/10.1099/jmm.0.025494-0
Brown N, Heelan M, Kavanagh K (2013) An analysis of the structural and functional similarities of insect hemocytes and mammalian phagocytes. Virulence 4:597–603. https://doi.org/10.4161/viru.25906
Li DD, Hu GH, Zhao LX, Hu DD, Jing YY, Wang Y (2013) Using Galleria mellonella-Candida albicans infection model to evaluate antifungal agents. Biol Pharm Bull 36:1482–1487
Ullah I, Khan AL, Ali L, Khan AR, Wagas M, Lee IJ, Shin JH (2014) An insecticidal compound produced by an insect-pathogenic bacterium suppresses host defenses through phenoloxidase inhibition. Molecules 19:20913–20928. https://doi.org/10.3390/molecules191220913
Champion OL, Wagley S, Titball RW (2016) Galleria mellonella as a model host for microbiological and toxin research. Virulence 7:840–845. https://doi.org/10.1080/21505594.2016.1203486
Tsai CJY, Loh JMS, Proft T (2014) Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7:214–229. https://doi.org/10.1080/21505594.2015.1135289
Rossi SA, Trevijano-Contador N, Scorzoni L, Mesa-Arango AC, de Oliveira HC, Werther K, de Freitas RT, Mendes-Giannini MJ, Zaragoza O, Fusco-Almeida AM (2016) Impact of resistance to fluconazole on virulence and morphological aspects of Cryptococcus neoformans and Cryptococcus gattii isolates. Front Microbiol 7:153. https://doi.org/10.3389/fmicb.2016.00153
Sowa-Jasiłek A, Zdybicka-Barabas A, Staczek S, Wydrych J, Skrzypiec K, Mak P, Deryło K, Tchórzewski M, Cytryńska M (2016) Galleria mellonella lysozyme induces apoptotic changes in Candida albicans cells. Microbiol Res 193:121–131. https://doi.org/10.1016/j.micres.2016.10.003
Gizińska M, Staniszewska M, Ochal Z (2019) Novel sulfones with antifungal properties: antifungal activities and interactions with Candida spp. virulence factors. Mini-Rev Med Chem 19:12–21. https://doi.org/10.2174/1389557518666180924121209
Gillum AM, Tsay EYH, Kirsch DR (1984) Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198:179–182. https://doi.org/10.1007/bf00328721
Fonzi WA, Irwin MY (1993) Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728
Ness F, Prouzet-Mauleon V, Vieillemard A, Lefebvre F, Noël T, Crouzet M, Doignon F, Thoraval D (2010) The Candida albicans Rgd1 is a RhoGAP protein involved in the control of filamentous growth. Fungal Genet Biol 47:1001–1011. https://doi.org/10.1016/j.fgb.2010.07.007
Wilson RB, Davis D, Enloe BM, Mitchell AP (2000) A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65–70. https://doi.org/10.1002/(SICI)1097-0061(20000115)16:1<65::AID-YEA508>3.0.CO;2-M
Wilson RB, Davis D, Mitchell AP (1999) Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol 181:1868–1874
Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJ (2000) Cip10, an efficient and convenient integrating vector for Candida albicans. Yeast 16:325–327. https://doi.org/10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#
Banville N, Browne N, Kavanagh K (2012) Effect of nutrient deprivation on the susceptibility of Galleria mellonella larvae to infection. Virulence 3:497–503. https://doi.org/10.4161/viru.21972
Kocisko DA, Caughey B (2006) Cell-based PrP-res assay and in vivo scrapie testing. In: Kheterpal J, Wetzel R (eds) Methods in enzymology, 2nd edn. Elsevier Inc, USA
Loh JM, Adenwalla N, Wiles S, Proft T (2013) Galleria mellonella larvae as an infection model for group A Streptococcus. Virulence 4:419–428. https://doi.org/10.4161/viru.24930
Majoros L, Kardos G, Szabó B, Sipiczki M (2005) Caspofungin susceptibility testing of Candida inconspicua: correlation of different methods with the minima fungicidal concentration. Antimicrob Agents Chemother 49:3486–43488. https://doi.org/10.1128/AAC.49.8.3486-3488.2005
Staniszewska M, Bondaryk M, Malewski T, Schaller M (2014) The expression of the Candida albicans gene SAP4 during hyphal formation in human serum and in adhesion to monolayer cell culture of colorectal carcinoma Caco-2 (ATCC). Cent Eur J Biol 9:796–810. https://doi.org/10.2478/s11535-014-0311-4
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Bader O, Krauke Y, Hube B (2008) Processing of predicted substrates of fungal Kex2 proteinases from Candida albicans, C. glabrata, Saccharomyces cerevisiae and Pichia pastoris. BMC Microbiol 8:116
Bader O, Schaller M, Klein S, Kukula J, Haack K, Muhlschlegel F, Korting HC, Schafer W, Hube B (2001) The KEX2 gene of Candida glabrata is required for cell surface integrity. Mol Microbiol 41:1431–1444
Khandelwal NK, Kaemmer P, Förster TM, Singh A, Coste AT, Andes DR, Hube B, Sanglard D, Chauhan N, Kaur R, d’Enfert C, Mondal AK, Prasad R (2016) Pleiotropic effects of the vacuolar ABC transporter MLT1 of Candida albicans on cell function and virulence. Biochem J 473:1537–1552. https://doi.org/10.1042/BCJ20160024
Talibi D, Raymond M (1999) Isolation of a putative Candida albicans transcriptional regulator involved in pleiotropic drug resistance by functional complementation of a pdr1 pdr3 mutation in Saccharomyces cerevisiae. J Bacteriol 181:231–240
Pais P, Costa C, Cavalheiro M, Romão D, Teixeira MC (2016) Transcriptional control of drug resistance, virulence and immune system evasion in pathogenic fungi: a cross-species comparison. Front Cell Infect Microbiol 6:131. https://doi.org/10.3389/fcimb.2016.00131
Pierce CG, Uppuluri P, Tummala S, Lopez-Ribot JL (2010) A 96 well microtiter plate-based method for monitoring formation and antifungal susceptibility testing of Candida albicans biofilms. J Vis Exp 44:2287. https://doi.org/10.3791/2287
Ignasiak K, Maxwell A (2017) Galleria mellonella (greater wax moth) larvae as a model for antibiotic susceptibility testing and acute toxicity trials. BMC Res Notes 10:428. https://doi.org/10.1186/s13104-017-2757-8
Funding
MS and MG were supported by fundings from the National Science Centre (No. DEC-2011/03/D/NZ7/06198); MK was supported by Ministry of Science and Higher Education, Grant for Young Scientist 2016/17; RGH and HMMM were supported by Consejo Nacional de Ciencia y Tecnología (ref. CB2011/166860; PDCPN2014-247109, and FC 2015-02-834), Universidad de Guanajuato (ref. 0087/13; ref. 1025/2016; Convocatoria Institucional para Fortalecer la Excelencia Académica 2015), and Red Temática Glicociencia en Salud (CONACYT-México); MS and ZO were supported by Warsaw University of Technology, Faculty of Chemistry. The C. albicans kex2Δ/kex2Δ was obtained in the Department of Genetic Engineering of NIPH-NIH on the basis of previous permits (decision No. 196 of 29/12/2017; 04-66/2017; No. 53 of 27/04/2018; No. 01.2-24/2018).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Responsible Editor: Celia Maria de Almeida Soares
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Staniszewska, M., Gizińska, M., Kazek, M. et al. New antifungal 4-chloro-3-nitrophenyldifluoroiodomethyl sulfone reduces the Candida albicans pathogenicity in the Galleria mellonella model organism. Braz J Microbiol 51, 5–14 (2020). https://doi.org/10.1007/s42770-019-00140-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-019-00140-z