Abstract
In the recent COVID-19 pandemic, World Health Organization emphasized that early detection is an effective strategy to reduce the spread of SARS-CoV-2 viruses. Several diagnostic methods, such as reverse transcription-polymerase chain reaction (RT-PCR) and lateral flow immunoassay (LFIA), have been applied based on the mechanism of specific recognition and binding of the probes to viruses or viral antigens. Although the remarkable progress, these methods still suffer from inadequate cellular materials or errors in the detection and sampling procedure of nasopharyngeal/oropharyngeal swab collection. Therefore, developing accurate, ultrafast, and visualized detection calls for more advanced materials and technology urgently to fight against the epidemic. In this review, we first summarize the current methodologies for SARS-CoV-2 diagnosis. Then, recent representative examples are introduced based on various output signals (e.g., colorimetric, fluorometric, electronic, acoustic). Finally, we discuss the limitations of the methods and provide our perspectives on priorities for future test development.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Modern human DNA keeps the traces of ancient coronavirus infections from 25,000 years ago. Our history of mankind is a continuous battle against these stealthy viruses. In 2003, the first outbreak of SARS-CoV, which is known as severe acute respiratory syndrome coronavirus infected more than 8000 and killed over 800 people. In 2007, MERS-CoV of Middle East respiratory syndrome coronavirus affected > 2400 and killed more than 850 people. The outbreak of coronavirus disease 2019 (COVID-19) being sweeping the world is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1, 2]. Since its discovery, this virus quickly spread to most continents owing to its highly transmissible nature, fast evolution, and unavailability of appropriate therapy and diagnostics systems [3, 4]. Therefore, this incident soon evolved into the pandemic in March 2020 and continued to be the greatest global public health crisis to date. Based on publicly available data (COVID-19 Dashboard by the Center for Systems Science and Engineering at Johns Hopkins University), the number of confirmed cases of new coronary pneumonia rose to 517,888,062 cases (infection rate: ~ 6.7%) worldwide, with 6,253,145 deaths as of 8:00 BST on May 10th, 2022. About 3.4% Mortality rate was estimated by WHO as of March 3, 2022. Despite the tremendous developments of vaccines by pioneering pharmaceutical companies (e.g., Pfizer, Moderna, Sinovac Biotech, and others) and new discovered treatments (e.g., Cepharanthine [5], anti-VEGF [6]), there are still demands for great efforts since the ever-going emergence of new virus strains [7,8,9,10]. Data indicate total vaccine doses administered reached 11,367,262,812 doses, but there were still 72,798 deaths in the past 28 days. Therefore, particular emphasis must still be put on the development of diagnostic tools that could identify the infected people at a manageable early stage.
Computed tomography (CT) scan was the earliest diagnostic method used in clinical practice for COVID-19 determination. However, the excessive use of CT placed an extra burden on the radiology department. Currently, the gold standard diagnosis of COVID-19 is reverse transcription polymerase chain reaction (RT-PCR) by nucleic acid amplification. However, the whole process of this method needs a long detection time (usually 2–6 h) and complicated operation (requires a sterile environment and equipment such as PCR amplifiers) [11]. Methods based on CRISPR (clustered regularly interspaced short palindromic repeats) have been reported later, which typically takes a much shorter time (45–70 min). Apart from these laboratory-based, time-consuming, labor and reagent-demanding nucleic acid amplification tests (RT-PCR, and CRISPR as stated above), rapid serology-based detection methods have also been developed lately. Though these immunoassays are not as specific as the nucleic acid-based tests during the early stage of the infection, such a portable and facile operation greatly benefits on-site individual self-testing. Recently, more and more researchers from analytic science and material sciences fields also contributed novel techniques to COVID-19 diagnosis. And some newly developed methods have also received conditional approval under emergency use authorization (EUA). As such, hereby, we would focus on some newly emerging rapid diagnostic methods, especially from the material and chemical science perspective, to help make an appropriate decision and prompt public health actions.
In this review, we present recent advances in the development of in vitro diagnostics for SARS-CoV-2 (Fig. 1), driven by materials science progress. We first introduce the infection mechanism of SARS-CoV-2 and summarize the current diagnostic methods for patient identification. Next, we present representative examples of the novel detection techniques for visual sensing based on the modes of colorimetric, fluorometric, electronic, acoustic, etc. Finally, we give the current challenges of developing accurate, sensitive, portable, fast, and low-cost diagnostics, as well as future perspectives relying on the analysis of recent advances in materials, devices and artificial intelligence. We hope this review would stimulate more collaboration between medicine and materials to facilitate the efficient SARS-CoV-2 detection and relieve the impact of the epidemic to protect human health, consequently helping people to return to their normal lives.
SARS-CoV-2 Infection Mechanism and Diagnostic Methods
Rapid, low-cost and accurate viral detection is regarded as the most efficient strategy to intercept the COVID-19 infection. A series of methods or techniques for SARS-CoV-2 detection have been proposed since the outbreak of COVID-19 pandemic in 2019 (Fig. 1). For instance, RT-PCR for nucleic acid detection, testing kits for viral antigen and antibody detection etc. These methods of COVID-19 diagnosis generally are devised to detect the targeted substance based on the specificity of viral genome, antigen and antibody against SARS-CoV-2 virion. Consequently, to improve the accuracy, sensitivity and specificity of viral detection, it is vital to confirm and understand the SARS-CoV-2 structure, genome, antigen, replication in host cell, infection mechanism and immunoreaction of human body, as well as the mechanism of various detection assay of SARS-CoV-2 virion.
SARS-CoV-2 Structure, Replication and Infection Mechanism
In accordance with the diversity of genome sequence [12] and serological reactions, coronaviruses are divided into four genera [13, 14], namely, alpha, beta, gamma and delta. These viruses generally cause severe human respiratory diseases, such as MERS-CoV and SARS-CoV caused outbreak in the last decade [15]. SARS-CoV-2, one of coronaviruses is classified as beta-CoV by the World Health Organization (WHO), which has resulted in COVID-19 pandemic since 2019 [16]. SARS-CoV-2 virion consists of a vesicle and a large RNA genome. RNA genome with 26–32 kilobases in length enveloped by a vesicle constructs a unique structure of SARS-CoV-2 virion, which resembles a solar corona as shown in Fig. 2a. The specifical structure of SARS-CoV-2 was validated by electron microscope [14, 17, 18] and shown in Fig. 2b [17]. On the virion surface, four types of proteins were detected, including Trimeric spike (S) protein, Envelope (E) protein, Membrane (M) protein and Hemagglutinin-Esterase (HE) protein. In the vesicle, Nucleocapsid (N) protein is bound to the RNA genome of virus and protects viral genome [14].
Structure of SARS-CoV-2 and its replication process: a Scheme of SARS-CoV-2 structure; b transmission electron microscope images of SARS-CoV-2 virion; c the replicated process of SARS-CoV-2 in host cell. a, b Adapted with permission from Refs. [14, 17], Copyright 2021 Wiley-VCH. c is adapted with permission from Ref. [15], Copyright 2021 Elsevier

Adapted with permission from Ref. [22] Copyright 2022 Springer Nature
Genome organization of SARS and SARS-like CoVs.
The proteins in virion individually play different physiological roles in viral replication. Due to the presence of spike proteins on the viral surface, the infection of SARS-CoV-2 is significantly improved. In Fig. 2c, when SARS-CoV-2 enters human body through nose or mouth, the S protein on the outer surface of the virion enables recognize ACE2 receptors on the host cellular membrane. The S protein then binds to the host receptors, which accelerates the viral and host cell membrane fusion [14, 18]. Also, HE protein helps in the attachment and destruction of sialic acid receptors on the host cellular surface. Subsequently, the SARS-CoV-2 RNA is released into the host cell, and hijacks host ribosomes to translate polyproteins, which initiates viral RNA replication and transcription. More viral genomic RNA and proteins are then replicated, synthesized, and assembled in the endoplasmic reticulum and Golgi apparatus to generate large amount of mature progeny virions in host cell. In the end, the mature progeny virions are released through exocytosis and initiate a new round of infection [14, 19]. In the replication of virions, N protein helps virus breakdown the defense mechanism and deregulate the cellular cycle of host cell. The E and M proteins are responsible for the assembly and formation of new virions [14].
After the upper airway infection deriving from the replication of virions in host cell, the innate immune response of human body is triggered. In the immune response, IgM and IgG antibodies are made in succession and become detectable after 1–2 weeks of infection. IgM antibody enables maintain in the blood until the 3rd week of infection and IgG can remain in the blood to afford long-term immunity [13, 14]. According to the specificity of spike proteins on the viral surface, viral genome and the antibody during the immunity of body, the COVID-19 diagnosis, therefore, can be performed by detecting the antigen, viral nucleic acid in bodily secretions and the antibody in blood.
Diagnostic Techniques of COVID-19 and Applications
Chest computed tomography (CT) imaging detection technique, nucleic acid detection techniques, immunological detection techniques are the most common COVID-19 diagnostic methods [20]. Nucleic acid, antibody and antigen testing are more reliable and accurate than CT, allowing for the rapid identification of asymptomatic infected people [21]. The National Medical Products Administration (NMPA) has approved 94 COVID-19 detection reagents as of April 20, 2022, including 34 nucleic acid detection reagents, 31 antibody detection reagents, and 29 antigen detection reagents.
Nucleic Acid Detection Techniques
The entire genome sequence of SARS-CoV-2 was rapidly obtained thanks to advances in sequencing technology and published on the NCBI website on January 5, 2020, as shown in Fig. 3 [22]. Specific target sequences are chosen and employed in the construction of nucleic acid detection techniques based on the genomic sequence of SARS-CoV-2. For clinics, quarantine, rehabilitation, and discharge, nucleic acid testing is a useful diagnostic tool [23]. Current nucleic acid detection methods for SARS-CoV-2 include real-time quantitative polymerase chain reaction (RT-qPCR) [24], loop-mediated isothermal amplification (LAMP) [25, 26], and clustered regularly interspaced short palindromic repeats/Cas associated system (CRISPR/Cas) [27].
-
(1)
RT-qPCR Detection Technique
The RT-qPCR technology, which can amplify extremely minute amounts of viral target gene sequences in extraction solution, is the gold standard for SARS-CoV-2 accurate detection. The following is the RT-PCR technologic detection principle (as shown in Fig. 4): SARS-CoV-2 RNA is reversely transcribed into cDNA after being denatured at high temperatures; Low-temperature annealing is used to amplify the cDNA; Fluorescent dyes (SYBR Green I dye)/probes (TaqMan probe) that bind exclusively to the targeted segment (cDNA) are introduced during the amplification process to monitor cDNA amplification in real time [28, 29]. When the fluorescence signal surpasses the predetermined fluorescence threshold the number of amplification cycles (Ct value) can be used to determine the presence of the target nucleic acid molecule. Ct value < 35 is recorded as positive, according to the National Health Commission of China; no Ct value or Ct ≥ 35 is labeled as negative.
Fig. 4 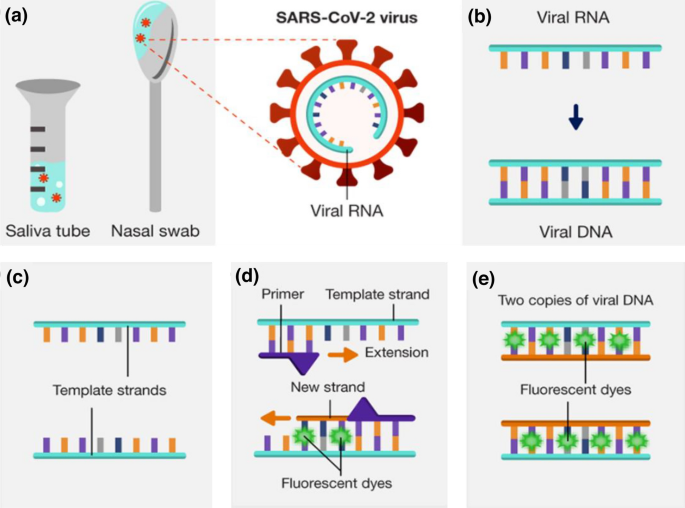
Reprinted with permission from Ref. [29], Copyright 2022 NHGRI
Schematic diagram of RT-qPCR: a sample collection, b reverse transcription, c separation, d duplication, and e end of cycle.
RT-qPCR detection method incorporates PCR amplification, product detection, and quantitative analysis [30]. Furthermore, before amplification, the primers and probes of this approach must be coupled with the target nucleic acid sequence, contributing to a far better specificity than traditional PCR [31]. Traditional PCR open-tube detection causes aerosol pollution and false-positive results. However, RT-qPCR requires professional and technical employees to operate, the equipment and reagents are relatively expensive, and the detection cost is high, all of which limit its widespread usage in community laboratories.
-
(2)
LAMP Detection Technique
In order to stop the virus from spreading, both community clinics and hospitals require a rapid testing technique for COVID-19 diagnosis. The extensively utilized isothermal amplification technology for SARS-CoV-2 nucleic acid detection is LAMP combined with reverse transcription (RT-LAMP). This approach, first reported by Notomi et al. [32], amplifies the target nucleic acids using the strand displacement activity of DNA polymerase and a set of inner and outer primers (four or six distinct primer sequences). The exponential amplification of nucleic acid can be achieved within 20–40 min of constant temperature amplification using strand-displacing DNA polymerase (Bst DNA polymerase) [33]. The presence of target genes can be detected after the reaction by looking at the precipitation in the reaction tube with the naked eye, and agarose electrophoresis or the addition of fluorescent dye can be employed to improve sensitivity and accuracy of detection [34, 35]. It should be noted that while employing the LAMP detection method to detect viruses, SARS-CoV-2 RNA is reversed transcription into cDNA. The detailed process of LAMP technology is shown in Fig. 5.
Fig. 5 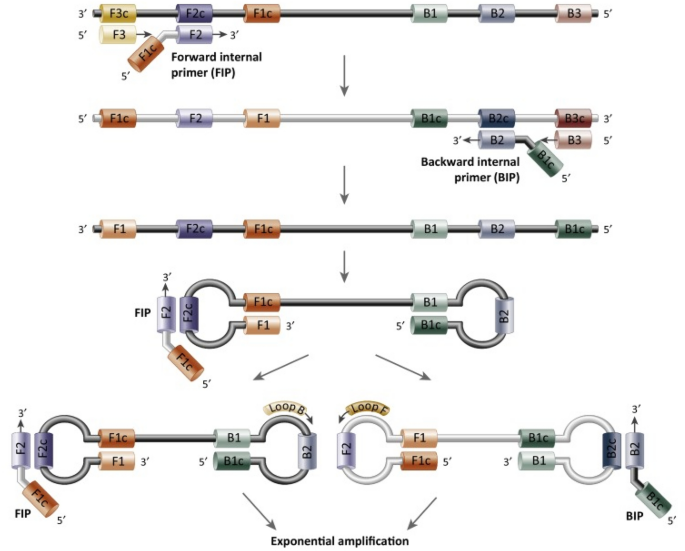
Adapted with permission from Ref. [36], Copyright 2015 Elsevier
Schematic diagram of LAMP. LAMP uses a set of 4–6 primers that recognize 6–8 different sequences of target DNA. Synthesis is initiated by strand-displacing DNA polymerase, and two of the primers form loop structures, which is helpful for the subsequent rounds of amplification.
Different from the RT-qPCR technique, the advantage of LAMP is that the nucleic acid can be swiftly amplified at a constant temperature without relying on a precise temperature cycle system, which considerably decreases the time of nucleic acid amplification and enhances detection performance [37]. For the COVID-19 pandemic diagnosis, Boao Biotechnology Co., Ltd. licensed “six respiratory virus nucleic acid testing kits (constant temperature amplification chip method)” that are based on LAMP technology and can detect six coronavirus targets. It can not only tell the difference between normal people and COVID-19 patients, but it can also tell the difference between mild COVID-19 patients and influenza patients, lowering the possibility of human-to-human transmission owing to a misdiagnosis. Isothermal amplification can only amplify the target efficiently. The amplified products are identified and quantified by fluorescent probes and the intensity of fluorescence. However, the specificity of these products is often affected by the target gene sequence and amplification specificity, resulting in inaccurate results or false-negative results.
-
(3)
CRISPR/Cas System
CRISPR is a potent gene-editing technique that has shown promise in clinical trials. Therefore, a nucleic acid detection technique combining CRISPR and isothermal amplification has been devised for the detection of SARS-CoV-2 virion in molecular diagnostic procedures [38,39,40].
Based on the auxiliary activity of the Cas enzyme, DNA endonuclease-targeted CRISPR trans reporter (DETECTR) and specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) and are utilized to detect SARS-CoV-2 [41, 42]. In the SHERLOCK technique, RNA is isothermally amplified into DNA using reverse transcription recombinase polymerase amplification (RT-RPA). Subsequently, the amplified product (DNA) is combined with T7 transcription and transformed into target RNA (as shown in Fig. 6). The target RNA is recognized and cleaved by the Cas13a-reRNA complex. The cleaved RNA reporters can be visualized by fluorescence signal or colored on the transverse lateral-flow strip [43]. DETECTR works in a similar way as SHERLOCK. Cas12 instead of Cas 13a is used to identify target DNA that can be identified following reverse transcription [44]. A detection sensitivity of 10 copies/µL can be achieved for the both CRISP/Cas techniques and COVID-19 diagnosis can be finished within 1 h [45].
Fig. 6 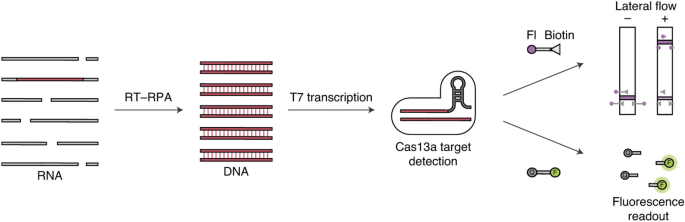
Adapted with permission from Ref. [43], Copyright 2020 Springer Nature
Schematic diagram of SHERLOCK.
The CRISPR-based assay has been used in clinical practice in China and elsewhere. The Food and Drug Administration of the United States (U.S. FDA) granted emergency approval CRISPR technology for the COVID-19 diagnosis in May 2020. (The Sherlock TM CRISPR SARS-CoV-2 Kit) [46]. SARS-CoV-2 DETECTR Reagent Kit devised by Jennifer’s team was also authorized by the U.S. FDA in August 2020. Hangzhou Zhongdao Biotechnology Co., Ltd. has received marketing approval for the SARS-CoV-2 nucleic acid detection kits (CRISPR immunochromatography), which are based on CRISPR technology and recombinant enzyme-mediated chain replacement nucleic acid amplification technology. Hangzhou Jieyi Biotechnology Co., LTD., Zhejiang Province, created a COVID-19 rapid test kit with CRISPR isothermal test, which had also been approved for usage. In terms of practical application, the CRISPR/Cas detection technique has some limitations. The accuracy of SARS-COV-2 nucleic acid detection is reduced due to point mutations, deletions, and inversions that cause off-target effects of CRISPR-based platform.
Immunological Detection Techniques
Antigen and antibody detection are two types of immunological detection. Antigen detection is based on the specifical selection and recognition of monoclonal antibodies to the antigen proteins of SARS-CoV-2 (e.g., N protein and S protein) [47]. When viral extraction solution containing SARS-CoV-2 goes through the testing strip due to capillarity, antigen on the surface of virions binds to the colloidal gold-labeled antibody on the conjugation pad to form a labeled antibody-antigen-capture antibody complex (as shown in Fig. 7). The complex then aggregates on the test line due to the specifical combination between capture antibody and antigen, and then the test line emerges with red color. Simultaneously, the antibody modified with gold nanoparticles also combines with recombinant antigen and the control line also presents, which is identified as a positive result [48]. If only the control line is presented, it means no SARS-CoV-2 virions in the extraction solution and indicates a negative result.
Schematic conception and dipstick assay of the LFIA test strips. Adapted with permission from Ref. [49], Copyright 2021 MDPI
Antibody testing is a method of detecting antibodies against SARS-CoV-2 virions produced by human body. When virus infects human body, it stimulates immune cells and immune cells to respond to virus by generating corresponding antibodies, which can be divided into two types: IgM and IgG antibodies [50]. Antibody detection works in a similar way to antigen detection. Figure 7 shows the mechanism of rapid antibody detection by testing kits. When IgM/IgG antibody attaching to the colloidal gold-labeled antigen on the conjugation pad forms a labeled antigen-antibody complex, the test line turns colored owing to the concentration of gold nanoparticles. Meanwhile, the control line reveals color signal. It indicates an infected case of the sample owner. If the test line does not exhibit color, it is concluded that the sample does not contain IgM/IgG antibodies, indicating a negative result. The presence of non-specific antibodies in normal serum competes with the IgM/IgG antibodies produced following immune stimulation, potentially leading to a false positive result. Furthermore, false negative results can occur when virus enters human body and no IgM/IgG antibodies or only a low number of IgM/IgG antibodies are generated in human body (window period).
In addition, enzyme-linked immunosorbent assay (ELISA) is another immunological detection technique for the detection of SARS-CoV-2 virus based on the specifical combination between viral antigen and antibody [51]. ELISA includes three detection methods, namely direct ELISA, indirect ELISA, and sandwich ELISA. Antigens or antibodies are used in both direct and indirect ELISA to identify the SARS-CoV-2 virus. For sandwich ELISA, two different antibodies (capture antibodies and detection antibodies) are used to recognize viral antigens. The detection mechanism of ELISA is as follows: viral antigen in extraction solution specifically combines with capture antibody, which is immobilized on the surface of test plate, and forms an antigen-antibody complex; Subsequently, the antigen–antibody complex is detected by an additional tracer antibody, which results in a color change on the plate and confirms the presence of virion [52, 53]. Virion can be quantified by the change of color intensity and test results generally can be afforded within 2–5 h [54]. ELISA technology, in comparison to nucleic acid detection methods, can detect SARS-CoV-2 virion in the early stages of infection (within 2 weeks).
CT imaging Detection Technique
CT imaging detection technique is a noninvasive medical imaging technology used in radiology diagnoses. Inflammation of the lungs is common in COVID-19 patients. COVID-19 individuals with multiple ground-glass opacity (GGO) in both lungs can exhibit the imaging features and can be observed by using a chest CT scan, which possesses the advantages of being rapid and having high resolution [55]. As a result, CT has become a significant diagnostic technique for COVID-19. It was a crucial tool for COVID-19 diagnosis early on the breakout of COVID-19. Even though CT is a crucial technique for diagnosing COVID-19, it has several limitations. According to the report by Hope et al., the imaging characteristics of pneumonia in COVID-19 individuals were not disease-specific [56]. It is also possible deriving from a variety of infectious and non-communicable disorders. The CT technique can couple with nucleic acid or other immunological detection methods to improve the accuracy of diagnosis. Consequently, it makes sense and provides reliable results when the highly suspected COVID-19 cases perform chest CT. Chest CT examination can be an auxiliary tool for the definite diagnosis of COVID-19.
Visual Detection of COVID-19 in Different Applications
To control the COVID-19 pandemic and prevent the continuous spread of SARS-CoV-2, reliable technologies for the rapid, simple, inexpensive and accurate point-of-care (POC) COVID-19 diagnosis are urgently required, especially, the visual detection of SARS-CoV-2 virions, which would significantly simplify the viral detection and shorten the mass screening cycle. In general, rapid and visual detection assay of virions involves simple equipment and the results enables be indicated by visible features or signals, such as, color change, fluorescent and luminescent output. Currently, varying approaches of visual detection have been performed in the rapid screening of nucleic acid, viral antigen and antibody [54, 57] as described in the last section. For instance, CRISPER/Cas based biosensing technology, IgM/IgG-targeted LFIA and chemiluminescence immunoassay (CLIA) based on colloidal gold immunoassays. These visual detection techniques have been used to fabricate various diagnostic equipment or tools, including viral antigen and antibody test kits, intelligent fabrics and chromogenic solution.
-
(1)
Viral Antigen and Antibody Test Kits
For rapid and visualizing on-spot mass screening of SARS-CoV-2 carriers, different commercial test kits have been devised based on the LFIA [54]. The LFIA detects viral antigen [58,59,60,61,62] or antibody [50] by an immune-chromatographic assay, which is similar to the commercial pregnancy test. As shown in Fig. 8, capture antibody against SARS-CoV-2 is immobilized on the chromatographic strip at the test line and anti-antibody is fixed at the control line. When viral extraction solution is added into the sample pad and triggers capillary action along the strip. To begin, the antigen on the viral surface enables specifically binding to antibody modified with fluorescent probe or Au nanoparticles and then forms an antigen-antibody complex at conjugated pad. The complex then aggregates at the test line due to the specific combination between viral antigen and capture antibody. Subsequently, the test line on the strip emerges red line or presents fluorescent signal due to the concentration of Au particles or fluorescent probe. Meanwhile, the antibody also combines with anti-antibody and then the control line presents, which illustrates a positive result of detection. If only the control line is displayed, it means a negative result. However, the suspected case with a positive result further needs to perform nucleic acid test by RT-PCR to make a definite diagnosis.
Fig. 8 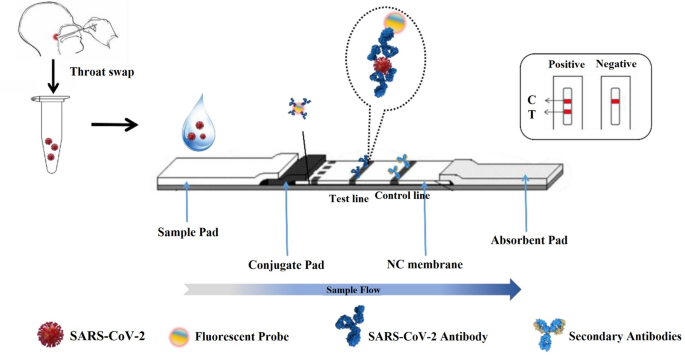
Adapted with permission from Ref. [57], Copyright 2022 Elsevier
Scheme of antigen-detection strip.
Similarly, the LFIA is also used for detection of antibody against SASR-CoV-2 and viral nucleic acid by replacing antibody with antigen or complementary gene sequences. The relative researches have been reported by Zhu et al. for nucleic acid detection [63] and Li et al. for antibody measurement [50]. Technology of LFIA provides a rapid, low cost and visual detection assay, which plays a crucial role in the early mass screening of SARS-CoV-2 carriers and significantly reduces the number of infected cases. However, antigen detection based on LFIA has a lower sensitivity than RT-qPCR, and the sensitivity and specificity of antigen detection can only be improved when the viral load of the sample is extremely high. At present, Guangzhou Wondfo Biotechnology Co., Ltd. and Beijing Kingwolf Biotechnology Co., Ltd. have created China’s first Novel Coronavirus antigen test kits, and COVID-19 diagnosis can be done within 20 min. When the viral load is high, the antigen test can quickly find positive cases and aid in the rapid diagnosis of suspected patients as well as the mass screening of relevant populations, allowing for early diagnosis and intervention [64, 65]. Furthermore, antigen testing can only be done in conjunction with other tests and cannot be used to diagnose COVID-19 pandemic on its own.
-
(2)
Visual Detection by Intelligent Fabrics
Various CRISPR/Cas biosensors have been designed for rapid on-site COVID-19 diagnosis [45, 66]. CRISPR/Cas biosensors combine guide RNA (CRISPR RNA or crRNA) with Cas enzyme[67]. When crRNA binds to targeted nucleic acid, the Cas enzyme is activated and then cleaves the quenched ssDNA fluorophore probes, resulting in the emission of a fluorescent signal. Combining with freeze-dried, cell-free (FDCF) reactions activated by hydration, a CRISPR-Cas12a wearable sensor was prepared by Nguyen and coworkers [68]. The CRISPR-Cas12a sensor enabled detect viral nucleic acid and metabolites, and report the presence of targeted subjects by color changes, fluorescent and luminescent outputs (see Fig. 9a). In Fig. 9b, the CRISPR based sensors were integrated with fibers and the fibers were used to weave intelligent fabrics. The intelligent fabrics were able to detect the nucleic acid of SARS-CoV-2 by emitting fluorescent signal when aqueous solution containing virions was splashed on the fabrics. Additionally, a face mask integrated the CRISP-Cas12a sensor was explored to detect SARS-CoV-2 for respiratory sample. In Fig. 9c, a large region on the face mask was made of aerosol as collection sample pad. A reservoir for hydration was set to trigger the lateral flow assay (LFA) by pressing the red button in Fig. 9d. According to the mechanism of CRISP-Cas12a sensor and LFA (see Fig. 9a), the visual on-site detection of SARS-CoV-2 can be performed by observing the strip on the left side of face mask. The facile design and excellent combination between fabrics and viral detection will dramatically improve the efficiency of COVID-19 diagnosis and reduce the time gap between sample collection and result acquisition.
Fig. 9 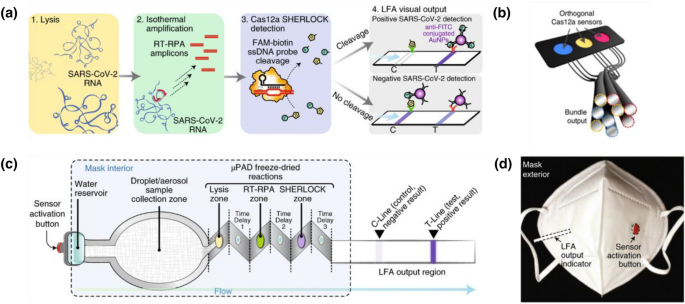
Adapted with permission from ref. [68], Copyright 2021 Springer Nature
a Mechanism of CRISP-Cas12a biosensor and the freeze-dried reactions. SARS-CoV-2 RNA is released from lysis and target gene amplifies at room temperature and then Cas12a results in the cleavage of fluorescent ssDNA probes. The fluorescent outcome of the reaction was purified by the LFA and confirmed the positive result of COVID-19 diagnosis. b The bundle of fibers modified with CRISP-Cas12a biosensor. c Schematic diagram of the SARS-CoV-2 sensor. Puncture of the water reservoir initiated flow through aerosol region and freeze-dried reactions. Subsequently, viral particles collected from respiration in aerosol region downstream integrated into a µPAD device. The visual output is displayed by an LFA strip. d Image of a face mask integrated a SARS-CoV-2 sensor.
-
(3)
Visual Detection by Chromogenic Solutions
Another visual detection assay for COVID-19 diagnosis depends on the chromatic change of solution, where the positive result is evaluated by the chromatic change of solution. The chromatic change of solution is attributed to the variation of solution features, including pH value, dispersity and aggregation of particles. The solution features are altered in different approaches. For instance, Moitra et al. modified N gene targeted antisense oligonucleotide (anti-gene) of SARS-CoV-2 on Au nanoparticles [69]. When samples containing SARS-CoV-2 virions was mixed with the anti-gene, the viral genomic sequences would bind to the anti-gene by complementary base pairing. The specific binding between viral RNA and N gene results in the agglomeration of Au nanoparticles, and then displays a color change of solution for COVID-19 diagnosis (see Fig. 10). In addition, Chow and coworkers reported a rapid and one-step chromogenic reverse transcriptional loop-mediated isothermal amplification assay (RT-LAMP) for the detection of SARS-CoV-2 virions [70]. When the target gene of SARS-CoV-2 presented in the RT-LAMP solution, the target gene initiated the amplification of RNA templates, resulting in the significant decrease of pH value of solution. Due to the decrease of pH value, the detected solution involved pH indictor would show chromatic change and COVID-19 assay was performed according to the color change of solution.
Fig. 10 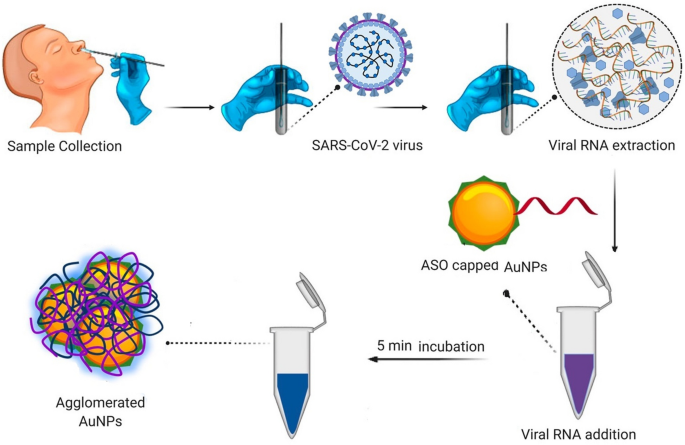
Adapted with permission from Ref. [69], Copyright 2020 American Chemical Society
COVID-19 diagnosis was performed by chromatic change of solution. Anti-gene sequence of SARS-CoV-2 was modified on the surface of gold nanoparticles. When the gold nanoparticles were added into viral extraction solution, SARS-CoV-2 target gene would combine with anti-gene resulting in the agglomeration of Au nanoparticles and showing chromatic change from violet to blue.
Material Systems and Equipment Applied for Visual Detection of SARS-CoV-2 Virions
COVID-19 Diagnosis by Colorimetric and Fluorescent Signals
For the rapid, simple, low-cost and on-site COVID-19 mass screening, various techniques for SARS-CoV-2 visible detection have been proposed. The detection is generally achieved by colorimetric or fluorescent signal. According to the colorimetric or fluorescence, the diagnostic results can be identified rapidly and easily by naked eye, and the techniques can be divided into three categories, including chromogenic effect (or chromatic change), fluorescent and chemiluminescent emission.
-
(1)
COVID-19 Diagnosis Based on Chromogenic Effect
Chromogenic effect means the color emergence in the detection of SARS-CoV-2 due to the aggregation of Au nanoparticles. Au nanoparticles were considered to possess unique advantages, such as excellent stability, eco-friendly and easy fabrication. Consequently, Au nanoparticles labeled antibody or viral antigen has been used for COVID-19 diagnosis and colloidal gold immunochromatography is developed based on antibody (antigen) labeled Au nanoparticles. Moreover, the testing result can be directly read out via the color change without using instruments. Now, different brands of test kits are available in the market. Viral antigens or anti‑SARS‑CoV‑2 human antibody can be detected based on colloidal gold immunochromatography.
The mechanism of SARS-CoV-2 antigen rapid test kit has been introduced in last section of “Viral antigen and antibody test kit” (see Fig. 7). The result of COVID-19 is identified according to the chromatic effect of Au nanoparticles at the test line (T-line) and quality control line (C-line). If the sample contains SARS-CoV-2 antigen, the antigen-antibody-colloidal gold sandwich will appear as a visible line (red or purple) at the T-line. The C-line is coated with goat anti-mouse antibody, which is combined with colloidal gold labeled antibody to form the visible C-line (red or purple). If the C-line does not show any color, the results are invalid and the patient needs to be tested again. The test result is always given in a few minutes. In general, C and T lines with the red or purple color means the positive result. If the T-line without the red or purple color and the C-line with the red or purple color means the negative result (see Fig. 7). The advantage of this method is real-time result feedback, easy operation and low-cost. However, the accuracy and specificity are mild. Thus, it can be used for mass screening.
-
(2)
COVID-19 Diagnosis Based on Fluorescent Signals
RT-PCR has been the gold standard for accurate detection of SARS-CoV-2. The more common method for SARS-CoV-2 detection is real-time quantitative PCR, which is used for detection, characterization, and quantitative of nucleic acid applications. Fluorescent labeling technique gives the signal in real time by adding a fluorescence reagent to the reaction system. Thus, the nucleic acid test actually determines the presence of viral nucleic acid in the sample by detecting the accumulation of fluorescent signals. The detail are as follows (see Fig. 4): the amplification of DNA is monitored in real time with fluorescent dyes or fluorescently labeled DNA probes. The amplification of the target gene produces a fluorescent signal that can be captured by the thermal cycler. If the fluorescence exceeds a defined threshold, the Ct is calculated. Ct values are sometimes used as a proxy for viral load. A low Ct value means more viral RNA in the sample, while a high Ct value means less viral load. In general, this method can be beneficial to show the result in a more safe and efficient style. The advantage of this method includes high accuracy and specificity. However, high-cost, time-consuming and equipment-assisted analysis are its limitations. [54, 57]
-
(3)
COVID-19 Diagnosis Based on Chemiluminescent Signals
CLIA is a typical assay method that combines immunoassay with chemiluminescence technique. It has been extensively developed in the past decades for its good specificity and sensitivity. The light is emitted when an electron transitions from an excited state to ground state. Hence, the chemiluminescence can avoid interference from the biological specimen, which is benefit from the advantage of light emission produced by the chemical reaction.
Wang and coauthors synthesized a novel type of chemiluminescent functionalized magnetic nanobeads under mild condition in two steps (Fig. 11) [71]. Specifically, with the help of amino magnetic beads (MBNH2), ABEI-AuNPs were synthesized by reducing HAuCl4 with N-(aminobutyl)-N-(ethylisoluminol) (ABEI). The ABEI-AuNPs were directly grown on the surface of MB-NH2. Finally, Co2+ was composited onto the surface to generate MB@ABEI-Au/Co2+ (MAA/Co2+). The MAA/Co2+ particles showed excellent chemiluminescence and magnetic properties. Moreover, the antibody can easily and directly connect with Au nanoparticles in the MAA/Co2+ nanobeads via Au-S bond, Au-N bond, and electrostatic adsorption. Hence, for the rapid diagnosis of COVID-19 by detecting the N protein in SARS-CoV-2 virions, MAA/Co2+ is used as a sensing platform to build a label-free immunoassay. The immunoassay exhibited excellent performance, and possessed a good linear correlation between the chemiluminescence intensity and the logarithm of the concentration of N protein within a range of 0.1 pg/mL–10 ng/mL. Besides, it showed a low detection limit of 69 fg/mL and good selectivity based on the magnetic separation function.
Fig. 11 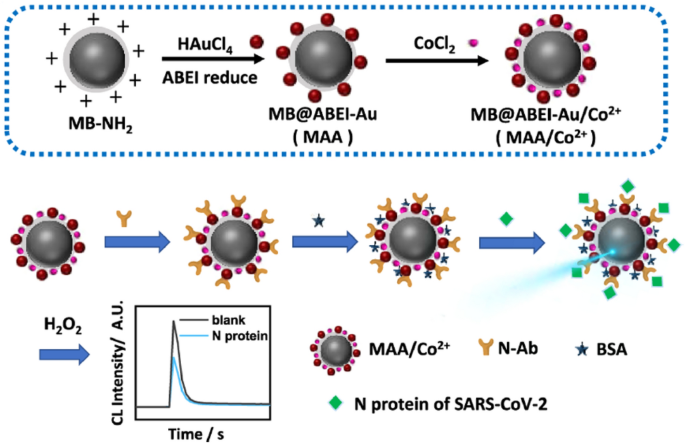
Adapted with permission from Ref. [71], Copyright 2021 American Chemical Society
Schematic illustration displays the synthesized procedures of MAA/Co2+ (up) and fabrication process of label-free chemiluminescent immunoassay for the rapid detection of the N protein (down).
Most of the reported approaches for N protein assays are immunoassays based on labeling methods, including nanomaterial labeling and molecular labeling. Liu and coworkers developed a novel nanozyme, Co–Fe@hemin-peroxidase nanozyme, which was modified with antibody. Using the antibody labeled nanozyme, a chemiluminescent paper was designed to detect the spike antigen of SARS-CoV-2 (Fig. 12) [72]. First of all, the nanozyme chemiluminescence probes were labeled with S-dAb and dispersed onto the conjugate pad of test strip. During the lateral flow of the sample, nanozyme probes specifically combine with S-cAb and S-RBD to formation of the sandwich complexes. Due to the high peroxidase activity, the nanozyme probes enable catalyze luminol substrates to produce chemiluminescence signals in the presence of H2O2 under alkaline conditions. This signal can be well recorded with a smartphone for further analysis. Chemiluminescent intensity at the T-line and C-line is quantified and the chemiluminescent ratio of T-line/C-line is calculated. It is noted that there is a right correlation between the concentration of S-RBD antigen and the chemiluminescent ratio of T-line to C-line. In a word, the Co-Fe@hemin-peroxidase nanozyme is the vital part of this chemiluminescence paper, which catalyzes chemiluminescence, thereby amplifying the immune signal. In addition, the detection limit of recombinant spike antigen of SARS-CoV-2 is 0.1 ng/mL, while the linear range is 0.2–100 ng/mL.
Fig. 12 Schematic illustration of the nanozyme chemiluminescence paper test for SARS-CoV-2 S-RBD antigen. Adapted with permission from Ref. [72], Copyright 2021 Elsevier
Surface-Enhanced Raman Scattering (SERS) Detection Methods
Raman spectroscopy is a technique used to detect the signals from vibrational, rotational, and other low-frequency modes in a system. It is often adopted in chemistry to obtain structural fingerprints to identify molecules. However, Raman signals arise from inelastic scattering processes and the intensity is generally weak, limiting its application for the detection of low-concentration samples. The phenomenon of Surface-Enhanced Raman Scattering (SERS) was first reported in 1973 [73]. Subsequently, the report of single-molecule SERS in 1997 arose a huge interest since it afforded an intensity of Raman signal matching that of a typical fluorescence [74, 75]. In a standard SERS technique, when the energy of the laser light matches the excitation energy of surface plasmon, the electromagnetic field around the metallic surface is greatly enhanced and thus leads to the amplification of characteristic Raman signals from the molecules located in the vicinity of metallic surfaces. SERS sensing techniques require a specific substrate that is tailored to the target. The substrate generally consists of plasmonic metallic nanoparticles, on which a specific ligand can bind with the target. Raman signals of ligand will change once the ligand is attached to the target, providing a clear signal of detection. Due to the high sensitivity, convenient accessibility and non-destructive detection, SERS has gained popularity in many clinical applications such as disease prognosis, drug delivery monitoring as well as diseases diagnosis with real-time therapeutic response [76, 77]. After the breakout of COVID-19, researchers have been developing various SERS-based sensing techniques for rapid detection of SARS-CoV-2 and important achievements have been made in different aspects [78,79,80,81,82,83,84].
Noble metal nanoparticles are most common samples used as SERS substrates for the easy preparation and enhanced electromagnetic effect. Especially, “hot spots” at the gap between nanoparticles gives an even higher enhancement factor and are widely adopted in practical substrates [85]. Huang et al. [82] reported a fishing developed Ag-coated SiO2 nanoparticles loading with dual-layers Raman molecule (DTNB). Then, the nanoparticles were used as SE mode device (see Fig. 13a) that can show response to the receptor-binding domain (RBD) of SARS-CoV-2 at the concentrations low to 0.625 ng/mL in various detection environments. In the device, aptamers and small molecules were attached to the surface of gold nanoparticles. When binding to specific RBD, stable hotspots would form and produce strong SERS signal response in less than 15 min. The results provide good research foundation for practically adopting SERS as a useful technique for the protein detection.
To further improve the accuracy of COVID-19 diagnosis, Liu et al. [83] RS tags for simultaneous detection of anti-SARS-CoV-2 IgM/IgG. Operating principle of the real-time two channel SERS-based lateral flow immunoassay (SERS-LFIA) strip was shown in Fig. 13b. The detection limit of the approach described above is 800 times higher than that of the standard Au-based LFIA method. Recently, Leong et al. [84] reported a hand-held SERS-based breathalyzer, by which an individual COVID-19 diagnosis could be accomplished in 5 min with a sensitivity of 95%. Their SERS-based breathalyzer established a robust, discriminant analysis model based on the vibrational fingerprint changes, which derived from the interactions between molecular receptors and breath metabolites. Furthermore, the SERS-based breathalyzer was capable for performing high throughput classifications. Their strategy would promote the research and application of next-generation human breath diagnostic toolkits, which can be available for mass screening.
SERS-based detection mechanism for SARS-CoV-2 assay. a Structure diagram of the fishing mode device (AgAN-Au-AptRBD-N-M-Au). b Operating mechanism of the anti-SARS-CoV-2 IgM/IgG via the SERS-LFIA strip. a Reproduced with permission from Ref. [82], Copyright 2020 American Chemical Society; b Reproduced with permission from Ref. [83], Copyright 2021 Elsevier
In spite of the high specificity, rapid and non-destructive detection, it is still a great challenge for the SERS techniques to perform accurate diagnosis because of the relatively low signal to noise ratio and the complicated change of Raman signals in multispecies environment. Additionally, the large-scale commercialization of SERS nanotags is also challenging because of the high cost to fabricate reproducible SERS substrates with high sensitivity, thermal and chemical stability, reusable features.
Electrochemical Detection Methods
Over the years, with the vigorously booming of nanomaterials and continuous emergence of various molecular biorecognition elements (BRE), electrochemical biosensors develop rapidly since the birth of glucose electrochemical sensor in 1962 [86]. In the electrochemical biosensors, solid electrode is used as the basic electrode, and the recognition elements or bioactive sensitive unit are fixed on the signal converter. Biometric reaction occurs when the target appears. Subsequently, the signal converter transforms the concentration signal of the target into measurable electrochemical signals, such as potential, current, impedance or capacitance, so as to realize the quantitative or qualitative analysis of the target. At present, electrochemical biosensors have become one of the most-used techniques for real-time detection of SARS-CoV-2 in clinical diagnosis [87,88,89]. In addition, the detection can be finished in a few minutes, which provides a good foundation to develop COVID-19 POC testing equipment [90]. Here, we introduce the representative progresses of electrochemical sensors in COVID-19 diagnosis mainly deriving from the perspective of material science.
In the past two years, lots of reports about SARS-CoV-2 detection through electrochemical biosensors, self-supported or fabricated on some kinds of substrates (e.g. paper, glass, and PDMS) have been done, in which the solid electrodes such as graphene, carbon, and gold are modified with nanomaterials [91,92,93,94,95]. Alafeef et al. [94] developed an electrochemical platform using modified graphene for selective and ultrasensitive monitoring of SARS-CoV-2 viral RNA. In the biosensor, specific designed thiol-modified antisense oligonucleotides (ssDNA)-capped gold nanoparticles (AuNPs) were employed as the sensing matrix (Fig. 14a). When contacting with SARS-CoV-2 RNA the specific RNA-DNA hybridization led to the variation of charge/electron mobility on the graphene surface, resulting in the out voltage change of the sensor. The fully integrated sensors successfully exhibited specific output signal of SARS-CoV-2 in less than 5 min using the incubation of RNA samples, and afforded a sensitivity of 231 copies/µL and a detection limit of 6.9 copies/µL.
Panat and co-workers [95] reported a 3D printed COVID-19 rapid test chip (Fig. 14b). The biosensing platform, which was prepared by 3D-printing Aerosol Jet (AJ) gold nanoparticles into gold-micropillar array electrode, and followed by coating with rGO (reduced graphene oxide) nanoflakes. In the end, the viral antigen was immobilized on the electrode surface via an amidation reaction. When antibodies were introduced on the electrode surface by microfluidic device, they specifically bonded with the antigens accompanied with a leap of impedance in the electrical circuit. The change of impedance could be monitored by electrical impedance spectroscopy (EIS), enabling SARS-CoV-2 to be diagnosed within seconds. Furthermore, a antibodies-detected sensitivity of 1 × 10− 12 M for spike S1 and 1 × 10− 15 M for RBD antigens of SARS-CoV-2 was achieved. Noteworthy, the sensor can be renewed within a minute simply through using low-pH chemistry in which eluting the antibodies from the antigens, and then successive detection with the same sensor is allowed.
Electrochemical biosensors for SARS-CoV-2 detection. a Graphene based electrochemical platform with AuNPs capped with ssDNA probes for the detection of SARS-CoV-2 virions. b The sketch-map of the test chip. c The configuration of a MolEMS g-FET and its sensing mechanism. a Reproduced with permission from Ref. [94], Copyright 2020 American Chemical Society; b reproduced with permission from ref. [95], Copyright 2021 Wiley-VCH; c reproduced with permission from Ref. [96], Copyright 2022 Springer Nature
Field-effect transistors (FETs) are a kind of signal amplification device, in which a tiny change of input signal will induce a pronounced change of output current. Liquid-gated FETs and electrochemical transistors, using liquid and electrolyte solution as gate electrode respectively, can also serve as electrochemical biosensors. Recently, both liquid-gated graphene FETs and organic electrochemical transistors (OECTs) exhibited rapid, ultrasensitive or/and ultraprecise performance in the detection of SARS-CoV-2 through different sensing response mechanisms [96, 97]. Wei and co-workers [96] reported series of liquid-gated graphene FETs (g-FETs) integrated with interface sensing design by multitarget DNA probes or antigens. Among them, a typical case was the construction and application of molecular electromechanical system (MolEMS), which was a DNA self-assembly system. In the DNA system, a flexible single-stranded DNA (ss-DNA) bounded with an aptamer probe cantilever was grafted to a stiff tetrahedral double-stranded DNA (ds-DNA) structure. Under electrostatic activation, the g-FET device modified with MolEMS (Fig. 14c) performed an ultrasensitive, specific and direct detection of SARS-CoV-2 RNA with a concentration limit below ~ 0.02 copies/µL. The detection of SARS-CoV-2 could be done within four minutes without RNA extraction. The limit of detection (LoD) was much lower than that of a typical RT-qPCR assay (0.2–1 copies/µL).
Although electrochemical analysis displays the advantages of rapid detection and high sensitivity, enabling the possibility of on-site and POC testing in various sites including airports, local clinics, hospitals and even at home, there are still many challenges that need to be further considered and improved for large-scale industrial applications: First, how to avoid the influence of external factors such as temperature, light, electromagnetism and basic solution on the signal response; Second, factors such as short shelf life and lower conductivity of nanomaterials used on sensor surfaces limit the biosensors performances. Last but not the least, stability, reliability and consistency of core sensor components should be focused, and standardized manufacturing process of sensor chips need to be established.
MS-Based COVID-19 Diagnostic Methods
Mass Spectrometry (MS) is a versatile characterization method that has been broadly applied in laboratory analysis. Possessing high sensitivity and excellent specificity, MS can not only identify a large variety of compounds, but also provide detailed information about molecular weight and structure. With the improvement of complementary instrumentations in recent years, MS can be used for the detection and quantification of relevant biomolecules in the dynamics of organisms, including proteins, carbohydrates, nucleic acids, and lipids [98]. Now, MS has been used for the identification of SARS-CoV-2 and analysis of infection phases with high flux and high accuracy [99]. Compared with PCR and LFIA, the sampling of MS is none or less invasive. Besides that, MS for COVID-19 diagnosis is rapid and can be done within 10 min. Also, MS eliminates the possibility of false-negative responses caused by insufficient sensitivity of other approaches. With the outbreak of COVID-19 pandemic at the end of 2019, the emergent need for the rapid and accurate detection of SARS-CoV-2 has accelerated the improvement of MS-based alternative diagnostic methods.
Like other diagnostic approaches, MS-based ones identify COVID-19 infections by detecting SARS-CoV-2 pathogens directly or by pathogen-induced biomolecular changes in human bodies. The typical workflow of MS-based omic methods is illustrated in Fig. 15 [100]. Specimens are collected from nasopharyngeal and oropharyngeal swabs, urines, blood, etc., and pretreated with a virus deactivation process. Different omic methods, including proteomics, glycomics, lipidomics and metabolomics, generally require pretreatment to extract relevant components. Subsequently, the extracted components are injected into liquid or gas chromatography (LC/GC) complemented with MS, which enables the detection of the target biomolecules in complex specimens. With multivariate statistical analysis, the biomolecules are identified and quantified by comparing MS spectral.

Reproduced with permission from Ref. [100], Copyright 2020 American Chemical Society
Workflow of MS-based test for COVID-19 diagnosis.
In addition, soft ionization techniques are quite reliable in characterization of biomolecules when they are incorporated with MS, and the techniques are practicable for the test of SARS-CoV-2. The typical ionizations are matrix-assisted laser desorption/ionization (MALDI) [101, 102] and electrospray ionization (ESI) [103, 104]. Both of the two techniques initiate the fragmentizing of macromolecules in gas phase by pulsed laser or high voltage sources. Due to their low efficiency of fragmentation, MALDI and ESI afford a analytical order of magnitude of 106 molecular weight, which covers most of biopolymers including proteins [105], peptides [106], carbohydrates [107], and nucleic acids [108]. At present, MALDI-MS and ESI-MS have been applied for clinical diagnosis of bacterial and viral infections. In viral detection, the two techniques now are the crucial complementary methods for the routine diagnostic tools, and extend the detection from the gene of just known viruses to known/unknown pathogens. The U.S. FDA issued an emergency use authorization for SARS-CoV-2 RNA detection based on PCR and MALDI-MS in September 2020, which was the first MS-based method approved for emergent COVID-19 diagnosis [109]. In another research, ESI-MS was coupled with PCR to identify viruses for respiratory specimens. The introduction of MS techniques into PCR can significantly decrease the possibility of false-positive responses [110].
As indicated above, the MS-based methods are still an indirect detection for SARS-CoV-2 assay, and cannot substitute the current diagnostic methods, such as PCR or LFIA. Indeed, MS requires a high-cost instruments, and is not easy to operate compared with other common clinical test methods. In addition, analytical results provided by MS are more complex than PCR or LFIA detection. It means more efforts and time are needed for the analysis of MS results, resulting in the inadaptability of MS-based techniques for rapid public COVID-19 diagnosis.
To adapt to the detection of public health emergencies, more efforts have been made to develop a portable and fast MS-based method and great progress has been achieved. Recently, a novel MS-based instrument for COVID-19 diagnosis was reported. A instrument, InspectIR COVID-19 Breathalyzer, based on gas chromatography-mass spectrometry (GC-MS), could separate and identify five volatile organic compounds (markers) related to SARS-CoV-2 virion in exhaled breath specimens [111]. Once the target compounds were detected, an unconfirmed positive response was got and then the target compounds could be verified by molecular identification for a definite diagnosis. In a test of 2409 specimens, the Breathalyzer afforded a sensitivity of 91.2% and specificity of 99.3%. This was the first COVID-19 diagnostic technique for breath samples and can evaluate about 160 specimens per day. The U.S. FDA issued an EUA for the MS-base technique on April 14, 2022, not U.S. FDA cleared yet, and the details about this instrument and its inventors have not been disclosed. Once approved for general use, 100 sets per week of the Breathalyzer are expected to be manufactured to meet the emergent requirement for the mass screening of COVID-19 diagnosis.
Nanofluidic Detection Methods
Nanofluidic devices based on biological nanopore techniques are powerful tools for the detection and analysis of biological molecules, especially for the long-chain biomolecules, such as proteins, peptides, and nucleic acids [112]. In the nanofluidic device, a thin membrane loaded with lipid bilayers and biological nanopores is located in the middle of the device to separate the buffer solutions into two wells, which are connected via bio-nanopores. When applying a voltage from one well to the other one ionic current flow via nanopores and generates a primary current signal across the membrane, which is measured by a high-resolution ammeter. Meanwhile, the applied voltage also drives the charged and long-chain biomolecules electrophoretically to “swim” through the nanopores. Therefore, some fraction of the across-membrane ionic current is intercepted by the biomolecules, and affected by the composition, charge, and sequence of the translocating biomolecule [113]. Recording the ionic current alteration can directly read and decode the sequence of residues, including amino acids in proteins or bases in DNA [114]. Thus, the nanofluidic analysis shows a promising potential for the identification and sequence reading of specific biological macromolecules without any replications. However, due to the fragile feature and instability of the nanofluidic devices, the technique has not been used for the COVID-19 diagnosis. To accelerate the application of nanofluidic devices or techniques, more efforts should be made on the improvements of nanopore sensitivity, the signal-noise ratio of electrochemical instruments, and stability of the platform materials.
Sound-Based Diagnosis of COVID-19
The development and application of rapid, affordable, and readily available testing methods is critical to effectively interrupt the spread of SARS-CoV-2 virus. The standard RT-PCR method, which tests nasopharyngeal and throat swab samples, requires chemical reagents and specialized analysts, and the testing process is invasive, resource-dependent and time consuming. Another commonly used antigen testing for COVID-19 diagnosis, although time-saving, still requires chemical reagents. SARS-CoV-2 virus has been shown to affect the respiratory system of the infected patients, changing the sound of coughing, breathing and speaking. The sound produced by our body can be used as a biomarker for early diagnosis of disease [115]. The patients infected with COVID-19, including those who are symptomatic and asymptomatic can be quickly screened out by the different sounds from healthy people. Therefore, sound-based testing for COVID-19 pandemic has been developed into a highly efficient and easy-to-implement method.
The first step in sound-based COVID-19 detection is acoustic data collection. The sound data is usually collected through crowdsourcing. The participants record their cough and breath sounds using devices, such as mobile phones and computers, and then upload them to a sound database. Several sound datasets have been developed, including Cambridge [116], NoCoCoDa [117], Coswara [118], COUGHVID [119], Virufy [120], and they have been widely used to validate the feasibility of using sound for COVID-19 detection [121, 122]. The collected data need to be analyzed and classified to determine whether someone has been infected with COVID-19 virus (Fig. 16). Pahar et al. developed seven classifiers based on machine learning to diagnose COVID-19 cases from cough sounds using Coswara dataset, and found that the Resnet50 architecture can distinguish the cough between a COVID-19 infected patient and a healthy person with an AUC (area under the curve) of 0.98 on the Coswara dataset, indicating that COVID-19 screening based on automatic classification of cough sounds is feasible [123].

Reproduced with permission from Ref. [120], Copyright 2020 University of California
Schematic showing the COVID-19 diagnostic method based on a crowdsourced cough sound database and artificial intelligence cough test.
Artificial intelligence (AI) has been widely used in the health sector, and deep learning and machine learning (ML) techniques also have been extensively applied for COVID-19 diagnosis (Fig. 17) [124]. Son et al. developed a diagnostic method of COVID-19 based on the COUGHVID cough dataset and AI cough test to distinguish the cough sound of COVID-19 patients from those of healthy people [125]. An AI deep learning model was trained on the features extracted from the cough sound data, endowing the diagnostic method up to 93% sensitivity and 94% specificity. A novel stacking approach with deep learning CNN (convolutional neural network) models was further proposed to improve the model performance [122]. The detection of COVID-19 for symptomatic and asymptomatic patients was achieved by using the spectrogram images of cough and breath sound, and the spectrograms from cough sound are more reliable than the spectrograms breath sound.
While there are many advantages to using sound to detect COVID-19, there are still some problems with this approach. Since cough is also a symptom of more than 30 other diseases, distinguishing cough sound caused by COVID-19 from that of other diseases remains a huge challenge. Imran et al. addressed this problem by investigating the differences between the pathomorphological changes in the respiratory system caused by COVID-19 infection and those caused by other diseases [115]. The APP they developed based on AI-powered screening solution and CNN classifiers can differentiate the COVID-19 associated cough from the coughs caused by pertussis and bronchitis, as well as non-contagious coughs. The impact of COVID-19 on the respiratory system is significantly different from other diseases, and cough can be used as a test marker for the diagnosis of COVID-19.

Reproduced with permission from Ref. [126], Copyright 2022 Springer Nature
Keywords network relationships related to COVID-19 research.
For sound-based COVID-19 testing, the following aspects need to be addressed in the future: (1) The sample sizes of the existing datasets for COVID-19 testing are relatively small, building crowdsourced datasets based on big data is a must for accurate detection. (2) AI-based algorithms for extracting sound features should be further developed, and classifiers based on deep neural networks should be more focused. (3) More attention should be paid to study the sound characteristics of COVID-19 patients at different disease stages and establish a dataset, so as to determine the disease progression of patients by analyzing the sound.
Applications of Smartphones in Visual Detection of COVID-19
Recently, POC testing techniques including lateral flow technologies [127, 128], paper-based microfluidic chips [129, 130], isothermal nucleic acid amplification devices [131, 132], etc, have been developed for visual detection of COVID-19. The popularization of mobile communications has greatly increased the efficiency of acquisition, processing and transmission of test data. Smartphones, as a kind of ubiquitous portable devices played an increasingly important role in visual detection of viruses due to their high-resolution cameras, powerful data processing and storage capabilities. Various smartphone-based visual detection techniques, such as optical biosensors, electrochemical biosensors, surface acoustic wave biosensors have been developed [133, 134]. In addition, the corresponding smartphone apps have also been developed to record, process and transmit the test data [135, 136].
-
(1)
Smartphone-Based Electrochemical Biosensors
Because of the advantages of simplicity, reliability and low cost, electrochemical technology has been widely used in quantitative detection of pathogens. In particular, electrochemical methods are very suitable for miniaturized and handheld devices, so smartphone-based electrochemical biosensors have attracted more and more attention.
Zhao et al. used electrochemical method in conjunction with smartphone to detect the SARS-CoV-2 virus for the first time (Fig. 18a) [137]. Their device is based on the supersandwich-type recognition strategy and does not require nucleic acid amplification and reverse transcription for detecting the RNA of COVID-19 virus. The biosensors exhibited high specificity, selectivity and low detection limit, and showed a higher detectable ratio than the RT-qPCR method in actual clinical assays.
Fig. 18 Smartphone-based biosensors for COVID-19 virus visual detection. a Schematic showing the smartphone-based electrochemical biosensors based on the supersandwich-type recognition strategy for RNA detection of COVID-19 virus [137]. b Digital image of a portable smartphone-based biosensor based on miniaturized electrochemical immunosensor made of laser-scribed graphene (LSG) and three-dimensional gold nanostructures (AuNS) for rapid POC COVID-19 diagnosis [138]. c Images acquired from a smartphone camera showing the improved signal-to-noise ratio due to the presence of modified agarose beads illuminated by a 450 nm laser diode [139]. d Digital image showing a smartphone camera was used to capture the capillary flow video of the tested samples. a Reproduced with permission from Ref. [137], Copyright 2021 Elsevier. b is reproduced with permission from Ref. [138], Copyright 2021 American Chemical Society. c Reproduced with permission from ref. [139], Copyright 2021 Royal Chemical Society. d Reproduced with permission from Ref. [129], Copyright 2022 Elsevier
Beduk et al. realized rapid POC COVID-19 diagnosis using a smartphone-based electrochemical biosensor (Fig. 18b) [138]. A miniaturized electrochemical immunosensor was first developed based on laser-scribed graphene (LSG) and three-dimensional (3D) gold nanostructures (AuNS). The LSG/AuNS electrode was then integrated into a reconfigurable multi-measurement polypotentiostat POC device, which had a dual connector and was able to connect to a smartphone via micro-USB port. The device showed a detection limit of 2.9 ng/mL and demonstrated its applicability to clinical testing by successfully diagnosing 23 blood serum samples from COVID-19 patients.
-
(2)
Smartphone-Based Optical Biosensors
Smartphone-based optical biosensors are simple in design and low in cost, enabling qualitative analysis of test results with the naked eye or quantitative analysis with an optical detector. The devices can use a smartphone to calculate the relative concentration of the analyte based on the color intensity of the samples being tested. Because the camera in smartphones can distinguish color tone variations, smartphone-based biosensors can visually detect and analyze viruses based on the absorption or reflection intensity of the analyte.
Adrover-Jaume et al. developed a smartphone-based colorimetric biosensors for detecting the IL-6 (a prognosis biomarker of COVID-19) level in blood and respiratory samples from the patients infected with COVID-19 [140]. The biosensors exhibit a low detection limit, wide dynamic semi-quantitative testing range, and low matrix interference, which combine with the low requirements of the mobile APP to the photographic conditions, making them a promising tool for the decentralized management of COVID-19 patients.
Fluorescence sensing is also a widely used optical detection technique, which has high sensitivity, good anti-interference, simple operation and fast signal acquisition speed [141]. Soares et al. designed a POC SARS-CoV-2 virus detection platform by combining LAMP, centrifugal microfluidics and smartphone-based fluorescence signal detection [139]. In this platform, modified agarose beads were used to remove the intrinsic background of primer dimers, and the fluorescence signal resolution was significantly improved (Fig. 18c). The platform can be used to detect viral RNA directly from nasopharyngeal swab samples, and the sensitivity and specificity were evaluated by analyzing 162 nasopharyngeal swab samples from the patients infected with SARS-CoV-2 virus. Although the sensitivity of this platform is not as good as that of PCR-based methods, the low cost allows it to be used more frequently on the field.
-
(3)
Other Smartphone-Based Sensors
In addition to electrochemistry and optics, several smartphone-based sensors have been developed based on other principles, such as acoustic wave biosensors, thermal sensors, and microfluidic sensors. Recently, a smartphone-based sensitive detection for SARS-CoV-2 virus through saliva tests has been developed to avoid the discomfort of nasopharyngeal swab tests [129]. The assay was performed using a smartphone, a paper microfluidic chip and an antibody-conjugated particle suspension. A Python script was developed and used to automatically measure the flow profile by a smartphone camera (Fig. 18d). The method demonstrated a low detection limit and high specificity, and achieved an overall accuracy of 89%.
In summary, series of conventional and novel detection methods have been proposed and applied for COVID-19 diagnosis as summarized in Table 1. The detection methods present different advantages and disadvantages deriving from the diversity of sensing mechanism and testing instruments. For example, RT-PCR method is comparatively accurate but time-consuming and labor demanding, while antigen/antibody test kit is easy and fast but suffers from inadequate cellular materials or errors in detection. Therefore, it is still challenging to develop a novel approach for the rapid, low-cost, point-of-care, accurate and visual mass screening of SARS-CoV-2. More attentions should be paid on the design and fabrication of materials, which should be used in the extraction of virions, the specific combination with viral antigen or antibody, the transcription and amplification of target genetic sequences, etc.
Table 1 Summary of varying detection methods for COVID-19 diagnosis
Common Challenges and Prospect
The rapid and visual detection techniques of SASR-CoV-2 virions have been devised based on the specific combination between antigen [59, 60, 62, 142] and antibody [50]. For instance, the commercial antigen test kits have been widely used in COVID-19 diagnosis in the world [61, 143]. However, the accuracy and specificity of the rapid detection techniques [58, 60, 144] are still lower than that of the nucleic acid test of RT-PCR performed in clinical laboratories due to the invalidation of antibody or antigen, the lack of standard sample collections [145] etc. Currently, nucleic acid test of RT-PCR is regarded as the golden standard for the COVID-19 diagnosis and provides the final medical result. Although PCR techniques possess excellent accuracy for SARS-CoV-2 assay, false-positive result also takes place occasionally in clinical detection. The false-positive results are attributed to the difference in the extracted efficiency of virions, the transportation of samples, the time gap between sample collections and detections, the approaches of sample collections and the diversity of different viral extraction solution [146]. Therefore, the accuracy of nucleic acid test is still a challenge for the COVID-19 diagnosis.
In spite of specificity and sensitivity of PCR techniques and antigen detection by test kits, the materials of swab and virial extraction solution are another two challenges for COVID-19 diagnosis. The materials used for the fabrication of nasopharyngeal swab are limited to polyester and dacron [146]. Also, the virial extraction solutions produced by different factories display obvious diversity in the extracted efficiency of virions, and standard to evaluate the viral extraction is lacking.
Rapid diagnosis has become an essential aspect of social involvement as a result of the COVID-19 pandemic’s severe impact on human life. CT imaging, RT-qPCR, and antibody/antigen technology are mostly used in the diagnosis of COVID-19. RT-qPCR assays, in particular, are commonly used in schools and workplaces to identify infected individuals and reduce COVID-19 transmission. The RT-qPCR testing method for COVID-19 diagnosis, on the other hand, is costly and time-consuming, and it cannot fully meet the enormous demand for the test. In the future, a faster and more easy-readable detection technology has the potential to become the quasi-gold standard for COVID-19 or next-generation virus diagnosis, however, there are still significant practical constraints.
Common Challenges
In general, based on the commercialized and reported emerging visualized detection approaches, we summarize four common challenges from materials aspects (Fig. 19):
-
(1)
Lacking new concepts in visual detection materials At present, commercial visual detection methods based on biochemical analysis heavily rely on colloidal gold technology (SPR), which is challenged in stability, accuracy, and especially cost control.
-
(2)
Limited to liquid-phase tests The current commercial visualization detection technology adopts the liquid sampling approach, but concerning the solid surface, air/aerosols in public environments (train stations, shopping malls, airports, etc.), there still lacks a viable technique to achieve the visual detection for solid surface and gas phase, which is crucial for controlling the spread of the pandemic in the first place.
-
(3)
Disposal of detection materials Till now, billions of test kits have been used for COVID-19 diagnosis worldwide and a huge amount of viral extracting solution, test strips, and nasopharyngeal swabs were used in viral screening assay [58, 147]. The disposable test kits are mainly composed of polymer materials and inorganic buffer. After detection, the test kits cannot be reused for further detection and therefore causes serious pollution and might generate health hazard. However, very few researches have focused on the recycling and the degradation of related polymer materials used in the detection of the SARS-CoV-2 virus. The enrichment and recycle of gold nanoparticles [148, 149], especially from the polymer waste should be studied. The reliability of biodegradable polymer materials (such as PGA poly(glycolic acid); PLA poly(lactic acid); PCL polycaprolactone; cellulose; and starch; etc.) should also be tested and hopefully put to real applications. [150, 151]
-
(4)
Difficult to meet the diversity of application scenarios Although various techniques have been explored for COVID-19 diagnosis, each of the techniques is only limited to one type of sample or single site. For instance, it is a great challenge to use one technique for the real-time detection of the SARS-CoV-2 virus for swap samples from patients, surface or aerosol samples nearby the crowds in the airport, train, market, etc.
Prospects
Developing alternative methods that can provide specific, sensitive, portable, fast-respond, and low-cost detection without complicated sample preparation is always essential for fighting against the epidemic. Here we propose some key factors that need to be considered to broaden the commercial applications (Fig. 20):
-
(1)
Reliability improvement on the working material New materials capable of enhancing the specificity and detection limit of the current immunoassay are of great importance. Meanwhile, improving the stability of materials is also crucial for the usage under different circumstances including special temperatures, humidity, and concentration of the samples.
-
(2)
Integrated detection and data collection Integrate the detection, data collection, and data report to realize the real-time, online, and remote crowd-control, thus would potentially shut down the virus transmission [152, 153].
-
(3)
Wearable devices The design and integration of wearable detectors would fulfill the real-time, personal detection [154]. There are already pioneering trials on self-protection gears such as masks and protective suits that can display real-time detection results.
-
(4)
Recyclization of detection materials The plastics, fibers, and membranes used in detection tool kits ought to be composed of degradable material or recyclable materials as much as possible [155]. Standard regulations for material recycling should be made to help realize the full life cycle of the detection materials.
Meanwhile, considering the highly transmissible nature of the COVID-19 in the environment, the development of self-sanitizing protective equipment, antiviral surface coating, or non-toxic aerosols with detection properties would surely be powerful tools for disburden clinical tests and isolating people from the direct contact [156], therefore reducing the risk of infection.
References
Fauci AS, Lane HC, Redfield RR. COVID-19—navigating the uncharted. N Engl J Med. 2020;382:1268–9.
Hu B, Guo H, Zhou P, Shi Z-L. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19:141–54.
Rajendran DK, Rajagopal V, Alagumanian S, Santhosh Kumar T, Sathiya Prabhakaran SP, Kasilingam D. Systematic literature review on novel corona virus SARS-CoV-2: a threat to human era. Virusdisease. 2020;31:161–73.
Kim D, Lee J-Y, Yang J-S, Kim JW, Kim VN, Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–21.
Scientists in our country have discovered a new drug for the treatment of the coronavirus and obtained the authorization of the invention patent. Sci Tech Daily. 2022. http://digitalpaper.stdaily.com/http_www.kjrb.com/kjrb/html/2022-05/13/node_2.htm. Accessed 20 May 2022.
Cao Y. The impact of the hypoxia-VEGF-vascular permeability on COVID-19-infected patients. Explor (Beijing China). 2021;1:20210051.
Creech CB, Walker SC, Samuels RJ. SARS-CoV-2 vaccines. J Am Med Assoc. 2021;325:1318–20.
Amanat F, Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52:583–9.
Castells MC, Phillips EJ. Maintaining safety with SARS-CoV-2 vaccines. N Engl J Med. 2020;384:643–9.
Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–27.
Wernike K, Keller M, Conraths FJ, Mettenleiter TC, Groschup MH, Beer M. Pitfalls in SARS-CoV-2 PCR diagnostics. Transbound Emerg Dis. 2021;68:253–7.
Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol. 2020;81:104260.
Mirzaei R, Karampoor S, Sholeh M, Moradi P, Ranjbar R, Ghasemi F. A contemporary review on pathogenesis and immunity of COVID-19 infection. Mol Biol Rep. 2020;47:5365–76.
Kumar V, Doshi KU, Khan WH, Rathore AS. COVID-19 pandemic: mechanism, diagnosis, and treatment. J Chem Technol Biotechnol. 2020;96:299–308.
Kumar S, Saxena SK. Structural and molecular perspectives of SARS-CoV-2. Methods. 2021;195:23–8.
Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zhang L, Ge J, Zheng L, Zhang Y, Wang H, Zhu Y, Zhu C, Hu T, Hua T, Zhang B, Yang X, Li J, Yang H, Liu Z, Xu W, Guddat LW, Wang Q, Lou Z, Rao Z. Structure of the RNA-dependent RNA polymerasefrom COVID-19 virus. Science. 2020;368:779–82.
Bagrov DV, Glukhov GS, Moiseenko AV, Karlova MG, Litvinov DS, Zaitsev Pcapital AC, Kozlovskaya LI, Shishova AA, Kovpak AA, Ivin YY, Piniaeva AN, Oksanich AS, Volok VP, Osolodkin DI, Ishmukhametov AA, Egorov AM, Shaitan KV, Kirpichnikov MP, Sokolova OS. Structural characterization of beta-propiolactone inactivated severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) particles. Microsc Res Tech. 2022;85:562–9.
Mendonca L, Howe A, Gilchrist JB, Sheng Y, Sun D, Knight ML, Zanetti-Domingues LC, Bateman B, Krebs AS, Chen L, Radecke J, Li VD, Ni T, Kounatidis I, Koronfel MA, Szynkiewicz M, Harkiolaki M, Martin-Fernandez ML, James W, Zhang P. Correlative multi-scale cryo-imaging unveils SARS-CoV-2 assembly and egress. Nat Commun. 2021;12:4629.
Yang H, Rao Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat Rev Microbiol. 2021;19:685–700.
Fernandes Q, Inchakalody VP, Merhi M, Mestiri S, Taib N, El-Ella DMA, Bedhiafi T, Raza A, Al-Zaidan L, Mohsen MO, Al-Nesf MAY, Hssain AA, Yassine HM, Bachmann MF, Uddin S, Dermime S. Emerging COVID-19 variants and their impact on SARS-CoV-2 diagnosis, therapeutics and vaccines. Ann Med. 2022;54:524–40.
Ren L, Li C. Recent progress on the diagnosis of 2019 novel coronavirus. Transbound Emerg Dis. 2020;67:1485–91.
Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–9.
Kang T, Lu J, Yu T, Long Y, Liu G. Advances in nucleic acid amplification techniques (NAATs): COVID-19 point-of-care diagnostics as an example. Biosens Bioelectron. 2022;206:114109.
Li D, Zhang J, Li J. Primer design for quantitative real-time PCR for the emerging coronavirus SARS-CoV-2. Theranostics. 2020;10:7150–62.
Huang WE, Lim B, Hsu C-C, Xiong D, Wu W, Yu Y, Jia H, Wang Y, Zeng Y, Ji M, Chang H, Zhang X, Wang H, Cui Z. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb Biotechnol. 2020;13:950–61.
Augustine R, Hasan A, Das S, Ahmed R, Mori Y, Notomi T, Kevadiya BD, Thakor AS. Loop-Mediated isothermal amplification (LAMP): a rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. Biology-Basel. 2020;9:182.
Li S, Huang J, Ren L, Jiang W, Wang M, Zhuang L, Zheng Q, Yang R, Zeng Y, Luu LDW, Wang Y, Tai J. A one-step, one-pot CRISPR nucleic acid detection platform (CRISPR-top): application for the diagnosis of COVID-19. Talanta. 2021;233:122591.
Kim J, Lim J, Lee C. Quantitative real-time PCR approaches for microbial community studies in wastewater treatment systems: applications and considerations. Biotechnol Adv. 2013;31:1358–73.
Understanding COVID-19 PCR testing. Natl Hum Genome Res Inst. 2022. https://www.genome.gov/about-genomics/fact-sheets/Understanding-COVID-19-PCR-Testing. Accessed 11 May 2022.
Guo J, Ge J, Guo Y. Recent advances in methods for the diagnosis of corona virus disease 2019. J Clin Lab Anal. 2022;36:e24178.
Ahmadzadeh M, Vahidi H, Mahboubi A, Hajifathaliha F, Nematollahi L, Mohit E. Different respiratory samples for COVID-19 detection by standard and direct quantitative RT-PCR: a literature review. Iran J Pharm Res. 2021;20:285–99.
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63.
Lim B, Ratcliff J, Nawrot DA, Yu Y, Sanghani HR, Hsu C-C, Peto L, Evans S, Hodgson SH, Skeva A, Adam M, Panopoulou M, Zois CE, Poncin K, Vasudevan SR, Dai S, Ren S, Chang H, Cui Z, Simmonds P, Huang WE, Andersson MI. Clinical validation of optimised RT-LAMP for the diagnosis of SARS-CoV-2 infection. Sci Rep. 2021;11:16193.
Nawattanapaiboon K, Pasomsub E, Prombun P, Wongbunmak A, Jenjitwanich A, Mahasupachai P, Vetcho P, Chayrach C, Manatjaroenlap N, Samphaongern C, Watthanachockchai T, Leedorkmai P, Manopwisedjaroen S, Akkarawongsapat R, Thitithanyanont A, Phanchana M, Panbangred W, Chauvatcharin S, Srikhirin T. Colorimetric reverse transcription loop-mediated isothermal amplification (RT-LAMP) as a visual diagnostic platform for the detection of the emerging coronavirus SARS-CoV-2. Analyst. 2021;146:471–7.
Reynes B, Serra F, Palou A. Rapid visual detection of SARS-CoV-2 by colorimetric loop-mediated isothermal amplification. Biotechniques. 2021;70:218–25.
Alhassan A, Li Z, Poole CB, Carlow CKS. Expanding the MDx toolbox for filarial diagnosis and surveillance. Trends Parasitol. 2015;31:391–400.
Li B, Zou B, Ma X, Wu H, Zhang Y, Zhou G. Research progress in technologies based on isothermal amplification of nucleic acids for detection of SARS-CoV-2. Chin J Virol. 2021;37:191–200.
Zhang W, Liu K, Zhang P, Cheng W, Li L, Zhang F, Yu Z, Li L, Zhang X. CRISPR-based approaches for efficient and accurate detection of SARS-CoV-2. Lab Med. 2021;52:116–21.
Wang Z, Xiang L, Lin F, Cai Z, Ruan H, Wang J, Liang J, Wang F, Lu M, Cui W. Inhaled ACE2-engineered microfluidic microsphere for intratracheal neutralization of COVID-19 and calming of the cytokine storm. Matter. 2022;5:336–62.
Zhang Y, Ding D. Portable and visual assays for the detection of SARS-CoV‐2. View. 2022;3:20200138.
de Puig H, Lee RA, Najjar D, Tan X, Soekensen LR, Angenent-Mari NM, Donghia NM, Weckman NE, Ory A, Ng CF, Nguyen PQ, Mao AS, Ferrante TC, Lansberry G, Sallum H, Niemi J, Collins JJ. Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci Adv. 2021;7:eabh2944.
Wang Z, Zhong C. Cas12c-DETECTOR: a specific and sensitive Cas12c-based DNA detection platform. Int J Biol Macromol. 2021;193:441–9.
Patchsung M, Jantarug K, Pattama A, Aphicho K, Suraritdechachai S, Meesawat P, Sappakhaw K, Leelahakorn N, Ruenkam T, Wongsatit T, Athipanyasilp N, Eiamthong B, Lakkanasirorat B, Phoodokmai T, Niljianskul N, Pakotiprapha D, Chanarat S, Homchan A, Tinikul R, Kamutira P, Phiwkaow K, Soithongcharoen S, Kantiwiriyawanitch C, Pongsupasa V, Trisrivirat D, Jaroensuk J, Wongnate T, Maenpuen S, Chaiyen P, Kamnerdnakta S, Swangsri J, Chuthapisith S, Sirivatanauksorn Y, Chaimayo C, Sutthent R, Kantakamalakul W, Joung J, Ladha A, Jin X, Gootenberg JS, Abudayyeh OO, Zhang F, Horthongkham N, Uttamapinant C. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng. 2020;4:1140–9.
Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–9.
Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, Miao X, Streithorst JA, Granados A, Sotomayor-Gonzalez A, Zorn K, Gopez A, Hsu E, Gu W, Miller S, Pan CY, Guevara H, Wadford DA, Chen JS, Chiu CY. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38:870–4.
Guglielmi G. First CRISPR test for the coronavirus approved in the United States. Nature. 2020. https://doi.org/10.1038/d41586-020-01402-9.
Ye Q, Shao W, Meng H. Performance and application evaluation of SARS-CoV-2 antigen assay. J Med Virol. 2022. https://doi.org/10.1002/jmv.27798.
Aguilar-Shea AL, Vera-Garcia M, Guerri-Fernandez R. Rapid antigen tests for the detection of SARS-CoV-2: a narrative review. Aten Prim. 2021;53:102127.
Hsieh W-Y, Lin C-H, Lin T-C, Lin C-H, Chang H-F, Tsai C-H, Wu H-T, Lin C-S. Development and efficacy of lateral flow point-of-care testing devices for rapid and mass COVID-19 diagnosis by the detections of SARS-CoV-2 antigen and anti-SARS-CoV-2 antibodies. Diagnostics. 2021;11:1760.
Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, Sun R, Wang Y, Hu B, Chen W, Zhang Y, Wang J, Huang B, Lin Y, Yang J, Cai W, Wang X, Cheng J, Chen Z, Sun K, Pan W, Zhan Z, Chen L, Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020;92:1518–24.
Dong G-P, Guo X-J, Sun Y-A, Zhang Z, Du L-P, Li M-Y. Diagnostic techniques for COVID-19: a mini-review of early diagnostic methods. J Anal Test. 2021;5:314–26.
Nguyen NNT, McCarthy C, Lantigua D, Camci-Unal G. Development of diagnostic tests for detection of SARS-CoV-2. Diagnostics. 2020;10:905.
Hicks SM, Pohl K, Neeman T, McNamara HA, Parsons KM, He J-s, Ali SA, Nazir S, Rowntree LC, Nguyen THO, Kedzierska K, Doolan DL, Vinuesa CG, Cook MC, Coatsworth N, Myles PS, Kurth F, Sander LE, Mann GJ, Gruen RL, George AJ, Gardiner EE, Cockburn IA. C SA-C-TES. A dual-antigen enzyme-linked immunosorbent assay allows the assessment of severe acute respiratory syndrome coronavirus 2 antibody seroprevalence in a low-transmission setting. J Infect Dis. 2021;223:10–4.
Ilkhani H, Hedayat N, Farhad S. Novel approaches for rapid detection of COVID-19 during the pandemic: a review. Anal Biochem. 2021;634:114362.
Wang K, Kang S, Tian R, Zhang X, Wang Y. Imaging manifestations and diagnostic value of chest CT of coronavirus disease 2019 (COVID-19) in the Xiaogan area. Clin Radiol. 2020;75:341–7.
Hope MD, Raptis CA, Henry TS. Chest computed tomography for detection of coronavirus disease 2019 (COVID-19): Don’t rush the science. Ann Intern Med. 2020;173:147–8.
Wu X, Chen Q, Li J, Liu Z. Diagnostic techniques for COVID-19: a mini-review. J Virol Methods. 2022;301:114437.
Kim HY, Lee JH, Kim MJ, Park SC, Choi M, Lee W, Ku KB, Kim BT, Changkyun Park E, Kim HG, Kim SI. Development of a SARS-CoV-2-specific biosensor for antigen detection using scFv-Fc fusion proteins. Biosens Bioelectron. 2021;175:112868.
Mak GC, Cheng PK, Lau SS, Wong KK, Lau CS, Lam ET, Chan RC, Tsang DN. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J Clin Virol. 2020;129:104500.
Scohy A, Anantharajah A, Bodeus M, Kabamba-Mukadi B, Verroken A, Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol. 2020;129:104455.
Verma N, Patel D, Pandya A. Emerging diagnostic tools for detection of COVID-19 and perspective. Biomed Microdevices. 2020;22:83.
Weitzel T, Legarraga P, Iruretagoyena M, Pizarro G, Vollrath V, Araos R, Munita JM, Porte L. Comparative evaluation of four rapid SARS-CoV-2 antigen detection tests using universal transport medium. Travel Med Infect Dis. 2021;39:101942.
Zhu X, Wang X, Han L, Chen T, Wang L, Li H, Li S, He L, Fu X, Chen S, Xing M, Chen H, Wang Y. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens Bioelectron. 2020;166:112437.
Ghasemi S, Harmooshi NN, Rahim F. Diagnostic utility of antigen detection rapid diagnostic tests for COVID-19: a systematic review and meta-analysis. Diagn Pathol. 2022;17:36.
Fabiani L, Caratelli V, Fiore L, Scognamiglio V, Antonacci A, Fillo S, De Santis R, Monte A, Bortone M, Moscone D, Lista F, Arduini F. State of the art on the SARS-CoV-2 toolkit for antigen detection: one year later. Biosensors-Basel. 2021;11:310.
Ackerman CM, Myhrvold C, Thakku SG, Freije CA, Metsky HC, Yang DK, Ye SH, Boehm CK, Kosoko-Thoroddsen TF, Kehe J, Nguyen TG, Carter A, Kulesa A, Barnes JR, Dugan VG, Hung DT, Blainey PC, Sabeti PC. Massively multiplexed nucleic acid detection with Cas13. Nature. 2020;582:277–82.
Li Y, Li S, Wang J, Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37:730–43.
Nguyen PQ, Soenksen LR, Donghia NM, Angenent-Mari NM, de Puig H, Huang A, Lee R, Slomovic S, Galbersanini T, Lansberry G, Sallum HM, Zhao EM, Niemi JB, Collins JJ. Wearable materials with embedded synthetic biology sensors for biomolecule detection. Nat Biotechnol. 2021;39:1366–74.
Moitra P, Alafeef M, Dighe K, Frieman MB, Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14:7617–27.
Chow FW, Chan TT, Tam AR, Zhao S, Yao W, Fung J, Cheng FK, Lo GC, Chu S, Aw-Yong KL, Tang JY, Tsang CC, Luk HK, Wong AC, Li KS, Zhu L, He Z, Tam EWT, Chung TW, Wong SCY, Que TL, Fung KS, Lung DC, Wu AK, Hung IF, Woo PC, Lau SK. A rapid, simple, inexpensive, and mobile colorimetric assay COVID-19-LAMP for mass on-site screening of COVID-19. Int J Mol Sci. 2020;21:5380.
Wang S, Shu J, Lyu A, Huang X, Zeng W, Jin T, Cui H. Label-free immunoassay for sensitive and rapid detection of the SARS-CoV-2 antigen based on functionalized magnetic nanobeads with chemiluminescence and immunoactivity. Anal Chem. 2021;93:14238–46.
Liu D, Ju C, Han C, Shi R, Chen X, Duan D, Yan J, Yan X. Nanozyme chemiluminescence paper test for rapid and sensitive detection of SARS-CoV-2 antigen. Biosens Bioelectron. 2021;173:112817.
Fleischmann M, Hendra PJ, Mcquillan AJ. Raman spectra of pyridine adsorbed at a silver electrode. Chem Phys Let. 1974;26:163–6.
Katrin Kneipp Y, Wang H, Kneipp, Lev T, Perelman I, Itzkan, Ramachandra R. Dasari MSF. Single molecule detection using surface-enhanced Raman scattering (SERS). Phys Rev Lett. 1997;78:1667–70.
Shuming Nie, Emory SR. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science. 1997;275:1102–6.
Langer J, Jimenez de Aberasturi D, Aizpurua J, Alvarez-Puebla RA, Auguie B, Baumberg JJ, Bazan GC, Bell SEJ, Boisen A, Brolo AG, Choo J, Cialla-May D, Deckert V, Fabris L, Faulds K, Garcia de Abajo FJ, Goodacre R, Graham D, Haes AJ, Haynes CL, Huck C, Itoh T, Kall M, Kneipp J, Kotov NA, Kuang H, Le Ru EC, Lee HK, Li JF, Ling XY, Maier SA, Mayerhofer T, Moskovits M, Murakoshi K, Nam JM, Nie S, Ozaki Y, Pastoriza-Santos I, Perez-Juste J, Popp J, Pucci A, Reich S, Ren B, Schatz GC, Shegai T, Schlucker S, Tay LL, Thomas KG, Tian ZQ, Van Duyne RP, Vo-Dinh T, Wang Y, Willets KA, Xu C, Xu H, Xu Y, Yamamoto YS, Zhao B, Liz-Marzan LM. Present and future of surface-enhanced Raman scattering. ACS Nano. 2020;14:28–117.
Joseph MM, Narayanan N, Nair JB, Karunakaran V, Ramya AN, Sujai PT, Saranya G, Arya JS, Vijayan VM, Maiti KK. Exploring the margins of SERS in practical domain: An emerging diagnostic modality for modern biomedical applications. Biomaterials. 2018;181:140–81.
Yadav S, Sadique MA, Ranjan P, Kumar N, Singhal A, Srivastava AK, Khan R. SERS based lateral flow immunoassay for point-of-care detection of SARS-CoV-2 in clinical samples. ACS Appl Bio Mater. 2021;4:2974–95.
Srivastav S, Dankov A, Adanalic M, Grzeschik R, Tran V, Pagel-Wieder S, Gessler F, Spreitzer I, Scholz T, Schnierle B, Anastasiou OE, Dittmer U, Schlucker S. Rapid and sensitive SERS-based lateral flow test for SARS-CoV2-specific IgM/IgG antibodies. Anal Chem. 2021;93:12391–9.
Peng Y, Lin C, Long L, Masaki T, Tang M, Yang L, Liu J, Huang Z, Li Z, Luo X, Lombardi JR, Yang Y. Charge-transfer resonance and electromagnetic enhancement synergistically enabling mxenes with excellent SERS sensitivity for SARS-CoV-2 S protein detection. Nanomicro Lett. 2021;13:52.
Yang Y, Peng Y, Lin C, Long L, Hu J, He J, Zeng H, Huang Z, Li ZY, Tanemura M, Shi J, Lombardi JR, Luo X. Human ACE2-functionalized gold “virus-trap” nanostructures for accurate capture of SARS-CoV-2 and single-virus SERS detection. Nanomicro Lett. 2021;13:109.
Huang G, Zhao H, Li P, Liu J, Chen S, Ge M, Qin M, Zhou G, Wang Y, Li S, Cheng Y, Huang Q, Wang J, Wang H, Yang L. Construction of optimal SERS hotspots based on capturing the spike receptor-binding domain (RBD) of SARS-CoV-2 for highly sensitive and specific detection by a fish model. Anal Chem. 2021;93:16086–95.
Liu H, Dai E, Xiao R, Zhou Z, Zhang M, Bai Z, Shao Y, Qi K, Tu J, Wang C, Wang S. Development of a SERS-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-SARS-CoV-2 IgM/IgG in clinical samples. Sens Actuators B Chem. 2021;329:129196.
Leong SX, Leong YX, Tan EX, Sim HYF, Koh CSL, Lee YH, Chong C, Ng LS, Chen JRT, Pang DWC, Nguyen LBT, Boong SK, Han X, Kao YC, Chua YH, Phan-Quang GC, Phang IY, Lee HK, Abdad MY, Tan NS, Ling XY. Noninvasive and point-of-care surface-enhanced raman scattering (SERS)-based breathalyzer for mass screening of coronavirus disease 2019 (COVID-19) under 5 min. ACS Nano. 2022;16:2629–39.
Han XX, Rodriguez RS, Haynes CL, Ozaki Y, Zhao B. Surface-enhanced Raman spectroscopy. Nat Rev Methods Primers. 2021;1:87.
Leland C, Clark, Lyons C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann N Y Acad Sci. 1962;102:29–45.
Mahshid SS, Flynn SE, Mahshid S. The potential application of electrochemical biosensors in the COVID-19 pandemic: A perspective on the rapid diagnostics of SARS-CoV-2. Biosens Bioelectron. 2021;176:112905.
Luong AD, Buzid A, Vashist SK, Luong JHT. Perspectives on electrochemical biosensing of COVID-19. Curr Opin Electrochem. 2021;30:100794.
Peng R, Pan Y, Li Z, Qin Z, Rini JM, Liu X. SPEEDS: A portable serological testing platform for rapid electrochemical detection of SARS-CoV-2 antibodies. Biosens Bioelectron. 2022;197:113762.
Kaushik AK, Dhau JS, Gohel H, Mishra YK, Kateb B, Kim NY, Goswami DY. Electrochemical SARS-CoV-2 sensing at point-of-care and artificial intelligence for intelligent COVID-19 management. ACS Appl Bio Mater. 2020;3:7306–25.
Tian J, Liang Z, Hu O, He Q, Sun D, Chen Z. An electrochemical dual-aptamer biosensor based on metal-organic frameworks MIL-53 decorated with Au@Pt nanoparticles and enzymes for detection of COVID-19 nucleocapsid protein. Electrochim Acta. 2021;387:138553.
de Lima LF, Ferreira AL, Torres MDT, de Araujo WR, de la Fuente-Nunez C. Minute-scale detection of SARS-CoV-2 using a low-cost biosensor composed of pencil graphite electrodes. Proc Natl Acad Sci USA. 2021;118:e2106724118.
Eissa S, Zourob M. Development of a low-cost cotton-tipped electrochemical immunosensor for the detection of SARS-CoV-2. Anal Chem. 2021;93:1826–33.
Alafeef M, Dighe K, Moitra P, Pan D. Rapid, Ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2020;14:17028–45.
Ali MA, Hu C, Jahan S, Yuan B, Saleh MS, Ju E, Gao SJ, Panat R. Sensing of COVID-19 antibodies in seconds via aerosol jet nanoprinted reduced-graphene-oxide-coated 3D electrodes. Adv Mater. 2021;33:2006647.
Wang L, Wang X, Wu Y, Guo M, Gu C, Dai C, Kong D, Wang Y, Zhang C, Qu D, Fan C, Xie Y, Zhu Z, Liu Y, Wei D. Rapid and ultrasensitive electromechanical detection of ions, biomolecules and SARS-CoV-2 RNA in unamplified samples. Nat Biomed Eng. 2022;6:276–85.
Guo K, Wustoni S, Koklu A, Diaz-Galicia E, Moser M, Hama A, Alqahtani AA, Ahmad AN, Alhamlan FS, Shuaib M, Pain A, McCulloch I, Arold ST, Grunberg R, Inal S. Rapid single-molecule detection of COVID-19 and MERS antigens via nanobody-functionalized organic electrochemical transistors. Nat Biomed Eng. 2021;5:666–77.
Griffin JH, Downard KM. Mass spectrometry analytical responses to the SARS-CoV2 coronavirus in review. Trac-Trend Anal Chem. 2021;142:116328.
Lima NM, Fernandes BLM, Alves GF, de Souza JCQ, Siqueira MM, do Nascimento MP, Moreira OBO, Sussulini A, de Oliveira MAL. Mass spectrometry applied to diagnosis, prognosis, and therapeutic targets identification for the novel coronavirus SARS-CoV-2: a review. Anal Chim Acta. 2022;1195:339385.
Mahmud I, Garrett TJ. Mass spectrometry techniques in emerging pathogens studies: COVID-19 perspectives. J Am Soc Mass Spectrom. 2020;31:2013–24.
Rocca MF, Zintgraff JC, Dattero ME, Santos LS, Ledesma M, Vay C, Prieto M, Benedetti E, Avaro M, Russo M, Nachtigall FM, Baumeister E. A combined approach of MALDI-TOF mass spectrometry and multivariate analysis as a potential tool for the detection of SARS-CoV-2 virus in nasopharyngeal swabs. J Virol Methods. 2020;286:113991.
Nachtigall FM, Pereira A, Trofymchuk OS, Santos LS. Detection of SARS-CoV-2 in nasal swabs using MALDI-MS. Nat Biotechnol. 2020;38:1168–73.
Delafiori J, Navarro LC, Siciliano RF, de Melo GC, Busanello ENB, Nicolau JC, Sales GM, de Oliveira AN, Val FFA, de Oliveira DN, Eguti A, dos Santos LA, Dalçóquio TF, Bertolin AJ, Abreu-Netto RL, Salsoso R, Baía-da-Silva D, Marcondes-Braga FG, Sampaio VS, Judice CC, Costa FTM, Durán N, Perroud MW, Sabino EC, Lacerda MVG, Reis LO, Fávaro WJ, Monteiro WM, Rocha AR, Catharino RR. COVID-19 automated diagnosis and risk assessment through metabolomics and machine learning. Anal Chem. 2021;93:2471–79.
Cardozo KHM, Lebkuchen A, Okai GG, Schuch RA, Viana LG, Olive AN, Lazari CdS, Fraga AM, Granato CFH, Pintão MCT, Carvalho VM. Establishing a mass spectrometry-based system for rapid detection of SARS-CoV-2 in large clinical sample cohorts. Nat Commun. 2020;11:6201.
Li J, Guo M, Tian X, Wang X, Yang X, Wu P, Liu C, Xiao Z, Qu Y, Yin Y, Wang C, Zhang Y, Zhu Z, Liu Z, Peng C, Zhu T, Liang Q. Virus-host interactome and proteomic survey reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. Medicine. 2021;2:99–112.
Bezstarosti K, Lamers MM, Doff WAS, Wever PC, Thai KTD, van Kampen JJA, Haagmans BL, Demmers JAA. Targeted proteomics as a tool to detect SARS-CoV-2 proteins in clinical specimens. PLoS One. 2021;16:e0259165.
Zhao P, Praissman JL, Grant OC, Cai Y, Xiao T, Rosenbalm KE, Aoki K, Kellman BP, Bridger R, Barouch DH, Brindley MA, Lewis NE, Tiemeyer M, Chen B, Woods RJ, Wells L. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe. 2020;28:586–601.
Wandernoth P, Kriegsmann K, Groh-Mohanu C, Daeumer M, Gohl P, Harzer O, Kriegsmann M, Kriegsmann J. Detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by mass spectrometry. Viruses-Basel. 2020;12:849.
Emergency use authorization (EUA) summary: ethos laboratories SARS-CoV-2 MALDI-TOF assay (ethos laboratories). US Food Drug Adm. 2020. https://www.fda.gov/media/140780/download. Accessed 11 May 2022.
Shih H-I, Wang H-C, Su I-J, Hsu H-C, Wang J-R, Sun HFS, Chou C-H, Ko W-C, Hsieh M-I, Wu C-J. Viral respiratory tract infections in adult patients attending outpatient and emergency departments, Taiwan, 2012–2013: A PCR/electrospray ionization mass spectrometry study. Medicine. 2015;94:e1545.
Coronavirus (COVID-19) update: FDA authorizes first COVID-19 diagnostic test using breath samples. US Food Drug Adm. 2022. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-covid-19-diagnostic-test-using-breath-samples. Accessed 11 May 2022.
Hu ZL, Huo MZ, Ying YL, Long YT. Biological nanopore approach for single-molecule protein sequencing. Angew Chem Int Ed. 2021;60:14738–49.
Brinkerhoff H, Kang ASW, Liu JQ, Aksimentiev A, Dekker C. Multiple rereads of single proteins at single-amino acid resolution using nanopores. Science. 2021;374:1509–13.
Laszlo AH, Derrington IM, Ross BC, Brinkerhoff H, Adey A, Nova IC, Craig JM, Langford KW, Samson JM, Daza R, Doering K, Shendure J, Gundlach JH. Decoding long nanopore sequencing reads of natural DNA. Nat Biotechnol. 2014;32:829–33.
Imran A, Posokhova I, Qureshi HN, Masood U, Riaz MS, Ali K, John CN, Hussain MI, Nabeel M. AI4COVID-19: AI enabled preliminary diagnosis for COVID-19 from cough samples via an app. Inf Med Unlocked. 2020;20:100378.
Brown C, Chauhan J, Grammenos A, Han J, Hasthanasombat A, Spathis D, Xia T, Cicuta P, Mascolo C. Exploring automatic diagnosis of COVID-19 from crowdsourced respiratory sound data. In: Proc 26th ACM SIGKDD Int Confer Knowl Discov Data Mining. 2020. 3474-84.https://doi.org/10.1145/3394486.3412865.
Cohen-McFarlane M, Goubran R, Knoefel F. Novel coronavirus cough database: NoCoCoDa. IEEE Access. 2020;8:154087–94.
Sharma N, Krishnan P, Kumar R, Ramoji S, Chetupalli SR, Nirmala R, Ghosh PK, Ganapathy S. Coswara-a database of breathing, cough, and voice sounds for COVID-19 diagnosis. In: Proc Interspeech. 2020; p. 4811–5. https://doi.org/10.21437/Interspeech.2020-2768.
Orlandic L, Teijeiro T, Atienza D. The COUGHVID crowdsourcing dataset, a corpus for the study of large-scale cough analysis algorithms. Sci Data. 2021;8:156.
Chaudhari G, Jiang X, Fakhry A, Han A, Xiao J, Shen S, Khanzada A. Virufy. Global applicability of crowdsourced and clinical datasets for AI detection of COVID-19 from cough. arXiv. 2020. https://doi.org/10.48550/arXiv.2011.13320.
Chowdhury NK, Kabir MA, Rahman MM, Islam SMS. Machine learning for detecting COVID-19 from cough sounds: an ensemble-based MCDM method. Comput Biol Med. 2022;145:105405.
Rahman T, Ibtehaz N, Khandakar A, Hossain MSA, Mekki YMS, Ezeddin M, Bhuiyan EH, Ayari MA, Tahir A, Qiblawey Y, Mahmud S, Zughaier SM, Abbas T, Al-Maadeed S, Chowdhury MEH. QUCoughScope: an intelligent application to detect COVID-19 patients using cough and breath sounds. Diagnostics. 2022;12:920.
Pahar M, Klopper M, Warren R, Niesler T. COVID-19 cough classification using machine learning and global smartphone recordings. Comput Biol Med. 2021;135:104572.
Alyasseri ZAA, Al-Betar MA, Doush IA, Awadallah MA, Abasi AK, Makhadmeh SN, Alomari OA, Abdulkareem KH, Adam A, Damasevicius R, Mohammed MA, Zitar RA. Review on COVID-19 diagnosis models based on machine learning and deep learning approaches. Expert Syst. 2021;39:e12759.
Son M-J, Lee S-P. COVID-19 diagnosis from crowdsourced cough sound data. Appl Sci-Basel. 2022;12:1795.
Wu Z, Xue R, Shao M. Knowledge graph analysis and visualization of AI technology applied in COVID-19. Environ Sci Pollut Res. 2022;29:26396–408.
Hsiao WW, Le TN, Pham DM, Ko HH, Chang HC, Lee CC, Sharma N, Lee CK, Chiang WH. Recent advances in novel lateral flow technologies for detection of COVID-19. Biosensors-Basel. 2021;11:295.
Wang Y, Chen H, Wei H, Rong Z, Wang S. Tetra-primer ARMS-PCR combined with dual-color fluorescent lateral flow assay for the discrimination of SARS-CoV-2 and its mutations with a handheld wireless reader. Lab Chip. 2022;22:1531–41.
Akarapipad P, Kaarj K, Breshears LE, Sosnowski K, Baker J, Nguyen BT, Eades C, Uhrlaub JL, Quirk G, Nikolich-Zugich J, Worobey M, Yoon JY. Smartphone-based sensitive detection of SARS-CoV-2 from saline gargle samples via flow profile analysis on a paper microfluidic chip. Biosens Bioelectron. 2022;207:114192.
Kim S, Akarapipad P, Nguyen BT, Breshears LE, Sosnowski K, Baker J, Uhrlaub JL, Nikolich-Zugich J, Yoon JY. Direct capture and smartphone quantification of airborne SARS-CoV-2 on a paper microfluidic chip. Biosens Bioelectron. 2022;200:113912.
Day AS, Ulep TH, Safavinia B, Hertenstein T, Budiman E, Dieckhaus L, Yoon JY. Emulsion-based isothermal nucleic acid amplification for rapid SARS-CoV-2 detection via angle-dependent light scatter analysis. Biosens Bioelectron. 2021;179:113099.
Papadakis G, Pantazis AK, Fikas N, Chatziioannidou S, Tsiakalou V, Michaelidou K, Pogka V, Megariti M, Vardaki M, Giarentis K, Heaney J, Nastouli E, Karamitros T, Mentis A, Zafiropoulos A, Sourvinos G, Agelaki S, Gizeli E. Portable real-time colorimetric LAMP-device for rapid quantitative detection of nucleic acids in crude samples. Sci Rep. 2022;12:3775.
Xiao M, Tian F, Liu X, Zhou Q, Pan J, Luo Z, Yang M, Yi C. Virus detection: from state-of-the-art laboratories to smartphone-based point-of-care testing. Adv Sci. 2022;9: 2105904.
Laghrib F, Saqrane S, El Bouabi Y, Farahi A, Bakasse M, Lahrich S, El Mhammedi MA. Current progress on COVID-19 related to biosensing technologies: new opportunity for detection and monitoring of viruses. Microchem J. 2021;160:105606.
Panpradist N, Kline EC, Atkinson RG, Roller M, Wang Q, Hull IT, Kotnik JH, Oreskovic AK, Bennett C, Leon D, Lyon V, Gilligan-Steinberg SD, Han PD, Drain PK, Starita LM, Thompson MJ, Lutz BR. Harmony COVID-19: a ready-to-use kit, low-cost detector, and smartphone app for point-of-care SARS-CoV-2 RNA detection. Sci Adv. 2021;7:eabj1281.
Schary W, Paskali F, Rentschler S, Ruppert C, Wagner GE, Steinmetz I, Deigner HP, Kohl M. Open-source, adaptable, all-in-one smartphone-based system for quantitative analysis of point-of-care diagnostics. Diagnostics. 2022;12:589.
Zhao H, Liu F, Xie W, Zhou TC, OuYang J, Jin L, Li H, Zhao CY, Zhang L, Wei J, Zhang YP, Li CP. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sens Actuators B Chem. 2021;327:128899.
Beduk T, Beduk D, de Oliveira Filho JI, Zihnioglu F, Cicek C, Sertoz R, Arda B, Goksel T, Turhan K, Salama KN, Timur S. Rapid point-of-care COVID-19 diagnosis with a gold-nanoarchitecture-assisted laser-scribed graphene biosensor. Anal Chem. 2021;93:8585–94.
Soares RRG, Akhtar AS, Pinto IF, Lapins N, Barrett D, Sandh G, Yin X, Pelechano V, Russom A. Sample-to-answer COVID-19 nucleic acid testing using a low-cost centrifugal microfluidic platform with bead-based signal enhancement and smartphone read-out. Lab Chip. 2021;21:2932–44.
Adrover-Jaume C, Alba-Patino A, Clemente A, Santopolo G, Vaquer A, Russell SM, Baron E, Gonzalez Del Campo MDM, Ferrer JM, Berman-Riu M, Garcia-Gasalla M, Aranda M, Borges M, de la Rica R. Paper biosensors for detecting elevated IL-6 levels in blood and respiratory samples from COVID-19 patients. Sens Actuators B Chem. 2021;330:129333.
Yuan K, de la Asuncion-Nadal V, Cuntin-Abal C, Jurado-Sanchez B, Escarpa A. On-board smartphone micromotor-based fluorescence assays. Lab Chip. 2022;22:928–35.
Matsuda EM, de Campos IB, de Oliveira IP, Colpas DR, Carmo A, Brigido LFM. Field evaluation of COVID-19 antigen tests versus RNA based detection: potential lower sensitivity compensated by immediate results, technical simplicity, and low cost. J Med Virol. 2021;93:4405–10.
Yuce M, Filiztekin E, Ozkaya KG. COVID-19 diagnosis-a review of current methods. Biosens Bioelectron. 2021;172:112752.
Liu G, Rusling JF. COVID-19 antibody tests and their limitations. ACS Sens. 2021;6:593–612.
Mo X, Qin W, Fu Q, Guan M. Understanding the influence factors in viral nucleic acid test of 2019 novel coronavirus (2019-nCoV). Chin J Lab Med. 2020;43:213–6.
Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn. 2020;20:453–4.
Azzi L, Maurino V, Baj A, Dani M, d’Aiuto A, Fasano M, Lualdi M, Sessa F, Alberio T. Diagnostic salivary tests for SARS-CoV-2. J Dent Res. 2021;100:115–23.
Liu Z, Frasconi M, Lei J, Brown ZJ, Zhu Z, Cao D, Iehl J, Liu G, Fahrenbach AC, Botros YY, Farha OK, Hupp JT, Mirkin CA, Fraser Stoddart J. Selective isolation of gold facilitated by second-sphere coordination with alpha-cyclodextrin. Nat Commun. 2013;4:1855.
Pati P, McGinnis S, Vikesland PJ. Waste not want not: life cycle implications of gold recovery and recycling from nanowaste. Environ Sci Nano. 2016;3:1133–43.
Panchal SS, Vasava DV. Biodegradable polymeric materials: synthetic approach. ACS Omega. 2020;5:4370–9.
Vroman I, Tighzert L. Biodegradable polymers. Materials. 2009;2:307–44.
Sun L, Huang H, Ding Q, Guo Y, Sun W, Wu Z, Qin M, Guan Q, You Z. Highly transparent, stretchable, and self-healable ionogel for multifunctional sensors, triboelectric nanogenerator, and wearable fibrous electronics. Adv Fiber Mater. 2021;4:98–107.
Yang J, Li X-L, Zhou J-W, Wang B, Cheng J-L. Fiber-shaped supercapacitors: advanced strategies toward high-performances and multi-functions. Chin J Poly Sci. 2020;38:403–22.
Shi Q, Sun J, Hou C, Li Y, Zhang Q, Wang H. Advanced functional fiber and smart textile. Adv Fiber Mater. 2019;1:3–31.
Chen H, Dong X, Zhao Y, Wang D. Recycling and chemical upcycling of waste disposable medical masks. Acta Polym Sin. 2020;51:1295–306.
Zhou J, Hu Z, Zabihi F, Chen Z, Zhu M. Progress and perspective of antiviral protective material. Adv Fiber Mater. 2020;2:123–39.
Acknowledgements
This work was partially supported by the National Key Research and Development Program of China (2021YFA1201301/2021YFA1201300), Science and Technology Commission of Shanghai Municipality (20JC1414900, 19ZR1470600).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
There are no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, G., Wang, L., Meng, Z. et al. Visual Detection of COVID-19 from Materials Aspect. Adv. Fiber Mater. 4, 1304–1333 (2022). https://doi.org/10.1007/s42765-022-00179-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42765-022-00179-y