Abstract
Zea mays L corps apport to human consumption, complex matrices of compounds such as free and bounded phenolics, flavonoids and anthocyanins with high nutritional values and proved health benefits, which are dynamically synthesized since sprouting of grains. This study presents for the first time a Nuclear Magnetic Resonance (NMR) non-targeted metabolomics study of aqueous methanolic extracts of Mexican native blue and red Zea mays L. sprouts, produced with a specific germination scheme, at three different controlled temperatures. The proposed model comprises the rationalization of (poly)-phenolics metabolism dynamics as a function of sprouting time and temperature, which can be identified by more than thirty 1H NMR discriminant resonances at a chemical shift range between 7.7 and 6.3 ppm -mostly comprising typical hydroxyphenyl polyphenolic 1H frequencies- obtained with multivariate statistical analysis. Both principal component (PCA) and orthogonal projections to latent structures discriminant analysis (OPLS-DA) reveal a unique maize strain-, temperature- and time-dependent mapping of polyphenolic machinery during sprouting that might serve for optimizing germination schemes. Strengths and limitations of PCA and OPLS-DA analysis of non-targeted (poly)-phenolics NMR data matrix obtained from different blue and red maize sprouts’ methanolic extracts are discussed. Furthermore, a clear inverse correlation between temperature- and time-dependent signal intensity modulation of phenolic resonances and water line widths at half height is observed, suggesting a mechanism of how solvent could participate within the complex metabolomic matrix formation during germination. Finally, non-targeted NMR metabolomics data from sprouts’ methanolic extracts are contrasted with temperature- and time-dependent total phenolic contents obtained with standard quantitative methods.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Synthesis de novo of phytochemicals in plants often starts during the first stages of germination. Their importance to human health has been extensively shown [1, 2]. Epidemiological experiments have demonstrated the benefits of consuming phenolics and flavonoids from food sources, for reducing the risks of ailing chronic and degenerative diseases, due to their condition as natural antioxidants and anti-aging agents [3, 4]. Phenolics in maize are well known to exhibit antioxidant and prebiotic activity, as well as relevant chemo-preventive properties [5,6,7]. However, different phenotypes of maize present different bioactivities, due to their different phenolic and flavonoid profiles. For instance, purple, blue and red pigmented maize inhibits colorectal carcinogenesis in rats [8]. In terms of Mexican strains, the antioxidant activity of white and Mexican—American blue Zea mays L. has been reported [9,10,11], whereas the antioxidant activity is strongly correlated with the content of free and bounded phenolics. The later represents roughly 80% of the Total Phenolic Content (TPC) not only for maize, but also for wheat, barley, rice, oats and corn [12]. Those ratios can be highly modulated during germination [4], wherein in one hand, a seven-days sprouting process increases TPC and Total Flavonoid Content (TFC) in buckwheat [13] and in the other hand germination may decrease both levels of TPC and TFC in kidney beans [14], showing that mechanisms of phytochemical development during germination varies between plant species. As a consequence, the potential health benefits of sprouted grains have gain powerful attention [15]. Thus, the need to propose novel analytical methods for tracking biochemistry machinery during sprouting could shed light in strategies for optimizing accurate germination conditions in order to maximize natural antioxidant production, in an easy, straightforward and controlled way. In this sense, the multivariate patterns embedded in multiple patterns that define latent parameters in food stuff such as geographical origin, variety or even more specific such as metabolites related to beneficial properties to human health are coded within their nuclear magnetic resonance (NMR) spectra as rather simple resonance markers. In consequence, the proton NMR-based metabolomics profiling in combination with multivariate statistical analysis (MSA) comprise a robust non-invasive methodology for establishing biochemical associations amongst present metabolites within an extract. Present metabolomics study proposes a non-invasive, feasible, fast and highly reproducible approach to find specific chemical markers associated to imposed sprouting conditions that can reveal the most favorable germination conditions for expression of phenolic moieties with potential antioxidant activities.
2 Materials and methods
2.1 Materials
2.1.1 Seed rinsing
Blue and red pigmented maize seeds were collected from Chilchotla, Puebla, Mexico. All seeds were washed with distilled water prior to be immersed in a 1.4 ppm solution of colloidal silver solution (at 65%w/v, CAS No. 7440–22-4) during 15 min. Washing procedure finishes with a last rinse with distilled water for full silver removal.
2.1.2 Sprouting of pigmented maize seeds
A set of fifty pigmented maize seeds were settled in a Petri dish of 100 × 15 mm surface, as a function of selected sampling: two strains (blue and red maize), three different temperatures (10 °C, 25 °C and 30 °C), seven sprouting days (D + 1, D + 2, D + 3, D + 4, D + 5, D + 6 and D + 7 for samples germinated at 25 °C and 30 °C; D + 7, D + 14, D + 21, D + 28, D + 35, D + 42 and D + 49 for samples germinated at 10 °C) giving a total of 21 Petri dishes per maize strain. Each Petri dish had a cotton bed that was in turn moisten with a 0.175 ppm solution of 65%w/v colloidal silver. Seeds were carefully settled over the cotton bed and sprayed with distilled water every 48 h. Moisture was controlled to be in a range of around 80%. Sprouting essays at 25 and 30 °C, were carried out in a standard sprouting machine, and 10 °C essays were conducted in a controlled fridge. In all cases, sprouting was left at constant generic lighting conditions.
2.1.3 Maize’s germ flours
Once sprouts were collected after finishing each assigned germination day, samples were immediately frozen at −80 °C for 24 h. Afterwards they were cryogenized with liquid nitrogen for 10 min and immediately lyophilized during 72 h (Labconco, Corp. KS, USA) with a pressure of 0.015 mbar and a temperature of −50 °C to permanently quench metabolism. After dehydration, samples were milled and obtained flours were stored in a desiccator cabinet until its use.
2.1.4 Extraction of Phenolic compounds in aqueous methanol
Phenolic extraction was adapted from previous reports [16], with the following modifications: 1 g of maize flour was suspended in 15 mL of aqueous methanol (80% v/v) with exhaustive shake with the use of a vortex at 3500 × g for 5 min. Once the solution was homogenized, a water-bath at 90 °C with manual shaking was applied during 20 min. After full homogenization, samples were stored at −80 °C until its use.
3 Methods
3.1 Determination of total phenolic content in methanolic extracts
Folin-Ciocalteu method was adapted from Yang and coworkers [17] as follows: 20 µL of each phenolic extract were added to a solution containing 50 µL of Folin-Ciocalteu reagent (Sigma Aldrich), 250 µL of 7.5%w/v solution of Na2CO3 (Sigma Aldrich, CAS No. 497–19-8) and 280 µL of distilled water. Total phenolic content of blue and red maize sprouts expressed as milligrams of gallic acid equivalents per 100 g of dry flour weight (mg GAE / 100 g DW) were obtained by means of classical 756 nm absorbance readings with a 96-wells Microplate Reader Spectrophotometer (Varioskan Flash Spectral Scanning Multimode Reader, Thermo Scientific) based on a calibration curve with a solution containing variable concentrations of gallic acid (3,4,5-trihydroxybenzoic acid, CAS No. 149–91-7) running from 0 to 600 µg gallic acid/mL, 50 µL of Folin-Ciocalteu reagent and 250 µL of Na2CO3 (7.5%w/v).
3.2 UV–VIS spanning
UV–VIS absorbance spans from 200 to 800 nm of all methanolic extracts treated with Folin-Ciocalteu / carbonate solution (Sect. 2.2.1.) at different pH and temperature conditions were carried out in a 96-wells Microplate Reader Spectrometer (Varioskan Flash Spectral Scanning Multimode Reader, Thermo Scientific, Oregon, USA), with an increment of 2 nm per scan. Raw (A) and first derivative (ΔA) Absorbance spectra were processed and analyzed with the Skanlt Software (V 2.4.5, Thermo Scientific, Oregon, USA). Relevant absorbance data for quantifications were deduced from A/ΔA analysis.
3.3 Nuclear magnetic resonance spectroscopy
Solution-state NMR spectroscopy were carried with a Bruker Avance-III HD spectrometer (Bruker Biospin, Rheinstetten, Germany), operating at 14.1 T of magnetic field (equivalent to 600 MHz of proton frequency). Sample preparation comprised a mixture of 600 µL of each phenolic extract (a set of 21 samples per pigmented maize strain, a total of 42 NMR samples, see Sect. 2.1.4), with 75 µL of deuterated d4-methanol (Cambridge Isotope Labs). The following NMR schemes were acquired for the full set of blue and red maize sprouts samples:
-
a
Standard direct-excitation proton one dimensional NMR experiments {1D-1H-NMR} were acquired with 64 transients of 36 K complex points, having recycling delays of 4 s and acquisition time of 2.3 s, produced in turn experimental times per sample of 6′28′’. No apodization function was applied during Fourier Transform.
-
b
{1Hwater_presat NMR}: 1D single pulse NOESY experiments with an off-resonance shaped-pulse water presaturation during both relaxation delay (10 s) and mixing time (100 ms) and a 36.3 × 10–5 W power level irradiation [18,19,20], were acquired for all samples at the following conditions: a total of 64 transients were collected within 36 K complex data points, with a spectral width of 7812.5 Hz and acquisition times of 2.31 s, produced experimental times of 6′ 26′’.
3.4 Multivariate statistical analysis (MSA)
NMR post-processing for producing the MSA input variables was carried out as follows: ppm calibration and manual phase corrections were conducted with the use of Bruker TopSpin 4.0.8 software. Global and intermediate baseline corrections, least-squares NMR alignments, variable size bucketing for untargeted profiling and data matrix normalization were carried out with NMRProcFlow software [21]. Scaling and statistical analysis workflow for obtaining the Principal Component (PCA) and the Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA), from the constant sum normalized NMR data matrix, were developed with the BioStatFlow 2.9.2 software. In all cases, T2 Hotelling’s regions depicted by ellipses in score plots of each model define a 95% confidence interval [22]. R2X, R2Y and Q2 statistical parameters that define the quality of each model are expressed in all cases [23].
4 Results and discussion
4.1 NMR non-targeted metabolomics profiling
De novo synthesis of phytochemicals with antioxidant activity such as polyphenolic compounds [9, 10] in Zea Mays strains have gain important attraction, mostly during germination of plant seeds. Determination of said antioxidants have been mostly carried out by colorimetric and/or chromatographic approaches, whereas despite their robustness they present certain complexity in terms of sample preparation and chemical manipulations that could be time-consuming, laborious and costly. For that it is highly desirable to propose a set of cutting-edge techniques for determining and quantify antioxidants during germination in an easy and straightforward way, mostly when an important number of variables play a significant role during sprouting such as temperature, time or type of strain. Highly reproducible proton one-dimensional NMR schemes with a multipresaturation module for elimination of intense solvent signals that allow an important gain in the signal-to-noise ratio of produced metabolites during germination seems an attractive alternative for elucidation of temperature- and time-dependent variables involved in biosynthesis of antioxidants at said conditions.
Figure S.I.1 schemes the complete workflow for obtaining the methanolic extracts of maize’s sprouts needed for the NMR non-targeted metabolomics study: i) temperature- and time-controlled sprouting of pigmented maize seeds (Figure S.I.1A, see Sect. 2.1.2); ii) obtention of maize’s germ flours (Figure S.I.1.B, see Sect. 2.1.3) and iii) extraction of phenolic compounds in aqueous methanol (Figure S.I.1.C, see Sect. 2.1.4). The full data set comprising 42 samples (2 maize strains × 3 germination temperatures × 7 sprouting days) were profiled with standard 1D-1H-NMR direct irradiation and 1D-{1Hwater_presat NMR} water-to-methanol multi-presaturation schemes.
Standard 1D-1H-NMR spectra depicted in Fig. 1. throw up the following observations: When no special pulse sequence is applied for the full set of methanolic red and blue maize sprouts solutions, their major components comprise the observed 3.36 and 4.85 ppm resonances of respectively methanol and water. Frequency at 3.36 ppm does not suffer any significant change as a function of the complex matrix formed during sprouting or germination temperature for both Zea mays strains. However, interesting temperature- and time-dependent line-shape patterns are observed for the 4.85 ppm water resonances at each selected condition. For germinates carried out at 10 °C, water line widths at half height (LWHH, see also Table S.I.1) retain constant at around 200–250 Hz, with a doublet type signal for methanolic solutions comprising both strains sprouted during the first 21 days. An important LWHH decay due to a doublet to singlet line-shape modification of water signal is observed for the methanolic blue maize germ solution at D + 28 (LWHH = 206.1 Hz) and furthermore, a similar time-dependent LWHH behavior was observed for the red pigmented strain corresponding to the D + 42 sprout solution (LWHH = 145.67 Hz). A more drastic LWHH decay as a function of sprouting time for both maize strains were observed at 25 and 30 °C. Regarding line widths at 25 °C, a LWHH decay (from 248.01 Hz [D + 1] to 80.46 Hz [D + 7]) associated to a water line-shape modification since D + 2 (199.72 Hz) is observed for blue maize strains. More drastically, sampling from red maize extracts present a 242.83 Hz doublet signal at D + 2, a broad singlet signal (192.27 Hz) and sharper water signal singlets since D + 3 (88.7 Hz), with a minimum LWHH of 37.07 Hz at D + 6. Comprising LWWH water signal evolutions at T = 30 °C, generally they present milder time-dependent signal decay and line sharpening with respect observations described at T = 25 °C. For blue maize sprouts, doublet-to-singlet water signal coalescence occurs at D + 2 (209.07 Hz), likewise its lower temperature counterpart. Singlets’ line-sharpening begin from D + 3 (232.61 Hz), falling down to a LWHH value of 113.97 Hz at D + 7. In the same sense, doublet-to-singlet water signal coalescence begins at D + 3 (70.41 Hz) for red maize sprouts’ extracts at T = 30 °C, finding its sharpest LWHH value at D + 5 (51.86 Hz). Signal evolutions of 4.85 ppm water resonance has to be contrasted with the metabolomics profiles of temperature- and time-dependent blue and red pigmented maize sprouts’ methanolic extracts obtained with the solvent multi-presaturation {1Hwater_presat} NMR methodology (Fig. 2).
Temperature- and time-dependent standard 1D-1H-NMR overlaid (top) and stacked (bottom) spectra of blue (left) and red (right) maize sprouts’ extracts. Stacked spectra present the full frequency range that contain metabolites’ signals from methanolic extracts (Fig. 2A). Overlaid projections only show resonances from solvents: water (4.85 ppm) and methanol (3.36 ppm) that in turn represent the zoomed area of stacked plots (blue and red dotted lines). Color codes for overlaid spectra depend on sprouting times per controlled-temperature germination: D + 1 to D + 7 for sprouting essays at T = 25 and 30 oC; D + 7 to D + 49 for sprouting at 10 oC, whereas each color code (black, gray, brown, magenta, blue green and red) are specified in a top centered legend. All spectra were plotted at identical conditions in terms of signal intensity
One dimensional {1Hwater_presat NMR} stacked (A) and overlaid (B) spectra of methanolic blue (below dashed black line in A) and red (above dashed black line in A) pigmented maize sprouts solutions as a function of germination time and temperature. Possible resonances’ binning strategies are highlighted with gray boxes. However, data dimensionality for the present study was reduced by means of the NMR bucketing process of only 1H-aromatic resonances at a chemical shift range between 7.7 and 6.3 ppm (C)
As evidenced in Fig. 1, signal to noise ratio of metabolites’ proton resonances in standard 1D-1H-NMR are severely penalized due to the presence of intense water and methanol signals. For that, a solvent multi-suppression scheme is needed for obtaining a 16-fold signal-to-noise ratio gain of maize sprouts’ extracts [18,19,20], needed to prepare the output data matrix for multivariate statistical analysis.
4.2 Principal component and orthogonal partial least squares discriminant analysis
Figure 2 resumes the workflow to prepare the NMR output data matrix needed to PCA and OPLS-DA analysis: i) spectra processing (ppm calibrations, base-line corrections and frequency alignments) and ii) reduction of data dimensionality by frequency binning. The NMR bucketing strategy in the present work comprises an intelligent binning algorithm [24] for a frequency selection of only the 7.7–6.3 ppm region that concentrate (poly)-phenolics 1H-aromatic resonances. Two type of data matrix are proposed for multivariate statistical analysis:
-
Full data matrix (2 pigmented strains × 3 temperatures × 7 spouting days, Fig. 3)
-
Temperature-independent reduced data matrix (2 pigmented strains × 7 sprouting days, Fig. 4).
OPLS-DA score plot depicting a discrimination model derived from scatter loadings related to sprouting time (Figure S.I.2. bottom), generated from the 1H NMR full data (2 × 3 × 7) matrix, constructed from signal bucketing of solely aromatic proton resonances (7.7 – 6.3 ppm, bottom). Within the OPLS-DA plot, each outlier comprising the temperature and strain dependency are highlighted along the data delimited by T2 Hotelling’s ellipses as “10C” (scores from data at T = 10 °C), “25C” (scores from data at T = 25 °C) and “30C” (scores from data at T = 30 °C), whereas the strain dependency (blue and red Zea Mays L.) is indicated with the corresponding blue or red color. The color code for spouting times’ T2 Hotelling’s ellipses is indicated at the top right of the figure
OPLS-DA multivariate statistical analysis score plots generated from three temperature-independent (2 × 7) reduced NMR data matrices, constructed from signal bucketing of solely aromatic proton resonances (7.7 – 6.3 ppm, bottom). Color code used for both T2 Hotelling’s ellipses and scores plots are in agreement with NMR non-targeted metabolomics observables from blue and red Zea Mays L. strains
Unsupervised principal component analysis is generally used for organizing NMR data matrix and for determining correlations between selected sampling (maize pigmentation and sprouting’s time / temperature dependency) and outliers (discriminant resonances of (poly)-phenolic NMR observables). Complementarily, in order to maximize separations amongst samples, supervised orthogonal partial least squares discriminant analysis (OPLS-DA) was applied to reduced NMR data matrix. OPLS-DA permits to obtain optimal information from the dataset by identification of a more refined multivariate subspace for maximum group separations. PCA and OPLS-DA applied to the full data matrix, revealed several discriminant resonances of (poly)-phenolics observables during sprouting at the selected controlled conditions. Most relevant scatter loadings derived from non-targeted profiling comprise the following aromatic chemical shifts (Figure S.I.2):
-
a)
[PC1(-), PC2(-)]: 6.743, 6.73, 6.77, 7.111, 6.79, 6.713, 6.957, 6.869, 6.841, 7.039, 6.444 ppm
-
b)
[PC1(−), PC2( +)]: 7.3, 7.31, 7.17, 6.821, 7.063, 7.367, 7.324, 7.339, 7.165 ppm
-
c)
[PC1( +), PC2( +)]: 7.111, 6.929, 6.905, 6.841. 6.855, 6.743, 6.729, 6.781 ppm
-
d)
[PC1( +), PC2(−)]: 6.884, 6.722, 6.735 ppm
However, PCA models constructed from full data matrix (2 × 3 × 7), produce poor separations between groups (temperature, Figure S.I.3.A; sprouting time, Figure S.I.3.C) described by two-dimensional projections (PC1 = 19.55%, PC2 = 15.02%). For that, in order to significantly improve separations for providing pairwise comparisons between maize strains and their sprouting conditions, OPLS-DA modeling is applied for full data matrix. With a combination of one predictive and three orthogonal components in all cases, a pair of OPLS-DA models per screened factor (sprouting temperatures, Figure S.I.3.B; sprouting time, Fig. 3) were constructed by using the full data matrix. Discriminative analysis of NMR data matrix defined in terms of sprouting temperature factor (Figure S.I.3.B.) have improved separation between groups with respect its PCA counterpart (Figure S.I.3.A) but produce not satisfactory R2X, R2Y and Q2 statistical values (respectively 0.189, 0.247 and 0.014). In the other hand, OPLS-DA model describing sprouting time dependency (Fig. 3) present acceptable separations amongst groups (R2X = 0.854, R2Y = 0.947; Q2 = 0.646) with the limitation that said supervised multivariate statistical analysis model does not represent clear discriminations between maize varieties along the sprouting temperature dependency.
The best approach for describing the temperature-dependency factor of blue and red maize sprouting, series of reduced datasets were proposed for PCA/OPLS-DA multivariate analysis, by separating their temperature dependency in three independent (2 × 7) reduced matrices.
Figure 4 and Figure S.I.7 show respectively the OPLS-DA and PCA analysis, with the use of temperature-independent reduced data matrix (2 pigmented strains × 7 sprouting days), whereas variable separations that define blue and red maize strains’ subspace present the best agreements at T = 25 °C (PCA 1 = 95.55%, PCA 2 = 4.21%; R2X = 0.536, R2Y = 0.755; Q2 = 0.768), acceptable discriminations at T = 30 °C (PCA 1 = 91.74%, PCA2 = 6.24%; R2X = 0.39, R2Y = 0.758; Q2 = 0.661) and poor blue and red maize strains’ separations at T = 10 °C (PCA 1 = 67.75%, PCA 2 = 20.22%; R2X = 0.201, R2Y = 0.484; Q2 = 0.188), as expected.
4.3 Sprouts’ total phenolic contents and A/ΔA spans
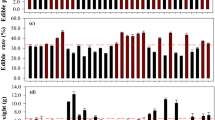
As can be observed in Fig. 5. and Table S.I.2., total phenolic contents obtained with standard UV–VIS method (Sect. 2.2.1.) of both blue and red maize sprouts as a function of time and temperature follow a similar trend as previously observed in buckwheat13, whereas increments of phenolic contents were directly correlated to an increase in spouting time (seven days), with the exception of sprouts at T = 10 °C. Deeper inspections of total phenolic content modulations for both strains as a function of time and temperature can be better conceived with a raw and first derivative (A/ΔA) absorbance span analysis (Sect. 2.2.2.). Figures S.I.5-S.I.6. respectively presents the raw and first derivative UV–VIS absorbance spectra of methanolic extracts of both blue (Top in both figures) and red (bottom in figures) sprouts, treated with Folin-Ciocalteu solution for phenolics determination. Relevant bands are highlighted at every condition: i) 275 nm: maximum of all Absorbance spectra (data not shown for visualization purposes); ii) inflexion point within Absorbance spans localized at 426 nm; iii) local maximum in absorbance spectra at 756 nm and iv) two zero crossing points within the ΔA spectra located at 426 and 756 nm. Interestingly, slopes from these zero crossing points increase with respect germination time for sprouts obtained from D + 1 to D + 7 at 25 and 30 °C. A/ΔA data from 10 °C sprouts reveal deficient phenolics production at such thermodynamic conditions, regardless the selected long periods for germination (up to 49 days). A/ΔA span analysis also reveal that 756 nm wavelength is the precise value to proceed with absorbances’ readings for obtaining phenolic contents.
UV–VIS quantifications of phenolic content of blue and red maize strains in methanolic extract expressed in mg gallic acid equivalents (GAE) / 100 g of dry flour weight (DW) as a function of sprouting time (from D + 1 to D + 7 for strains sprouted between 25 and 30 °C, from D + 7 to D + 49 for strains sprouted at 10 °C). Color code agrees with each pigmented strain. For visualization purposes, continuous lines connect data obtained from sprouts at T = 25 °C, dashed lines connect data obtained from sprouts at T = 30 °C and pointed lines connect those obtained at T = 10 °C. Error bars depict the standard deviation as well reported in Table S.I.2
In resume, UV–VIS absorbance quantifications as well as A/ΔA spans analysis, scheme some trends of time-dependent phenolic increments of both pigmented sprouts herein analyzed. Clear distinctions between phenolic contents obtained during 49 days at T = 10 °C, with respect the rest of dataset are revealed. However, distinctions between “close data” (i.e. time-dependent phenolic content produced between 25 and 30 °C, see also Figure S.I.5.) cannot be disentangled with absorbance data. In clear contrast, PCA and mostly OPLS-DA analysis of reduced NMR data (2 × 7) matrix (Fig. 5) allow a net discrimination between aromatic proton resonances even between T = 25 °C and T = 30 °C.
5 Conclusions
A set of novel spectroscopic approaches are proposed for analyzing relevant phytochemical production during sprouting of blue and red Mexican maize strains. Samples were subjected to a specific germination scheme, included temperature variations and once inactive, solutions for spectroscopic analysis were prepared with a simple methanolic aqueous mixture. Three spectroscopic observables were used for analyzing relevant variables involved in temperature- and time-controlled sprouting: UV–Vis A/ΔA spans, 1H-NMR water LWHHs and combined PCA/OPLS-DA multivariate statistical analysis applied to aromatic proton resonances’ data matrices. Phenolic contents obtained with 756 nm absorbance readings and A/ΔA spans provide the expected phenolics modulation as a function of sprouting temperature and time. However, subtle differences between T = 25 °C and 30 °C and maize strains are not distinguishable with the spectrophotometric technique. 4.86 ppm NMR linewidths at half heights of sprouts’ methanolic extracts is proposed as a novel observable for providing specific temperature- and time-dependent line shapes and are proposed as an additional sprouting fingerprint, but is not conclusive if said parameter could be used for disentangling maize strains’ differences. Two types of NMR data matrix modelling are proposed for multivariate statistical analysis: temperature- and time-dependent (2 × 3 × 7) and temperature-independent (2 × 7) arrangements, both constructed with a reduction of data dimensionality by frequency binning from uniquely resonances coming from aromatic NMR frequencies (7.7–6.3 ppm). Series of thirty discriminative resonances define the NMR data matrix, The OPLS-DA analysis from the former dataset accurately describes a subspace for sprouting time variable separations. Both PCA and OPLS-DA applied to the reduced temperature-independent NMR data matrix accurately describes a subspace for disentangling phenolics’ modulations from each maize strain, with the best discriminations at T = 25 °C. The NMR / OPLS-DA non-targeted analysis seems an attractive and fast method to be used for optimizing pro-health germination schemes. Targeted 1H NMR / OPLS-DA polyphenolic metabolomics would be the natural extension of the present work in order to identify and quantify the nature of each discriminant chemical marker herein reported, on behalf of obtaining keen and high-selective sprouting conditions, that can favor the biosynthesis of specific and desired hydoxyphenyl phytochemicals.
Abbreviations
- TPC:
-
Total Phenolic Content
- NMR:
-
Nuclear Magnetic Resonance
- MSA:
-
Multivariate Statistical Analysis
- PCA:
-
Principal Component
- OPLS-DA:
-
Orthogonal Projections to Latent Structures Discriminant Analysis
- LWHH:
-
Water Line Widths at Half Height
- UV–Vis A/ΔA:
-
Ultra-Violet Visible raw and first derivative Absorbance
- GAE:
-
Gallic Acid Equivalents
- DW:
-
Dry (flour) Weight
References
Liu RH (2004) Potential synergy of phytochemicals in cancer prevention: Mechanism of action. J Nutr 134:3979S-S3485. https://doi.org/10.1093/jn/134.12.3479S
Pientaweeratch S, Panapisal V, Tansirikongkol A (2016) Antioxidant, anti-collagenase and anti-elastase activities of Phyllanthus emblica, Manilkara zapota and silymarin: an in vitro comparative study for anti-aging applications. Pharma Biol 54:1865–1872. https://doi.org/10.3109/13880209.2015.1133658
Xiang XN, Guo X, Liu F, Li Q, Hu J, Brennan S (2017) Effect of light- and dark-germination on the phenolic biosynthesis, phytochemical profiles and antioxidant activities in sweet corn (Zea Mays L) sprouts. Int J Mol Sci 18:1246. https://doi.org/10.3390/ijms18061246
Guo X, Li T, Tang K, Liu RH (2012) Effect of germination on phytochemical profiles and antioxidant activity of mung bean sprouts (Vigna radiata). J Agric Food Chem 60:11050–11055. https://doi.org/10.1021/jf304443u
Christen S, Woodall AA, Shingenaga MK, Southwell-Keely P, Duncan MW, Ames BN (1997) γ-Tocopherol traps mutagenic electrophiles such as NOx and complements α-tocopherol: physiological implications. Proc Natl Acad Sci USA 94:3217–3222. https://doi.org/10.1073/pnas.94.7.3217
Fresco P, Borges F, Diniz C, Marques MPM (2006) New insights on the anticancer properties of dietary polyphenols. Med Res Rev 26:747–766. https://doi.org/10.1002/med.20060
Kanski J, Aksenova M, Stoyanova A, Butterfield DA (2002) Ferulic acid antioxidant protection against hydroxyl and peroxyl radical oxidation in synaptosomal and neuronal cell culture systems in vitro: structure-activity studies. J Nutr Biochem 13:273–281. https://doi.org/10.1016/S0955-2863(01)00215-7
Hagiwara A, Miyashita K, Nakanishi K, Sano M, Tamano S, Kadota T (2001) Pronounced inhibition by a natural anthocyanin, purple corn color of 2-amino-1-methyl-6-phenylimidazol [4,5b]-pyridine (PhIP)-associated colorectal carcinogenesis in male F344 rats pretreated with 1,2-dimethylhydrazine. Cancer Lett 171:17–25. https://doi.org/10.1016/S0304-3835(01)00510-9
Del Pozo-Insfran D, Brenes CH, Serna S, Talcott ST (2007) Polyphenolics and antioxidant capacity of white and blue corns processed into tortillas and chips. Cereal Chem 84:162–168. https://doi.org/10.1094/CCHEM-84-2-0162
Del Pozo-Insfran D, Brenes CH, Serna S, Talcott ST (2006) Polyphenolic and antioxidant content of white and blue corn (Zea Mays L.) products. Food Res Int 39:696–703. https://doi.org/10.1016/j.foodres.2006.01.014
López-Martínez LX, Oliart-Ros RM, Valerio-Alfaro G, Lee CH, Parkin KL, García HS (2009) Antioxidant activity, phenolic compounds and anthocyanins content of eighteen strains of Mexican maize. LWT-Food Sci Technol 42:1187–1192. https://doi.org/10.1016/j.lwt.2008.10.010
Adom KK, Liu RH (2002) Antioxidant activity of grains. J Agric Food Chem 50:6182–6187. https://doi.org/10.1021/jf0205099
Ren SC, Sun JT (2014) Changes in phenolic content, phenylalanine ammonia-lyase (PAL) activity and antioxidant capacity of two buckwheat sprouts in relation to germination. J Funct Foods 7:298–304. https://doi.org/10.1016/j.jff.2014.01.031
Dueñas M, Martínez-Villaluenga C, Limón RI, Penas E, Frías J (2015) Effect of germination and elicitation on phenolic composition and bioactivity of kidney beans. Food Res Int 70:55–63. https://doi.org/10.1016/j.foodres.2015.01.018
Swieca M, Dziki D (2015) Improvement in sprouted wheat flour functionally: Effect of time, temperature and elicitation. Int J Food Sci Technol 50:2135–2142. https://doi.org/10.1111/ijfs.12881
Bakan BA, Bily AC, Melcion D, Cahagnier B, Regnault-Roger C, Philogene BJR, Richard-Molard D (2003) Possible role of plant phenolics in the production of trichothecenes by Fusarium graminearum strains on different fractions of maize kernels. J Agric Food Chem 51:2826–2831. https://doi.org/10.1021/jf020957g
Yang L, Zhang H, Cheng L, Gu Z, Hua D, Qi X, Qian H, Wang L (2014) Effect of extrusion on the hydrophilic antioxidant capacity of four whole grains. J Food Nutr Res 2:80–87. https://doi.org/10.12691/jfnr-2-2-4
Vázquez- Leyva S, Vallejo-Castillo L, López-Morales CA, Herbert-Pucheta JE, Zepeda-Vallejo LG, Velasco- Velázquez M, Pavón L, Pérez-Tapia SM, Medina-Rivero E (2019) Identity profiling of complex mixtures of peptide products by structural and mass mobility orthogonal analysis. Anal Chem 91:14392–14400. https://doi.org/10.1021/acs.analchem.9b02873
López-Morales CA, Vázquez- Leyva S, Vallejo-Castillo L, Carballo-Uicab G, Munoz-Garcia L, Herbert-Pucheta JE, Zepeda-Vallejo LG, Velasco- Velázquez M, Pavón L, Pérez-Tapia SM, Medina-Rivero E (2019) Determination of peptide profile consistency and safety of collagen hydrolysates as quality attributes. J Food Sci 84:430–439. https://doi.org/10.1111/1750-3841.14466
Herbert-Pucheta JE, Padilla-Maya G, Milmo-Brittingham D, Lojero D, Gilmore AM, Raventos-Llopart L, Hernández-Pulido K, Zepeda-Vallejo LG (2019) Multivariate spectroscopy for targeting phenolic choreography in wine with A-TEEMTM and NMR crosscheck non-targeted metabolomics. Bio web Conf 15:02006. https://doi.org/10.1051/bioconf/20191502006
Jacob D, Deborde C, Lefevbre M, Maucourt M, Moing A (2017) NMRProcFlow: a graphical and interactive tool dedicated to 1D spectra processing for NMR-based metabolomics. Metabolomics 13:36. https://doi.org/10.1007/s11306-017-1178-y
Hotelling H (1931) The generalization of student’s ratio. Ann Math Statist 2:360–378. https://doi.org/10.1214/aoms/1177732979
Holmes E, Loo RL, Stalmer J, Bictash M, Yap IKS, Chan Q, Ebbels T, De Iorio M, Brown IJ, Veselkov KA, Daviglus ML, Kesteloot H, Ueshima H, Zhao L, Nicholson JK, Elliot P (2008) Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 47:396–400. https://doi.org/10.1038/nature06882
De Meyer T, Sinnaeve D, van Gasse B, Tsiporkova E, Rietzschel E, de Buyzere M, Gillebert T, Bekaert S, Martins JC, van Criekinge W (2008) NMR-based characterization of metabolic alterations in hypertension using an adaptive, intelligent binning algorithm. Anal Chem 80:3783–3790. https://doi.org/10.1021/ac7025964
Acknowledgements
Authors thank the Mexican Ministry of Science and Technology (CONACyT) for funding: J. E. H.-P. and E.V.-S. thanks CONACyT program No. 682 “Catedras CONACyT”. All authors acknowledge CONACyT program No. LN295321 “Laboratorios Nacionales” and CONACyT-INFRA 269012 (NMR instrumentation) and Instituto Politecnico Nacional FIDEICOMISO stipend for the acquisition of the 600 MHz NMR instrument.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Carreño-Carrillo, C.V., Sánchez, E.V., Verduzco, C.V. et al. Polyphenol-based nuclear magnetic resonance non-targeted metabolomics of temperature- and time-controlled blue and red maize sprouting. SN Appl. Sci. 3, 300 (2021). https://doi.org/10.1007/s42452-021-04171-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-021-04171-w