Abstract
The partitivirids feature an icosahedral protein coating accommodating both their dsRNA genome and RNA-dependent RNA polymerase (RdRP). This signifies that transcription and replication activities of the viral polymerase occur within the capsid, emphasizing that the viral cycle relies on polymerase incorporation. Particles lacking RdRP are defective and hence non-infectious. Encapsidation and replication are intricately linked for dsRNA viruses, to the extent that, for many of these, such as the cystovirids, the RdRP serves a dual role as a transcriptase/replicase and a pro-assembly factor, ensuring structural stability and overall capsid integrity. This work investigates if RdRP has a similar role within the capsid of Cannabis cryptic virus (CanCV), a betapartitivirus affecting Cannabis sativa. Utilizing reverse genetics in Nicotiana benthamiana, we conclusively established that RdRP expression is indispensable for CanCV’s virus-like particle formation. This study enhances our understanding of CanCV encapsidation, with RdRP serving a pivotal role as a pro-assembly factor. These preliminary findings contribute to the knowledge of viral assembly within the Partitiviridae family.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The term ‘cryptic’ is commonly used to describe numerous viruses within the Partitiviridae family (Boccardo et al. 1987). These multipartite cytoplasmatic viruses can infect their hosts persistently without inducing apparent symptoms, in a relationship that can be defined as symbiotic (Roossinck 2010). This viral family includes five genera that differ in host specificity: members of the genera Alphapartitivirus and Betapartitivirus infect both plants and fungi, members of the genus Gammapartitivirus exclusively infects fungi, those belonging to the genus Deltapartitivirus are found only in plants, and members of the genus Cryspovirus infect protozoa (Vainio et al. 2018). A recently discovered potential species of Partitiviridae, the galbut virus, has been surprisingly found to be common in wild populations of Drosophila melanogaster and Aedes aegypti, suggesting its prevalence in these insect populations (Cross et al. 2020, 2023). In plants, these viruses maintain a low viral titer and cannot be transmitted through mechanical means, grafting, or by vectors. However, they exhibit efficient transmission through seeds. When both parents are carriers, all offspring seedlings inherit the virus (Boccardo et al. 1987). In fungi, transmission depends on hyphal anastomosis and sporogenesis (Vainio et al. 2018). These transmission characteristics result from the absence of a viral protein responsible for movement (Vainio et al. 2018). Consequently, transmission can only take place during the division of host cells (Vainio et al. 2018).
The viral genome comprises two monocistronic double strand (ds)RNA fragments encoding the RdRP and the coat protein (CP). For some partitivirids, satellite dsRNA segments are sometimes found, although no replicative or structural function has been assigned to them (Compel et al. 1999; Kim et al. 2005). With a few exceptions (Shimura et al. 2022), these viruses do not encode silencing suppressors. Therefore, they are believed to rely on alternative strategies, such as coupling the replication process with encapsidation, to counteract the host’s gene silencing defense mechanism. Viral RdRP replicates and transcribes the dsRNA genome within the virion, ensuring that the dsRNA is never exposed to the host cytoplasmic DICER proteins. Semiconservative transcription results in the retention of the newly synthesized plus strand RNA (ssRNA(+)) inside the virion, while the template ssRNA(+) exits in the cytoplasm (Buck 1978). It either serves as a template for translation or becomes incorporated into new assembling viral particles, where RdRP utilizes it to reconstruct the dsRNA genome. Purified particles demonstrate in vitro replication activity (Pan et al. 2009), indicating the presence of RdRP molecules within the virion, presumed to interact with CPs through non-covalent bonds and to be anchored in proximity to one of the pores on the viral particle’s surface (Nibert et al. 2014). Viral particles are non-enveloped with T = 1 icosahedral symmetry, they range in size from 25 to 43 nm in diameter (Vainio et al. 2018). The capsid is formed by 120 CP units, arranged as 60 dimers, and dimeric surface protrusions are frequently observed. Diamond shaped CP tetramers (dimers of dimers) occur as intermediate of encapsidation (Vainio et al. 2018). Such structural features are also found for Picobirnavirus core capsids (Duquerroy et al. 2009) suggesting a shared evolutionary lineage, distinct from other dsRNA viruses (Tang et al. 2010). Within the Partitiviridae family, capsids exhibit significant variations in size and, to some extent, in shape. In fact, some gammapartitiviruses and betapartitiviruses share similar yet distinguishable structures. For instance, excluding surface protrusions, the primary capsid shells of gammapartitiviruses have smaller outer diameters, thinner average profiles, and more angular shapes, along with smaller interior-cavity volumes (Ochoa et al. 2008; Tang et al. 2010). Furthermore, the protruding domains of their coat protein dimers form two-fold-symmetrical arches, contrasting with the two fold-symmetrical “butte-like” structures formed by betapartitivirus CP dimers (Ochoa et al. 2008; Tang et al. 2010). Recent research has demonstrated that transient expression in Nicotiana benthamiana can serve as an efficient tool for the production and study of partitiviridae Virus-Like Particles. By transiently expressing the CP of the deltapartitivirus pepper cryptic virus 1 (PCV1), it was possible to characterize a highly disordered and hypervariable region within the capsid protrusions of the virus that is not required for VLP assembly (Byrne et al. 2021). This finding leads to the hypothesis that this hyper-variable region may have a role in the symbiotic relationship between deltapartitiviruses and various dicotyledonous hosts (Byrne et al. 2021). Another inherent deduction from the results is that the virus’s RdRP does not have structural roles, as the transient expression of CP alone is sufficient for VLP formation. This suggests, for deltapartitiviruses, at least for PCV1, RdRP inclusion within the capsid must occur through a mechanism separate from capsid formation.
With this work, our aim is to unravel the formation of particles of the betapartivirus Cannabis cryptic virus (CanCV), which is phylogenetically distant from PCV1. First identified in European Cannabis sativa plants a few years ago (Ziegler et al. 2012), it has since been detected in nearly every European variety tested without inducing any evident symptoms (Miotti et al. 2023; Righetti et al. 2018). Through the transient expression of its coding RNAs in N. benthamiana, we intend to examine the role of RdRP in the formation of CanCV VLPs, comparing this process with well-known models of other dsRNA viruses.
Materials and methods
VLPs purification and TEM observation and SDS page
Viral particles of CanCV were purified from 40 g of hemp infected leaves or patch area of agroinfiltrated N. benthamiana according to the protocol used by Turina et al. (2007). Visible fractions were isolated from a linear 7–50% (w/v) sucrose gradient and subjected to new pelletting after ultracentrifugation at 235’000 x g for 2 h. The resulting pellets were ultimately resuspended in 30 µl of RNAse-free water.
Two µl of the purification were used to visualize the presence of any VLPs with a TEM (Philips CM 10) after uranyl acetate staining (0.5-2%). Grids were prepared with the “drop” method (Bozzola and Russell 1999). Additionally, 10 µl of the purified VLPs were subjected to SDS-PAGE on a 12% polyacrylamide gel (Acrylamide/Bis-acrylamide 37.5/1) followed by Coomassie blue staining to analyze the detectable protein components.
RNA extraction
RNA was extracted from CanCV particles purified from leaves of infected C. sativa and N. benthamiana patch area using phenol:chloroform:isoamyl alcohol (25:24:1) phase extraction metod. Extracted RNA was then precipitated by adding two volumes of absolute ethanol with sodium acetate at a final concentration of 0.3 M. Subsequently, the resulting RNA pellets were resuspended in 50 µl of RNAse-free water.
Cloning of CanCV ssRNA1(+) and ssRNA2(+) into pJL89 binary vector
Full-length cDNA of both CanCV RNAs was synthesized using the RT Superscript™ IV kit from Thermo Fisher Scientific, following the manufacturer’s protocol with a modification involving the denaturation of the dsRNAs-primer mix at 99°C for 5 minutes. Sequence-specific reverse primers (CanCV_3’UTR_RdRP_R and CanCV_3’UTR_CP_R) were employed (Table 1), and the resulting single-stranded cDNAs were then amplified using the LongRange PCR kit from Promega, following the manufacturer’s instructions. We utilized primers designed to extend 5’ and 3’ of CanCV cDNAs with SmaI restriction site sequences (CanCV_RdRP_SmaI F/R and CanCV_CP_SmaI F/R). PCR products meeting the expected size of 2397 bp for RNA1 coding the RdRP and of 2266 for RNA2 coding the CP were isolated and purified using the Wizard SV gel and PCR clean-up system kit (Promega) and then digested with SmaI restriction enzyme (FastDigest from Thermo Fisher Scientific). After DNA purification from the agarose gel, the CanCV double-stranded cDNA fragments were ligated to the pJL89 binary vector (Delbianco et al. 2013) with T4 DNA ligase (Thermo Fisher Scientific). Blunt-end vector was generated through PCR amplification using the Phusion High-Fidelity PCR Kit (Thermo Fisher Scientific) with pJL89_Full_R and pJL89_Full_F primer using 10 ng of circular Dam-methylated plasmid as a template. Prior to gel purification of the amplicon with the Wizard SV gel and PCR clean-up system kit from Promega, the pJL89 circular plasmid was digested by DpnI (FastDigest from Thermo Fisher Scientific). After electroporation of Escherichia coli MC1022 cells, PCR screening of the recombinant colonies was performed to ensure the correct orientation of the plasmid fragments in order for the pJL89 plasmids to promote the expression of CanCV ssRNA(+) strands, referred to as pJL89_RdRP and pJL89_CP, respectively. This was accomplished using forward primers CanCV_RdRP_F or CanCV_CP_F (Table 1) in combination with the reverse primer pJL89_R (Table 1). Colony PCRs were carried out using the GoTaq® G2 Flexi DNA Polymerase kit from Promega. Plasmids were purified with PureYield™ Plasmid Miniprep System (Promega) and then Sanger sequenced. Subsequently, pJL89_RdRP and pJL89_CP were electroporated into Agrobacterium tumefaciens strain GV3101 (Holsters et al., 1980).
N. benthamiana agro-infiltrations
The desired agro-clones were cultivated in Luria-Bertani (LB) medium with appropriate antibiotics (Rifampicin 50 µM and Kanamycin 100 µM) for an overnight incubation at 28 °C. Saturated liquid cultures were then pelleted and resuspended in MES buffer containing 10mM MgCl2, 10mM 2-[N-morpholino] ethane sulfonic acid, and 200µM acetosyringone until a 600 nm optical density of 0.6 was attained. Different Agro-mixes were prepared with a 1:1:1 or 1:1 volume ratio, as detailed in Table 2. Fourteen-day-old N. benthamiana plants were agro-infiltrated with a needleless syringe and then grown with constant temperature and illumination (photoperiod 16/8 H day/night; temperature 22/18°C day/night; hygrometry 70%) for 7 days post infiltration (dpi), before patched area samplings.
Run-off in vitro transcription with CanCV PCR-amplified DNAs as a templates
Templates for the in vitro transcription of both negative and positive CanCV ssRNAs were generated via PCR using 10 ng of purified pJL89_RdRP and pJL89_CP as templates. The following pairs of primers were used: CanCV_RdRP_T7_F with CanCV_3’UTR_RdRP_R for ssRNA1(+); CanCV_RdRP_T7_R with CanCV_5’UTR_RdRP_F for ssRNA1(-); CanCV_CP_T7_F with CanCV_3’UTR_CP_R for ssRNA2(+) and CanCV_CP_T7_R with CanCV_5’UTR_CP_F for ssRNA2(-). Primer details and PCR annealing conditions are listed in Table 1. PCR products were purified using the PCR clean-up system kit from Promega and eluted in a volume of 25 µL of nuclease-free water. After quantification using a nanodrop (Thermo Fisher), 500 ng of each template was used for in vitro transcription with the RiboMAX™ Large Scale RNA Production System (Promega), in a volume of 20 µL, following the manufacturer’s instructions.
Strand specific RT-PCR with tagged primers
Complementary DNAs (cDNAs) of the CanCV genomic ssRNAs(-) were synthesized using tagged primers (CanCV_tag_RdRP/CP_F), with a 5’ end 20-nucleotide tag, unrelated to the virus (TGGACCCGGGATAAATTTGA). The reaction was carried out with 1 µL of particle purified RNAs using ImProm-II™ Reverse Transcriptase (Promega) following the manufacturer’s instructions with modifications involving the denaturation of the dsRNAs-primer mix at 99 °C for 5 min, and the cDNA synthesis temperature that was set at 42 °C for 1 h. To remove the tagged primers from the reaction, the reverse transcription product was purified using the PCR clean-up system kit from Promega and eluted in a volume of 50 µL of nuclease-free water. Following purification, 2 µL of the purified product was used as a template for PCR reactions with GoTaq Flexi G2 (Promega). These PCR reactions utilized the TAG primer in combination with either the CanCV_RdRP_R or CanCV_CP_R primer to detect ssRNA1(-) and ssRNA2(-), respectively. The reactions were conducted according to the manufacturer’s instructions. The primers used and RT and PCR annealing temperatures are listed in Table 1. To confirm the absence of any residues of tagged primers in the purified reverse RT product, PCRs using only the reverse primers were conducted. Additionally, to assess the specificity of the assay, it was also performed on the in vitro transcripts of positive and negative ssRNAs of CanCV. For the latter, 0.5 µL of the transcription product was used without the removal of the template DNA and performing RT step following manufacturer’s instructions, including the initial nucleic acid denaturation at 70 °C for 5 min (supplementary Fig. 1). Avoiding DNA denaturation, M-MLV reverse transcriptase is prevented from utilizing ssDNAs as templates (Gerard and D’Alessio 1993).
Results
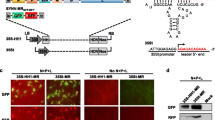
The potential role of CanCV RdRP in capsid assembly was investigated using a reverse genetics approach. By employing Agro-clones expressing the two viral ssRNA(+), transient and separate expression of these components allowed to assess whether VLP formation was dependent on functional complementation between RdRP and the structural protein CP. For this purpose, viral particles of CanCV were isolated from 40 g of infected C. sativa leaves. The genomic RNA extracted from these particles served as a template for the construction of the recombinant plasmids pJL89_RdRP and pJL89_CP. In these plasmids, the complete sequence of the two viral genomic segments was inserted between the double 35S expression promoter and the hepatitis delta virus ribozyme sequence to eliminate the polyadenylate tail from the transgenic RNAs expressed in the nucleus. This approach was taken based on the findings of Rigetti et al. (2018), which confirmed the absence of polyadenylation at the 3’ ends of the CanCV wild-type genomic segments. The simultaneous transient co-expression of ssRNA1(+) and ssRNA2(+) in N. benthamiana leaf agro-patches (Agromix 1, Table 2) did not result in the production of purified and observable VLPs under electron microscopy. This outcome was attributed to the sense PTGS induced by the transgenes (Parent et al. 2015) indicating that neither CanCV CP nor RdRP exhibits PTGS suppression activity in N. benthamiana. To address this limitation, the agroclone pBIN_p19_TBSV, expressing the silencing suppressor p19 from tomato bushy stunt virus (TBSV) (Peltier et al. 2012), was added to the infiltrating Agromix (Agromix 2, Table 2). After 7 dpi, in the agro-infiltrated areas, VLPs with sizes and morphological characteristics similar to those extracted from naturally infected hemp plants were successfully isolated (Fig. 1A1 and A2). SDS-PAGE analysis confirmed the presence of a protein with dimensions congruent to CanCV CP (74.88 kDa) (Fig. 1C). The slight visible band shift from the expected size could be attributed to post-translational modifications or altered detergent binding that modify CP electrophoretic properties (Rath et al. 2009; Shi et al. 2012). RdRP was not detectable, as observed in other studies on natural partitiviridae particles, where the presence of viral RdRP is presumed to be in quantities of a few molecules per particle and has been indirectly detected through in vitro enzymatic activity assays (Pan et al. 2009). A qualitative analysis of the genomic content within these particles, conducted using negative-strand-specific RT-PCR targeting CanCV RNA1 and RNA2, revealed the presence of these ssRNAs within the VLPs (Fig. 1B). Negative PCR amplification conducted with the primer pairs CanCV_CP_F / CanCV_CP_R and CanCV_RdRP_F / CanCV_RdRP_R confirmed the lack of Agroclone-derived T-DNAs in the purified particles. These results indicate that the RdRP likely converted the ssRNA(+) strands into dsRNA within CanCV VLPs. However, it’s important to specify that this analysis does not provide a quantitative assessment of ssRNA(-) relative to the purified VLPs, and the precise proportion of VLPs that encapsulate fragments of the bipartite virus genome remains to be determined. However, the expression of ssRNA2(+) alone, decoupled from the expression of RdRP, did not result in the purification of TEM-observable VLPs, indicating the necessity of the viral polymerase for their formation.
Observation of CanCV particles and analysis of protein and genomic content. TEM observation of purified particles from naturally infected C. sativa plants (A1) or N. benthamiana leaf patches infiltrated with Agromix 3 (A2). Electrophoretic analysis of the RT-PCR strand specific for ssRNA1(-) and ssRNA2(-) (B). The reactions were performed on RNA extracts from purified particles obtained from naturally infected C. sativa or N. benthamiana leaf patches infiltrated with Agromix 3. Additionally, reactions were conducted on total RNA extracts from Agromix 3 N. benthamiana leaf patches.” SDS page migration of Agromix 1VLPs protein content (C)
Discussion
In the context of reverse genetic studies, it is common practice to employ in vitro system (Le and Müller 2021; Asensio-Cob et al. 2023) or non-natural host organisms as bioreactors for viral particles (Bragard et al. 2000; Abrahamian et al. 2020; Yang et al. 2021), rather than the natural hosts of the investigated virus. Notably, N. benthamiana has been extensively employed in numerous studies to produce Virus-Like Particles (VLPs) from various viruses, including those of animal origin, for vaccines or nanocarrier development. (D’Aoust et al. 2008; Smith et al. 2023). A result, N. benthamiana can be used for the expression of both wild-type and mutated structural viral components, facilitating the characterization of encapsidation mechanisms in a diverse spectrum of viruses, including the partitiviridaes. In our investigation of complete virion formation, we have delved into the mechanisms characterizing this process, with a specific focus on the retention of RdRP molecules in viral particles of dsRNA viruses. The literature provides insights into various mechanisms for incorporating RdRPs, offering a clear perspective on the intricate dynamics of virion assembly. For instance, some viruses, such as yeast L-A virus, express their polymerase in the form of a fusion protein with the capsid protein (gag-pol), which is subsequently incorporated into viral particles as a minor structural component (Icho and Wickner 1989). However, in most cases, dsRNA viruses establish direct CP-RdRP interactions to anchor the replicase to the inner surface of the capsid. This interaction is essential for the activation of RdRP, which ensures the replication/transcription of the viral genome only once it is packaged inside the capsid (Patton et al. 2006).
Recently, exceptions to this previously assumed general model have been observed, particularly with the human picobirnavirus (hPBV). In hPBV, the RdRP appears to be enzymatically active without the presence of CP as a cofactor, and its packaging inside the virion does not seem to depend on a direct interaction with CP but rather on an interaction with the viral genome. The mechanism ensuring the structural integrity of the capsid of dsRNA viruses is also controversial, as experiments have shown that hPBV VLPs can form in the absence of RdRP expression (Collier et al. 2016). However, for the cystoviridae bacteriophage Φ6, it has been demonstrated that the polymerase has a role in procapsid assembly, accelerating the self-assembly process and stabilizing the compact conformation of the CP shell (Sun et al. 2018). Regarding partitiviridae viruses, the mechanism is still far from being fully understood. Experiments conducted in N. benthamiana have shown that the deltapartitivirus PCV-1 can assemble VLPs solely through the expression of its CP without RdRP. Nonetheless, the potential role of the ssRNA(+) CP genomic molecule cannot be ruled out, as it is expressed as a messenger RNA by the agro-clone 35S cassette, even if it is depleted of the UTRs at the 5’ and 3’ ends. In our preliminary study, we delved into the capsid assembly mechanisms of another partitivirus, specifically one belonging to the genus Betapartitivirus. Our findings suggest that the formation of the capsid in CanCV is contingent upon the expression of its RdRP. It is important to note that the potential formation of VLPs consisting solely of capsid proteins cannot be definitively ruled out, as their quantity may fall below the limit of detection of our particle purification and visualization methodology. Nevertheless, our results undeniably demonstrate that the coordinated co-expression of CP and RdRP in N. benthamiana is associated with a remarkably more efficient production of CanCV like particles. Based on our findings, we cannot definitively confirm whether the presence of genomic RNA (+) is essential for the assembly of VLPs. However, it is apparent that the sole presence of genomic RNA (+) is not sufficient to effectively guide the translated CPs into the cytoplasm to complete nucleation and the formation of the capsid envelope. This is highlighted by the absence of VLP formation in the presence of CanCV’s CP alone, despite its expression through transient agro-mediated transformation and the production of genomic RNA (+) as messenger RNA by the cellular RNA polymerase II. Although the strain-specific RT-PCR used to detect the negative strands of genomic dsRNAs within the purified particles cannot give us an indication of the amount of VLPs that retain the viral genome, it has determined that at least some of them possess negative-strand RNA, presumably part of the dsRNAs synthesized by the RdRP within the capsid. In vitro encapsidation studies of the segmented dsRNA blue tongue virus, a member of the Reoviridae family, have revealed that the formation of VLPs relies on the simultaneous presence of all ten genomic ssRNA(+) strands, along with the CP, and the non-structural proteins RdRP and RNA capping enzyme (CAP) (Lourenco and Roy 2011). It is conceivable that a fine mechanism that orchestrates the nucleation and assembly of the capsid, ensuring the preservation of genomic, structural, and functional integrity to minimize the formation of defective and non-infectious particles. Indeed, other members of the Reoviridae family, such as rotaviruses, are believed to assemble the CP shell around RdRP/CAP/ssRNA(+) complexes. This process is initiated by the non-covalent interaction between RdRP and the disordered N-terminal extensions protruding internally from the CP capsid. It is speculated that subsequent CP-CP homotypic contacts are then enhanced and stabilized by further ssRNA(+) interactions, ultimately leading to the assembly of the capsid inner core. Similar mechanisms may be suggested for CanCV, even if our data lack the capability to quantify the proportion of purified particles possessing genomic dsRNA. Indeed, transient expression of ssRNA1(+) and ssRNA2(+), stabilized by the expression of TBSV p19, ensures an adequate cytoplasmic presence of genomic ssRNAs (+). These, co-assisted by RdRP, may initiate the capsid nucleation process. Such speculations will remain unverified unless the presence of ssRNA(+) and RdRP in each purified particle from N. benthamiana is demonstrated. Future comparative experiments between VLPs purified from transiently transformed N. benthamiana and naturally infected C. sativa should employ sensitive protein identification and quantification techniques, such as liquid chromatography with tandem mass spectrometry. This approach will indicate the presence of RdRP in purified particles and provide the stoichiometric ratio between CP and RdRP, and therefore between VLPs and RdRP. The ability to finely quantify viral CPs will indeed help define the absolute quantity of purified particles in a solution and associate it with the quantification of genomic ssRNA(+).
References
Abrahamian P, Hammond RW, Hammond J (2020) Plant virus–derived vectors: applications in Agricultural and Medical Biotechnology. Annual Rev Virol 7(1):513–535. https://doi.org/10.1146/annurev-virology-010720-054958
Asensio-Cob D, Rodríguez JM, Luque D (2023) Rotavirus particle Disassembly and Assembly. Vivo Vitro Viruses 15(8):1750. https://doi.org/10.3390/v15081750
Boccardo G, Lisa V, Luisoni E, Milne RG (1987) Cryptic Plant Viruses. In Advances in Virus Research (Vol. 32, pp. 171–214). Elsevier. https://doi.org/10.1016/S0065-3527(08)60477-7
Bozzola JJ, Russell LD (1999) Electron Microscopy: principles and techniques for biologists. Jones & Bartlett Learning
Bragard C, Duncan GH, Wesley SV, Naidu RA, Mayo MA (2000) Virus-like particles assemble in plants and bacteria expressing the coat protein gene of Indian peanut clump virus. J Gen Virol 81(1):267–272. https://doi.org/10.1099/0022-1317-81-1-267
Buck KW (1978) Semi-conservative replication of double-stranded RNA by a virion-associated RNA polymerase. Biochem Biophys Res Commun 84(3):639–645. https://doi.org/10.1016/0006-291x(78)90753-2
Byrne M, Kashyap A, Esquirol L, Ranson N, Sainsbury F (2021) The structure of a plant-specific partitivirus capsid reveals a unique coat protein domain architecture with an intrinsically disordered protrusion. Commun Biology 4(1):1–8. https://doi.org/10.1038/s42003-021-02687-w
Collier AM, Lyytinen OL, Guo YR, Toh Y, Poranen MM, Tao YJ (2016) Initiation of RNA polymerization and polymerase encapsidation by a small dsRNA virus. PLoS Pathog 12(4):e1005523. https://doi.org/10.1371/journal.ppat.1005523
Compel P, Papp I, Bibó M, Fekete C, Hornok L (1999) Genetic interrelationships and genome organization of double-stranded RNA elements of Fusarium poae. Virus Genes 18(1):49–56. https://doi.org/10.1023/a:1008069318838
Cross ST, Maertens BL, Dunham TJ, Rodgers CP, Brehm AL, Miller MR et al (2020) Partitiviruses infecting Drosophila melanogaster and Aedes aegypti exhibit efficient Biparental Vertical Transmission. J Virol 94(20). https://doi.org/10.1128/jvi.01070-20
Cross ST, Brehm AL, Dunham TJ, Rodgers CP, Keene AH, Borlee GI, Stenglein MD (2023) Galbut Virus infection minimally influences Drosophila melanogaster Fitness traits in a strain and sex-dependent manner. Viruses 15(2):539. https://doi.org/10.3390/v15020539
D’Aoust M-A, Lavoie P-O, Couture MM-J, Trépanier S, Guay J-M, Dargis M et al (2008) Influenza virus-like particles produced by transient expression in Nicotiana benthamiana induce a protective immune response against a lethal viral challenge in mice. Plant Biotechnol J 6(9):930–940. https://doi.org/10.1111/j.1467-7652.2008.00384.x
Duquerroy S, Da Costa B, Henry C, Vigouroux A, Libersou S, Lepault J et al (2009) The picobirnavirus crystal structure provides functional insights into virion assembly and cell entry. EMBO J 28(11):1655–1665. https://doi.org/10.1038/emboj.2009.109
Gerard GF, D’Alessio JM (1993) Reverse transcriptase (EC 2.7.7.49). In: Burrell MM (ed) Enzymes of Molecular Biology. Humana, Totowa, NJ, pp 73–93. https://doi.org/10.1385/0-89603-234-5:73
Icho T, Wickner RB (1989) The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two Open Reading frames. J Biol Chem 264(12):6716–6723. https://doi.org/10.1016/S0021-9258(18)83488-3
Kim JW, Choi EY, Lee JI (2005) Genome Organization and expression of the Penicillium Stoloniferum Virus F. Virus Genes 31(2):175–183. https://doi.org/10.1007/s11262-005-1793-y
Le DT, Müller KM (2021) In Vitro Assembly of Virus-Like Particles and their applications. Life 11(4):334. https://doi.org/10.3390/life11040334
Lourenco S, Roy P (2011) In vitro reconstitution of Bluetongue virus infectious cores. Proc Natl Acad Sci USA 108(33):13746–13751. https://doi.org/10.1073/pnas.1108667108
Miotti N, Passera A, Ratti C, Dall’Ara M, Casati P (2023) A guide to Cannabis Virology: from the Virome Investigation to the development of viral biotechnological tools. Viruses 15(7):1532. https://doi.org/10.3390/v15071532
Nibert ML, Ghabrial SA, Maiss E, Lesker T, Vainio EJ, Jiang D, Suzuki N (2014) Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res 188:128–141. https://doi.org/10.1016/j.virusres.2014.04.007
Ochoa WF, Havens WM, Sinkovits RS, Nibert ML, Ghabrial SA, Baker TS (2008) Partitivirus structure reveals a 120-Subunit, Helix-Rich Capsid with Distinctive Surface arches formed by Quasisymmetric Coat-Protein Dimers. Structure 16(5):776–786. https://doi.org/10.1016/j.str.2008.02.014
Pan J, Dong L, Lin L, Ochoa WF, Sinkovits RS, Havens WM et al (2009) Atomic structure reveals the unique capsid organization of a dsRNA virus. Proc Natl Acad Sci USA 106(11):4225–4230. https://doi.org/10.1073/pnas.0812071106
Parent J-S, ´ E, Jauvion V, Bouché N, Bouché B, Eclin CB, ´ E, Hachet M, ´ et al (2015) Post-transcriptional gene silencing triggered by sense transgenes involves uncapped antisense RNA and differs from silencing intentionally triggered by antisense transgenes. Nucleic Acids Res 43(17):8464–8475. https://doi.org/10.1093/nar/gkv753
Patton JT, Carpio V-D, Tortorici R, M. A., Taraporewala ZF (2006) Coupling of Rotavirus Genome Replication and Capsid Assembly. In Advances in Virus Research (Vol. 69, pp. 167–201). Academic Press. https://doi.org/10.1016/S0065-3527(06)69004-0
Peltier C, Klein E, Hleibieh K, D’Alonzo M, Hammann P, Bouzoubaa S et al (2012) Beet necrotic yellow vein virus subgenomic RNA3 is a cleavage product leading to stable non-coding RNA required for long-distance movement. J Gen Virol 93(5):1093–1102. https://doi.org/10.1099/vir.0.039685-0
Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM (2009) Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc Natl Acad Sci 106(6):1760–1765. https://doi.org/10.1073/pnas.0813167106
Righetti L, Paris R, Ratti C, Calassanzio M, Onofri C, Calzolari D et al (2018) Not the one, but the only one: about Cannabis cryptic virus in plants showing ‘hemp streak’ disease symptoms. Eur J Plant Pathol 150(3):575–588. https://doi.org/10.1007/s10658-017-1301-y
Roossinck MJ (2010) Lifestyles of plant viruses. Philosophical Trans Royal Soc B: Biol Sci 365(1548):1899–1905. https://doi.org/10.1098/rstb.2010.0057
Shi Y, Mowery RA, Ashley J, Hentz M, Ramirez AJ, Bilgicer B et al (2012) Abnormal SDS-PAGE migration of cytosolic proteins can identify domains and mechanisms that control surfactant binding. Protein Science: Publication Protein Soc 21(8):1197–1209. https://doi.org/10.1002/pro.2107
Shimura H, Kim H, Matsuzawa A, Akino S, Masuta C (2022) Coat protein of partitiviruses isolated from mycorrhizal fungi functions as an RNA silencing suppressor in plants and fungi. Sci Rep 12(1):7855. https://doi.org/10.1038/s41598-022-11403-5
Smith T, O’Kennedy MM, Ross CS, Lewis NS, Abolnik C (2023) The production of Newcastle disease virus-like particles in Nicotiana benthamiana as potential vaccines. Frontiers in Plant Science, 14. https://www.frontiersin.org/articles/https://doi.org/10.3389/fpls.2023.1130910. Accessed 27 November 2023
Sun X, Ilca SL, Huiskonen JT, Poranen MM (2018) Dual role of a viral polymerase in viral genome replication and particle self-assembly. mBio 9(5). https://doi.org/10.1128/mbio.01242-18
Tang J, Ochoa WF, Li H, Havens WM, Nibert ML, Ghabrial SA, Baker TS (2010) Structure of Fusarium Poae virus 1 shows conserved and variable elements of partitivirus capsids and evolutionary relationships to picobirnavirus. J Struct Biol 172(3):363–371. https://doi.org/10.1016/j.jsb.2010.06.022
Turina M, Ricker MD, Lenzi R, Masenga V, Ciuffo M (2007) A severe disease of Tomato in the Culiacan Area (Sinaloa, Mexico) is caused by a New Picorna-Like viral species. Plant Dis 91(8):932–941. https://doi.org/10.1094/PDIS-91-8-0932
Vainio EJ, Chiba S, Ghabrial SA, Maiss E, Roossinck M, Sabanadzovic S et al (2018) ICTV virus taxonomy profile: Partitiviridae. J Gen Virol 99(1):17–18. https://doi.org/10.1099/jgv.0.000985
Yang J, Zhang L, Zhang C, Lu Y (2021) Exploration on the expression and assembly of virus-like particles. Biotechnol Notes 2:51–58. https://doi.org/10.1016/j.biotno.2021.08.003
Ziegler A, Matoušek J, Steger G, Schubert J (2012) Complete sequence of a cryptic virus from hemp (Cannabis sativa). Arch Virol 157(2):383–385. https://doi.org/10.1007/s00705-011-1168-8
Funding
Open access funding provided by Alma Mater Studiorum - Università di Bologna within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Contributions
M.D.A. and N.M. conceived the study and experiments. N.M., M.D.A. and D.B. performed the experiments. M.D.A., N.M., A.P., P.C. and C.R. wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.

42161_2024_1628_MOESM1_ESM.jpg
Supplementary Material 1. Electrophoretic analysis of strand-specific RT-PCR was conducted for CanCV ssRNA1(-) and ssRNA2(-). Reactions were performed using the in vitro transcript products ssRNA1(+), ssRNA2(+), ssRNA1(-), and ssRNA2(-) as templates. Moreover, RT products, derived from in vitro transcripts ssRNA1(-) and ssRNA2(-), served as templates for PCR, intentionally lacking primer Tag (ssRNA1(-) No Tag and ssRNA2(-) No Tag). As an additional negative control for the RT-PCR reactions, nuclease-free water (H2O) was used as template.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Miotti, N., Dall’Ara, M., Baldo, D. et al. The artificial production of viral-like particles in Nicotiana benthamiana suggests the pro-assembly role of the Cannabis cryptic virus RdRP. J Plant Pathol (2024). https://doi.org/10.1007/s42161-024-01628-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42161-024-01628-w